Safety, Immunogenicity, and Protective Efficacy of an H5N1 Chimeric Cold-Adapted Attenuated Virus Vaccine in a Mouse Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells, Proteins, and Viruses
2.2. Vaccine Rescued and Characteristics Analysed
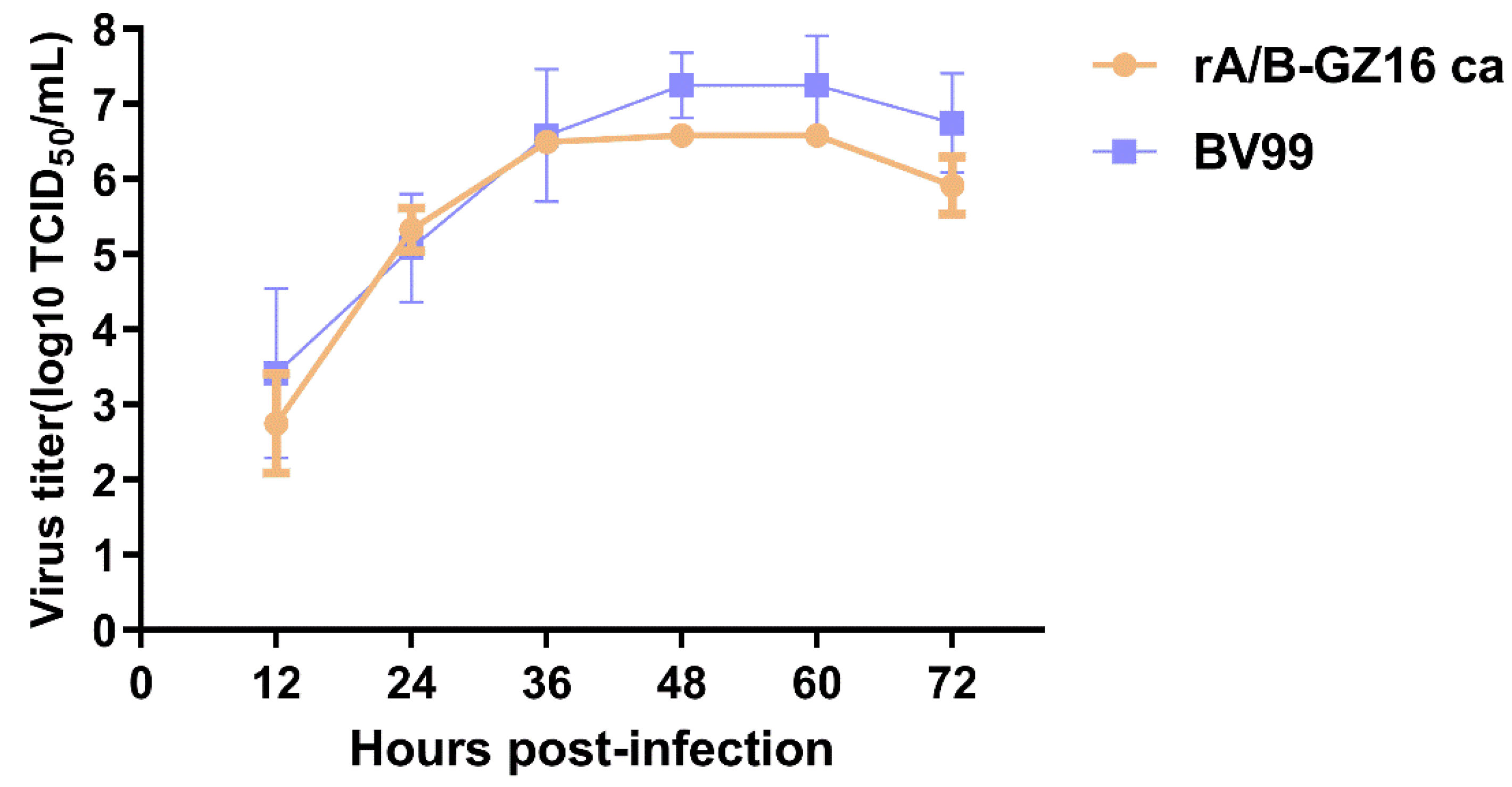
2.3. Viral Growth Kinetics in Cell Culture
2.4. Virus Titrations in MDCK Cells and Tissues
2.5. Mouse Immunization, Safety, and Protection
2.6. Hemagglutination Inhibition Antibody Assay
2.7. Micro-Neutralizing (MN) Assay
2.8. Enzyme-Linked Immunosorbent Assay (ELISA)
2.9. Protein Microarray Assay
2.10. ELISpot Assay
2.11. Luminex Assay
2.12. Statistical Analysis
3. Results
3.1. Biological Characteristics of the H5N1 Influenza Vaccine
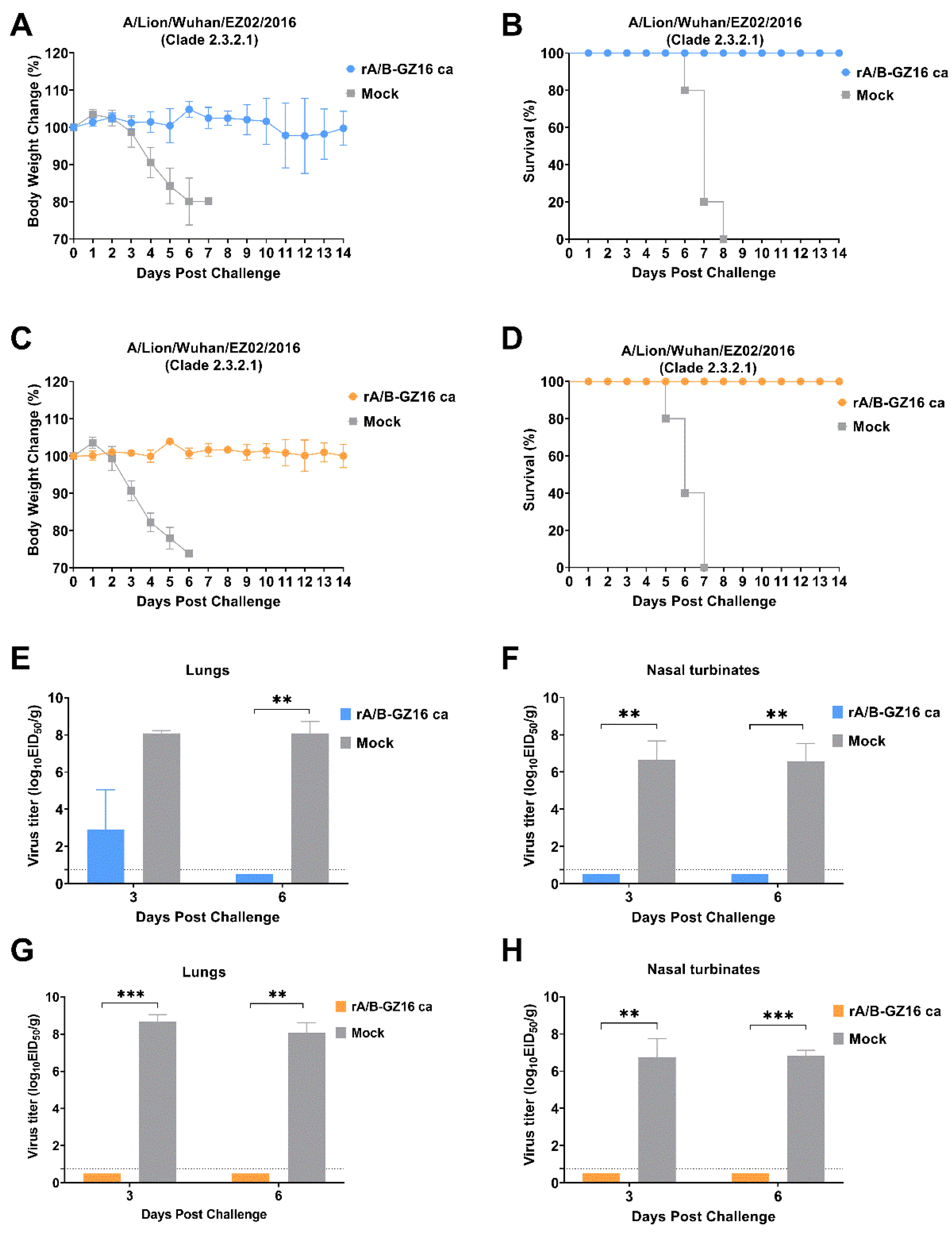
3.2. Safety of H5 Virus Vaccine
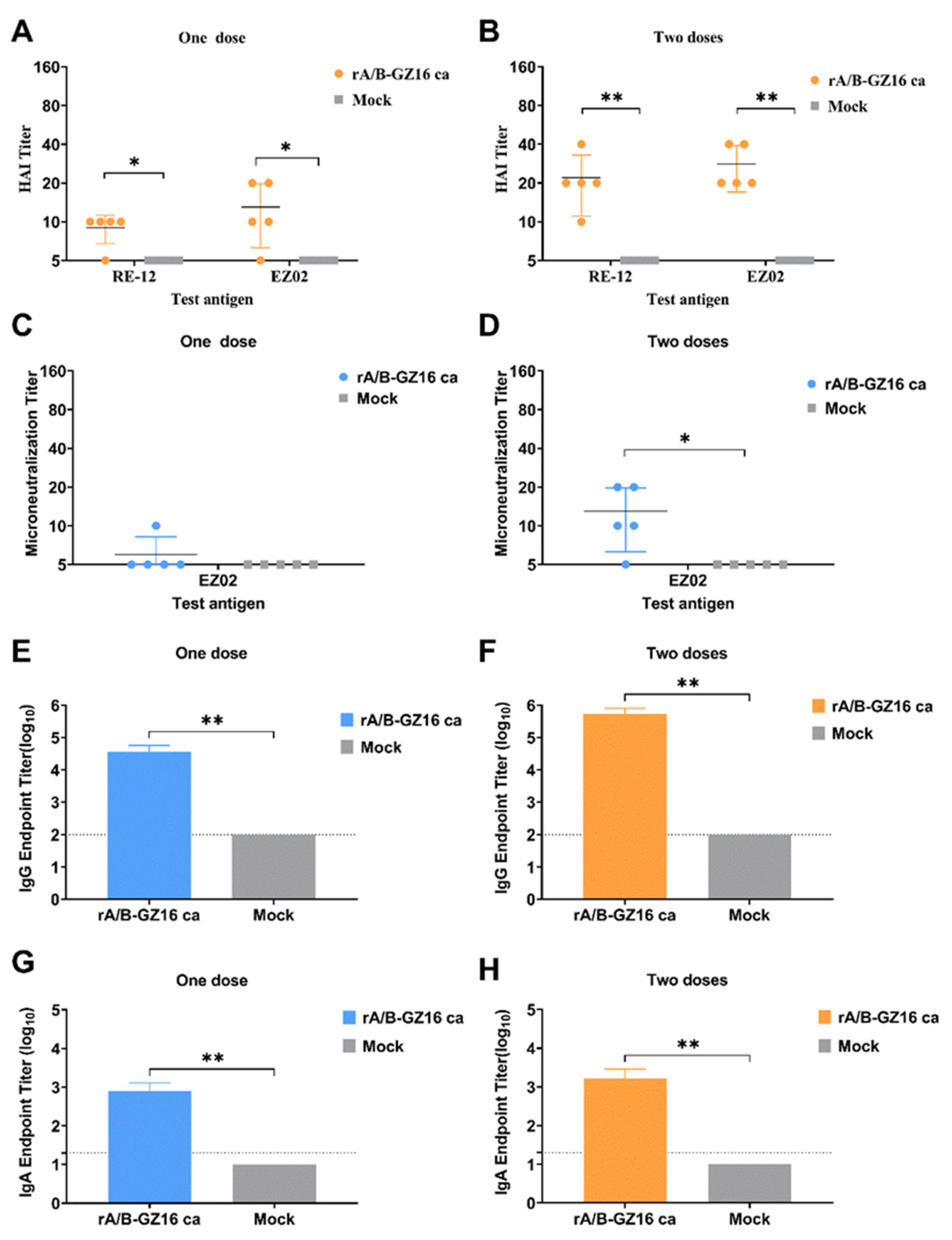
3.3. Antibody Immune Responses in Mice
3.3.1. Humoral Immune Responses
3.3.2. Mucosal Immune Responses
3.3.3. Protein Microarray Antibody Responses
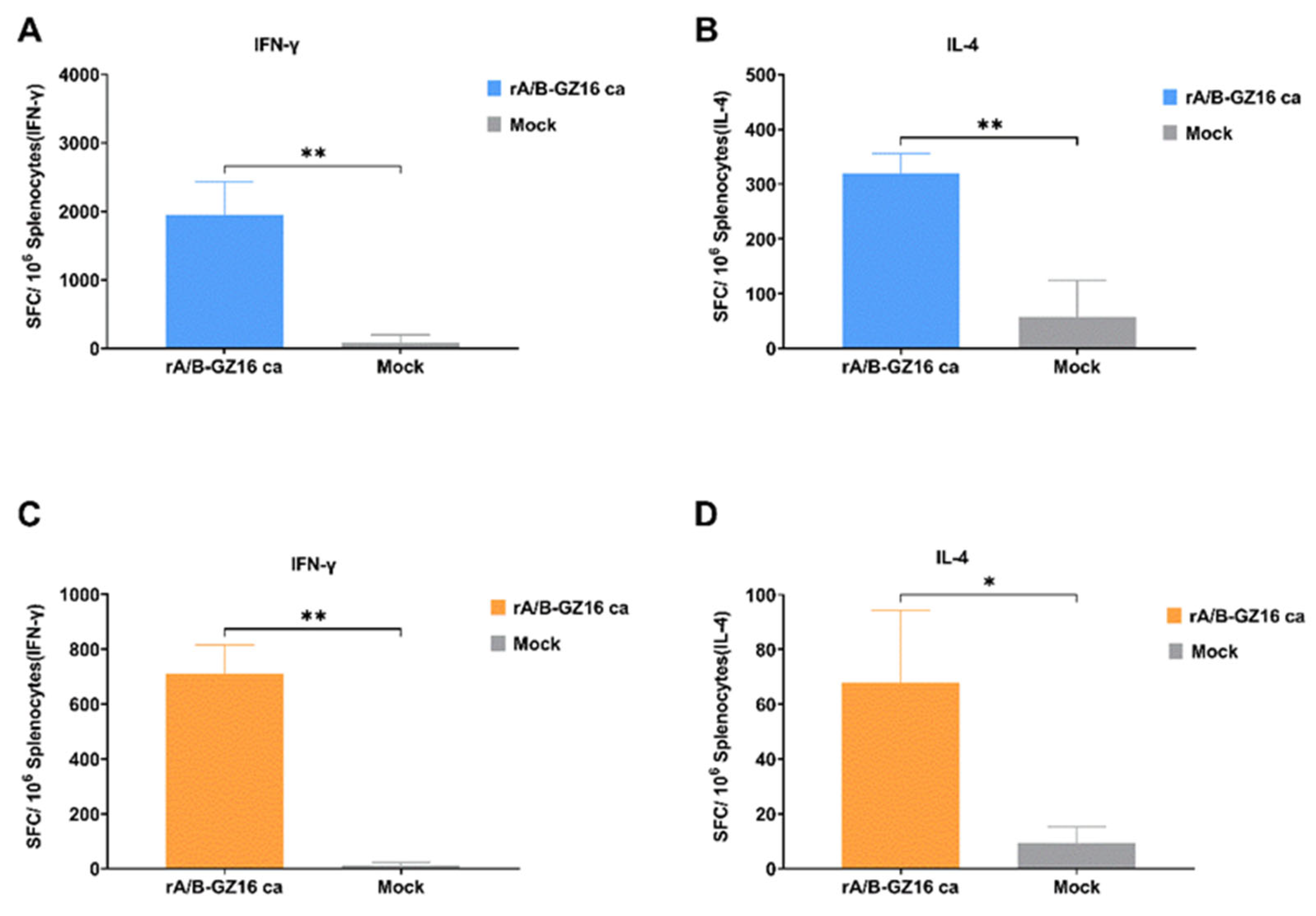
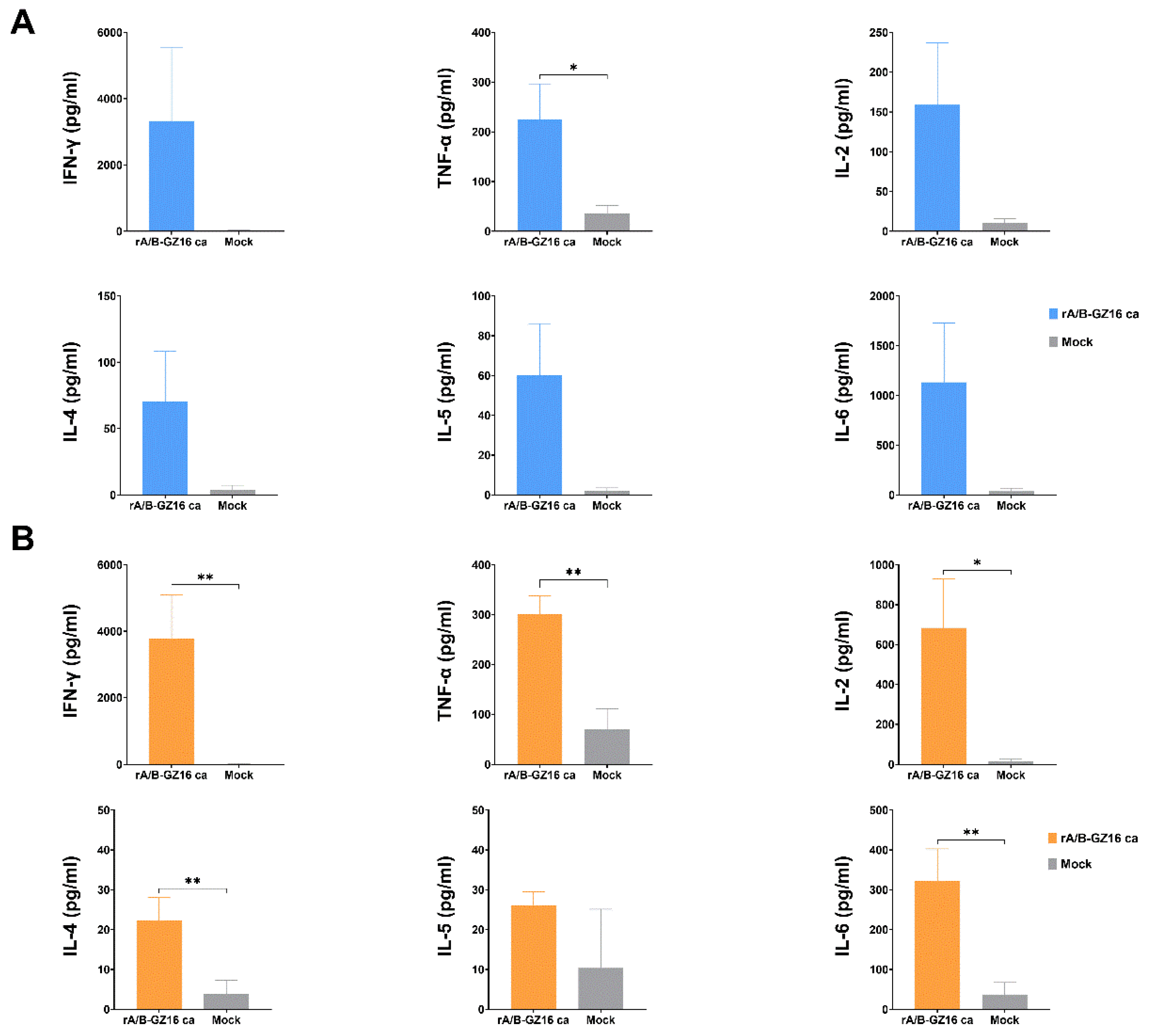
3.3.4. Mice Cellular Immunity
3.4. Protective Efficacy of H5 Influenza Vaccine
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO; OIE; FAO; H5N1 Evolution Working Group. Toward a unified nomenclature system for highly pathogenic avian influenza virus (H5N1). Emerg. Infect. Infect. Dis. 2008, 14, e1. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Subbarao, K.; Cox, N.J.; Guo, Y. Genetic characterization of the pathogenic influenza A/Goose/Guangdong/1/96 (H5N1) virus: Similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology 1999, 261, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Fasanmi, O.G.; Odetokun, I.A.; Balogun, F.A.; Fasina, F.O. Public health concerns of highly pathogenic avian influenza H5N1 endemicity in Africa. Vet. World 2017, 10, 1194–1204. [Google Scholar] [CrossRef] [PubMed]
- Dhingra, M.S.; Artois, J.; Robinson, T.P.; Linard, C.; Chaiban, C.; Xenarios, I.; Engler, R.; Liechti, R.; Kuznetsov, D.; Xiao, X.; et al. Global mapping of highly pathogenic avian influenza H5N1 and H5Nx clade 2.3.4.4 viruses with spatial cross-validation. ELife 2016, 5, e19571. [Google Scholar] [CrossRef]
- Lee, D.H.; Bahl, J.; Torchetti, M.K.; Killian, M.L.; Ip, H.S.; DeLiberto, T.J.; Swayne, D.E. Highly Pathogenic Avian Influenza Viruses and Generation of Novel Reassortants, United States, 2014–2015. Emerg. Infect. Dis. 2016, 22, 1283–1285. [Google Scholar] [CrossRef]
- Salzberg, S.L.; Kingsford, C.; Cattoli, G.; Spiro, D.J.; Janies, D.A.; Aly, M.M.; Brown, I.H.; Couacy-Hymann, E.; De Mia, G.M.; Dung do, H.; et al. Genome analysis linking recent European and African influenza (H5N1) viruses. Emerg. Infect. Dis. 2007, 13, 713–718. [Google Scholar] [CrossRef]
- Lipatov, A.S.; Evseenko, V.A.; Yen, H.L.; Zaykovskaya, A.V.; Durimanov, A.G.; Zolotykh, S.I.; Netesov, S.V.; Drozdov, I.G.; Onishchenko, G.G.; Webster, R.G.; et al. Influenza (H5N1) viruses in poultry, Russian Federation, 2005–2006. Emerg. Infect. Dis. 2007, 13, 539–546. [Google Scholar] [CrossRef]
- Chan, P.K. Outbreak of avian influenza A(H5N1) virus infection in Hong Kong in 1997. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2002, 34 (Suppl. 2), S58–S64. [Google Scholar] [CrossRef]
- El-Shesheny, R.; Kandeil, A.; Mostafa, A.; Ali, M.A.; Webby, R.J. H5 Influenza Viruses in Egypt. Cold Spring Harb. Perspect. Med. 2020, 11, a038745. [Google Scholar] [CrossRef]
- Lai, S.; Qin, Y.; Cowling, B.J.; Ren, X.; Wardrop, N.A.; Gilbert, M.; Tsang, T.K.; Wu, P.; Feng, L.; Jiang, H.; et al. Global epidemiology of avian influenza A H5N1 virus infection in humans, 1997–2015: A systematic review of individual case data. Lancet Infect. Dis. 2016, 16, e108–e118. [Google Scholar] [CrossRef]
- WHO Cumulative Number of Confirmed Human Cases for Avian Influenza A(H5N1) Reported to WHO, 2003–2021, 8 August 2021. Available online: https://www.who.int/publications/m/item/cumulative-number-of-confirmed-human-cases-for-avian-influenza-a(h5n1)-reported-to-who-2003-2021-8-august-2021 (accessed on 8 August 2021).
- Ambrose, C.S.; Wu, X.; Jones, T.; Mallory, R.M. The role of nasal IgA in children vaccinated with live attenuated influenza vaccine. Vaccine 2012, 30, 6794–6801. [Google Scholar] [CrossRef]
- Zhou, B.; Meliopoulos, V.A.; Wang, W.; Lin, X.; Stucker, K.M.; Halpin, R.A.; Stockwell, T.B.; Schultz-Cherry, S.; Wentworth, D.E. Reversion of Cold-Adapted Live Attenuated Influenza Vaccine into a Pathogenic Virus. J. Virol. 2016, 90, 8454–8463. [Google Scholar] [CrossRef]
- Suguitan, A.L., Jr.; McAuliffe, J.; Mills, K.L.; Jin, H.; Duke, G.; Lu, B.; Luke, C.J.; Murphy, B.; Swayne, D.E.; Kemble, G.; et al. Live, attenuated influenza A H5N1 candidate vaccines provide broad cross-protection in mice and ferrets. PLoS Med. 2006, 3, e360. [Google Scholar] [CrossRef]
- Kiseleva, I.; Larionova, N.; Rudenko, L. Live Attenuated Reassortant Vaccines Based on A/Leningrad/134/17/57 Master Donor Virus Against H5 Avian Influenza. Open Microbiol. J. 2017, 11, 316–329. [Google Scholar] [CrossRef]
- Fan, S.; Gao, Y.; Shinya, K.; Li, C.K.; Li, Y.; Shi, J.; Jiang, Y.; Suo, Y.; Tong, T.; Zhong, G.; et al. Immunogenicity and protective efficacy of a live attenuated H5N1 vaccine in nonhuman primates. PLoS Pathog. 2009, 5, e1000409. [Google Scholar] [CrossRef]
- Rudenko, L.; Isakova-Sivak, I. Pandemic preparedness with live attenuated influenza vaccines based on A/Leningrad/134/17/57 master donor virus. Expert Rev. Vaccines 2015, 14, 395–412. [Google Scholar] [CrossRef]
- Clegg, C.H.; Rininger, J.A.; Baldwin, S.L. Clinical vaccine development for H5N1 influenza. Expert Rev. Vaccines 2013, 12, 767–777. [Google Scholar] [CrossRef]
- Gerlach, T.; Elbahesh, H.; Saletti, G.; Rimmelzwaan, G.F. Recombinant influenza A viruses as vaccine vectors. Expert Rev. Vaccines 2019, 18, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Kittel, C.; Wressnigg, N.; Shurygina, A.P.; Wolschek, M.; Stukova, M.; Romanovskaya-Romanko, E.; Romanova, J.; Kiselev, O.; Muster, T.; Egorov, A. A genetically adjuvanted influenza B virus vector increases immunogenicity and protective efficacy in mice. Arch. Virol. 2015, 160, 2525–2534. [Google Scholar] [CrossRef] [PubMed]
- Kotomina, T.; Isakova-Sivak, I.; Kim, K.H.; Park, B.R.; Jung, Y.J.; Lee, Y.; Mezhenskaya, D.; Matyushenko, V.; Kang, S.M.; Rudenko, L. Generation and Characterization of Universal Live-Attenuated Influenza Vaccine Candidates Containing Multiple M2e Epitopes. Vaccines 2020, 8, 648. [Google Scholar] [CrossRef] [PubMed]
- Hai, R.; Martínez-Sobrido, L.; Fraser, K.A.; Ayllon, J.; García-Sastre, A.; Palese, P. Influenza B Virus NS1-Truncated Mutants: Live-Attenuated Vaccine Approach. J. Virol. 2008, 82, 10580–10590. [Google Scholar] [CrossRef]
- Hatta, Y.; Boltz, D.; Sarawar, S.; Kawaoka, Y.; Neumann, G.; Bilsel, P. M2SR, a novel live influenza vaccine, protects mice and ferrets against highly pathogenic avian influenza. Vaccine 2017, 35, 4177–4183. [Google Scholar] [CrossRef]
- Wang, P.; Zheng, M.; Lau, S.Y.; Chen, P.; Mok, B.W.; Liu, S.; Liu, H.; Huang, X.; Cremin, C.J.; Song, W.; et al. Generation of DelNS1 Influenza Viruses: A Strategy for Optimizing Live Attenuated Influenza Vaccines. MBio 2019, 10, e02180-19. [Google Scholar] [CrossRef]
- Talon, J.; Salvatore, M.; O’Neill, R.E.; Nakaya, Y.; Zheng, H.; Muster, T.; Garcia-Sastre, A.; Palese, P. Influenza A and B viruses expressing altered NS1 proteins: A vaccine approach. Proc. Natl. Acad. Sci. USA 2000, 97, 4309–4314. [Google Scholar] [CrossRef]
- Hai, R.; Garcia-Sastre, A.; Swayne, D.E.; Palese, P. A reassortment-incompetent live attenuated influenza virus vaccine for protection against pandemic virus strains. J. Virol. 2011, 85, 6832–6843. [Google Scholar] [CrossRef]
- Stepanova, E.; Krutikova, E.; Wong, P.F.; Matyushenko, V.; Bazhenova, E.; Isakova-Sivak, I.; Rudenko, L. Safety, Immunogenicity, and Protective Efficacy of a Chimeric A/B Live Attenuated Influenza Vaccine in a Mouse Model. Microorganisms 2021, 9, 259. [Google Scholar] [CrossRef]
- Flandorfer, A.; Garcia-Sastre, A.; Basler, C.F.; Palese, P. Chimeric influenza A viruses with a functional influenza B virus neuraminidase or hemagglutinin. J. Virol. 2003, 77, 9116–9123. [Google Scholar] [CrossRef][Green Version]
- Baker, S.F.; Nogales, A.; Finch, C.; Tuffy, K.M.; Domm, W.; Perez, D.R.; Topham, D.J.; Martinez-Sobrido, L. Influenza A and B virus intertypic reassortment through compatible viral packaging signals. J. Virol. 2014, 88, 10778–10791. [Google Scholar] [CrossRef]
- Yu, Z.; Cheng, K.; Sun, W.; Zhang, X.; Li, Y.; Wang, T.; Wang, H.; Zhang, Q.; Xin, Y.; Xue, L.; et al. A PB1 T296R substitution enhance polymerase activity and confer a virulent phenotype to a 2009 pandemic H1N1 influenza virus in mice. Virology 2015, 486, 180–186. [Google Scholar] [CrossRef]
- Katinger, H.; Egorov, A.; Ferko, B.; Romanova, J.; Katinger, D. Live Influenza Vaccine and Method of Manufacture. Patent WO 2002024876 A2, 28 March 2002. [Google Scholar]
- Li, Z.; Chen, H.; Jiao, P.; Deng, G.; Tian, G.; Li, Y.; Hoffmann, E.; Webster, R.G.; Matsuoka, Y.; Yu, K. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J. Virol. 2005, 79, 12058–12064. [Google Scholar] [CrossRef]
- Cheng, K.; Yu, Z.; Chai, H.; Sun, W.; Xin, Y.; Zhang, Q.; Huang, J.; Zhang, K.; Li, X.; Yang, S.; et al. PB2-E627K and PA-T97I substitutions enhance polymerase activity and confer a virulent phenotype to an H6N1 avian influenza virus in mice. Virology 2014, 468–470, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Yu, Z.; Cheng, K.; Sun, W.; Xin, Y.; Cai, J.; Ma, R.; Zhao, Q.; Li, L.; Huang, J.; Sang, X.; et al. Lowly pathogenic avian influenza (H9N2) infection in Plateau pika (Ochotona curzoniae), Qinghai Lake, China. Vet. Microbiol. 2014, 173, 132–135. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.J.; Bi, Y.; Wang, D.; Gao, G.F. On the Centenary of the Spanish Flu: Being Prepared for the Next Pandemic. Virol. Sin. 2018, 33, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Harfoot, R.; Webby, R.J. H5 influenza, a global update. J. Microbiol. 2017, 55, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Feng, Z.; Shu, Y.; Yu, H.; Zhou, L.; Zu, R.; Huai, Y.; Dong, J.; Bao, C.; Wen, L.; et al. Probable limited person-to-person transmission of highly pathogenic avian influenza A (H5N1) virus in China. Lancet 2008, 371, 1427–1434. [Google Scholar] [CrossRef]
- Ungchusak, K.; Auewarakul, P.; Dowell, S.F.; Kitphati, R.; Auwanit, W.; Puthavathana, P.; Uiprasertkul, M.; Boonnak, K.; Pittayawonganon, C.; Cox, N.J.; et al. Probable person-to-person transmission of avian influenza A (H5N1). N. Engl. J. Med. 2005, 352, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.; Streatfield, S.J.; Acosta, H.; Ganesan, S.; Fattom, A. A nanoemulsion-adjuvanted intranasal H5N1 influenza vaccine protects ferrets against homologous and heterologous H5N1 lethal challenge. Vaccine 2019, 37, 6162–6170. [Google Scholar] [CrossRef]
- Wong, S.S.; DeBeauchamp, J.; Zanin, M.; Sun, Y.; Tang, L.; Webby, R. H5N1 influenza vaccine induces a less robust neutralizing antibody response than seasonal trivalent and H7N9 influenza vaccines. NPJ Vaccines 2017, 2, 16. [Google Scholar] [CrossRef]
- Phan, H.T.; Pham, V.T.; Ho, T.T.; Pham, N.B.; Chu, H.H.; Vu, T.H.; Abdelwhab, E.M.; Scheibner, D.; Mettenleiter, T.C.; Hanh, T.X.; et al. Immunization with Plant-Derived Multimeric H5 Hemagglutinins Protect Chicken against Highly Pathogenic Avian Influenza Virus H5N1. Vaccines 2020, 8, 593. [Google Scholar] [CrossRef]
- Vemula, S.V.; Sayedahmed, E.E.; Sambhara, S.; Mittal, S.K. Vaccine approaches conferring cross-protection against influenza viruses. Expert Rev. Vaccines 2017, 16, 1141–1154. [Google Scholar] [CrossRef]
- Jang, Y.H.; Lee, E.Y.; Byun, Y.H.; Jung, E.J.; Lee, Y.J.; Lee, Y.H.; Lee, K.H.; Lee, J.; Seong, B.L. Protective efficacy in mice of monovalent and trivalent live attenuated influenza vaccines in the background of cold-adapted A/X-31 and B/Lee/40 donor strains. Vaccine 2014, 32, 535–543. [Google Scholar] [CrossRef]
- Kiseleva, I.V.; Voeten, J.T.; Teley, L.C.; Larionova, N.V.; Drieszen-van der Cruijsen, S.K.; Basten, S.M.; Heldens, J.G.; van den Bosch, H.; Rudenko, L.G. PB2 and PA genes control the expression of the temperature-sensitive phenotype of cold-adapted B/USSR/60/69 influenza master donor virus. J. Gen. Virol. 2010, 91, 931–937. [Google Scholar] [CrossRef]
- Gendon Iu, Z.; Markushin, S.G.; Tsfasman, T.M.; Akopova, I.I.; Akhmatova, N.K.; Koptiaeva, I.B. New cold-adapted donor strains for live influenza vaccine. Vopr. Virusol. 2013, 58, 11–17. [Google Scholar]
- Chen, Z.; Aspelund, A.; Kemble, G.; Jin, H. Genetic mapping of the cold-adapted phenotype of B/Ann Arbor/1/66, the master donor virus for live attenuated influenza vaccines (FluMist). Virology 2006, 345, 416–423. [Google Scholar] [CrossRef]
- Kim, H.; Schoofs, P.; Anderson, D.A.; Tannock, G.A.; Rockman, S.P. Cold adaptation improves the growth of seasonal influenza B vaccine viruses. Vaccine 2014, 32, 2474–2479. [Google Scholar] [CrossRef]
- Moser, M.J.; Hatta, Y.; Gabaglia, C.; Sanchez, A.; Dias, P.; Sarawar, S.; Kawaoka, Y.; Hatta, M.; Neumann, G.; Bilsel, P. Single-replication BM2SR vaccine provides sterilizing immunity and cross-lineage influenza B virus protection in mice. Vaccine 2019, 37, 4533–4542. [Google Scholar] [CrossRef]
- Dudas, G.; Bedford, T.; Lycett, S.; Rambaut, A. Reassortment between influenza B lineages and the emergence of a coadapted PB1-PB2-HA gene complex. Mol. Biol. Evol. 2015, 32, 162–172. [Google Scholar] [CrossRef]
- Baxter, R.; Eaton, A.; Hansen, J.; Aukes, L.; Caspard, H.; Ambrose, C.S. Safety of quadrivalent live attenuated influenza vaccine in subjects aged 2–49 years. Vaccine 2017, 35, 1254–1258. [Google Scholar] [CrossRef]
- Korenkov, D.; Isakova-Sivak, I.; Rudenko, L. Basics of CD8 T-cell immune responses after influenza infection and vaccination with inactivated or live attenuated influenza vaccine. Expert Rev. Vaccines 2018, 17, 977–987. [Google Scholar] [CrossRef]
- Schmidt, A.; Lapuente, D. T Cell Immunity against Influenza: The Long Way from Animal Models Towards a Real-Life Universal Flu Vaccine. Viruses 2021, 13, 199. [Google Scholar] [CrossRef] [PubMed]
- Hoft, D.F.; Babusis, E.; Worku, S.; Spencer, C.T.; Lottenbach, K.; Truscott, S.M.; Abate, G.; Sakala, I.G.; Edwards, K.M.; Creech, C.B.; et al. Live and inactivated influenza vaccines induce similar humoral responses, but only live vaccines induce diverse T-cell responses in young children. J. Infect. Dis. 2011, 204, 845–853. [Google Scholar] [CrossRef] [PubMed]
- Abreu, R.B.; Clutter, E.F.; Attari, S.; Sautto, G.A.; Ross, T.M. IgA Responses Following Recurrent Influenza Virus Vaccination. Front. Immunol. 2020, 11, 902. [Google Scholar] [CrossRef] [PubMed]
- Barria, M.I.; Garrido, J.L.; Stein, C.; Scher, E.; Ge, Y.; Engel, S.M.; Kraus, T.A.; Banach, D.; Moran, T.M. Localized mucosal response to intranasal live attenuated influenza vaccine in adults. J. Infect. Dis. 2013, 207, 115–124. [Google Scholar] [CrossRef]


| Virus | Virus Titer [log10(EID50 mL−1)] | RCT39 | RCT27 | Phenotype | ||
|---|---|---|---|---|---|---|
| 33 °C | 39 °C | 27 °C | ||||
| rA/B-GZ16 ca | 8.2 ± 0.2 | - | 6.9 ± 0.7 | 8.2 | 1.3 | ts, ca |
| BV99 | 7.7 ± 0.5 | - | 6.2 ± 0.2 | 7.7 | 1.5 | ts, ca |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, W.; Wang, Z.; Sun, Y.; Li, D.; Zhu, M.; Zhao, M.; Wang, Y.; Xu, J.; Kong, Y.; Li, Y.; et al. Safety, Immunogenicity, and Protective Efficacy of an H5N1 Chimeric Cold-Adapted Attenuated Virus Vaccine in a Mouse Model. Viruses 2021, 13, 2420. https://doi.org/10.3390/v13122420
Sun W, Wang Z, Sun Y, Li D, Zhu M, Zhao M, Wang Y, Xu J, Kong Y, Li Y, et al. Safety, Immunogenicity, and Protective Efficacy of an H5N1 Chimeric Cold-Adapted Attenuated Virus Vaccine in a Mouse Model. Viruses. 2021; 13(12):2420. https://doi.org/10.3390/v13122420
Chicago/Turabian StyleSun, Weiyang, Zhenfei Wang, Yue Sun, Dongxu Li, Menghan Zhu, Menglin Zhao, Yutian Wang, Jiaqi Xu, Yunyi Kong, Yuanguo Li, and et al. 2021. "Safety, Immunogenicity, and Protective Efficacy of an H5N1 Chimeric Cold-Adapted Attenuated Virus Vaccine in a Mouse Model" Viruses 13, no. 12: 2420. https://doi.org/10.3390/v13122420
APA StyleSun, W., Wang, Z., Sun, Y., Li, D., Zhu, M., Zhao, M., Wang, Y., Xu, J., Kong, Y., Li, Y., Feng, N., Wang, T., Zhao, Y., Yang, S., Gao, Y., & Xia, X. (2021). Safety, Immunogenicity, and Protective Efficacy of an H5N1 Chimeric Cold-Adapted Attenuated Virus Vaccine in a Mouse Model. Viruses, 13(12), 2420. https://doi.org/10.3390/v13122420





