A Novel Picornavirus Discovered in White Leg Shrimp Penaeus vannamei
Abstract
:1. Introduction
2. Materials and Methods
2.1. Shrimp Samples and Ethics Statement
2.2. Transmission Electron Microscopy (TEM) Analysis
2.3. Virus Purification
2.4. RNA Extraction, Library Construction and Virome Sequencing
2.5. Assembling and Phylogenetic Analysis
2.6. Viral RNA Extraction and cDNA Synthesis
2.7. PCR Amplification and Sequencing
2.8. Mass Spectrometry Analysis
3. Results
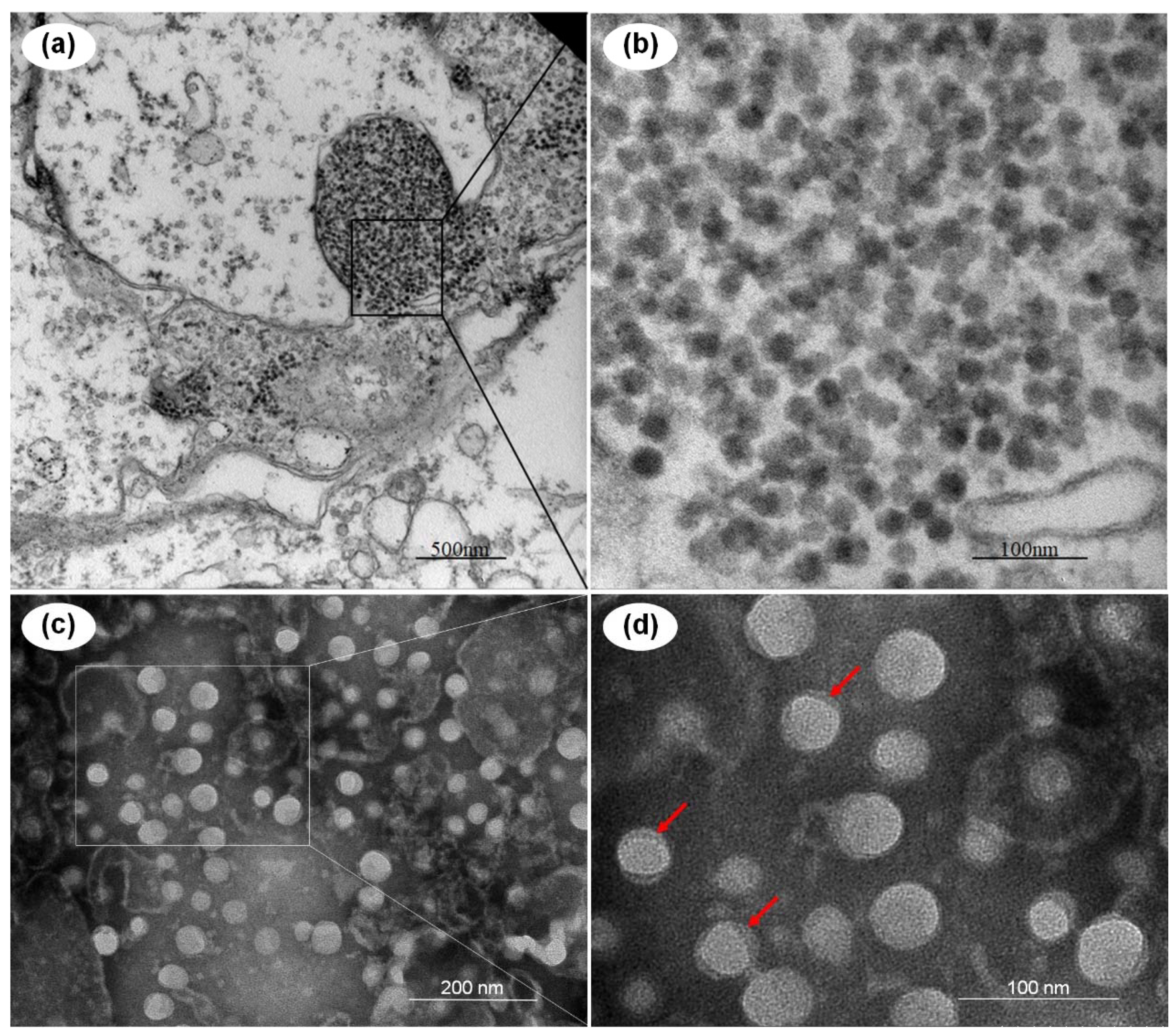
3.1. TEM Analysis of the Potential Virus and Morphology of the Purified Virions
3.2. Molecular Characterization of PvPV
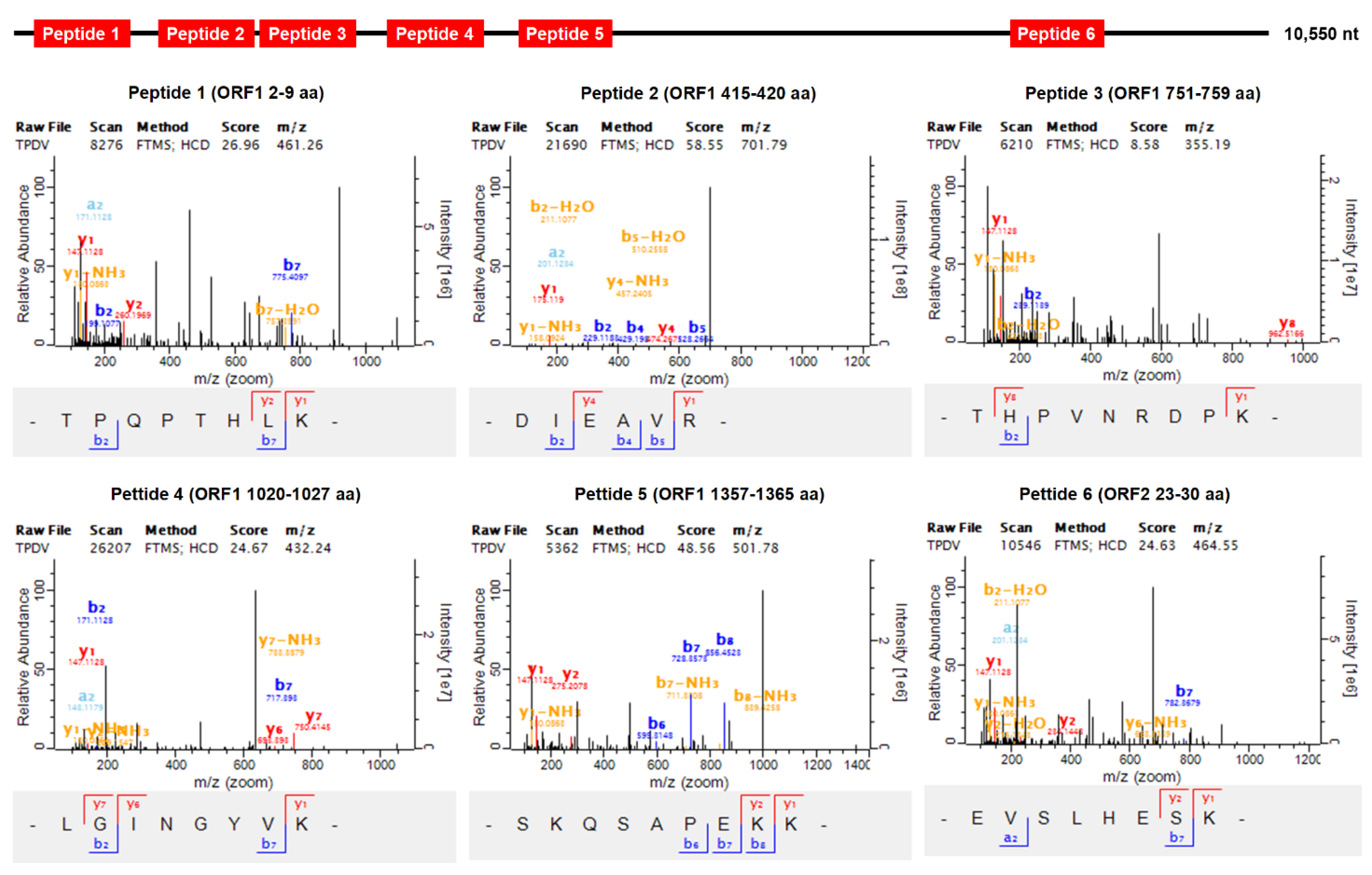
3.3. Identification of PvPV by Mass Spectrometry Analysis
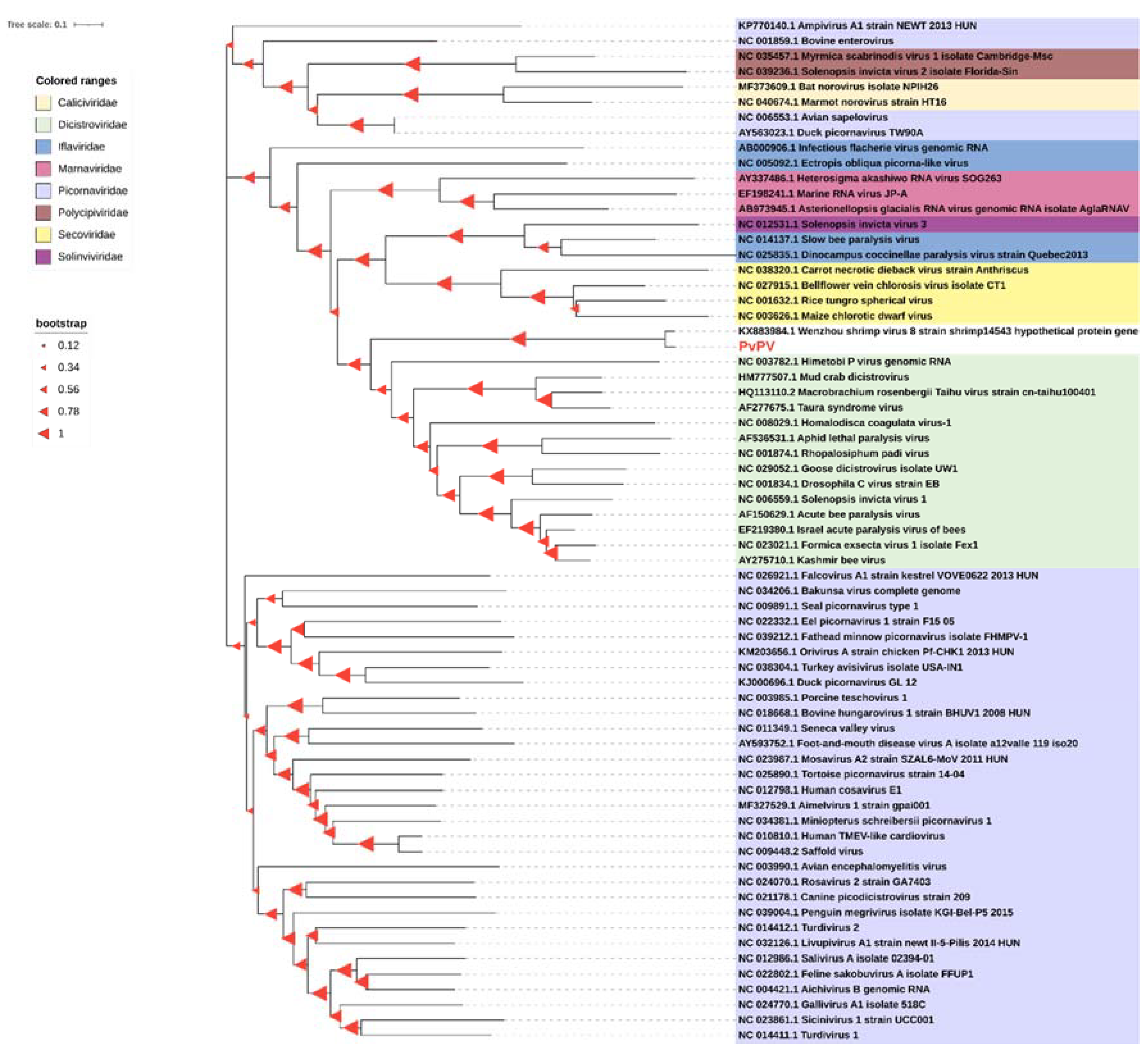
3.4. Phylogenetic Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Flegel, T.W.; Lightner, D.V.; Lo, C.F.; Owens, L. Shrimp Disease Control: Past, Present and Future. Dis. Asian Aquac. VI. Fish Health Sect. Asian Fish. Soc. Manila Philipp. 2008, 505, 355–378. [Google Scholar]
- Flegel, T.W. Historic emergence, impact and current status of shrimp pathogens in Asia. J. Invertebr. Pathol. 2012, 110, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Soto, M.A.; Lotz, J.M. Epidemiological Parameters of White Spot Syndrome Virus Infections in Litopenaeus vannamei and L. setiferus. J. Invertebr. Pathol. 2001, 78, 9–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soto, M.A.; Shervette, V.R.; Lotz, J.M. Transmission of white spot syndrome virus (WSSV) to Litopenaeus vannamei from infected cephalothorax, abdomen, or whole shrimp cadaver. Dis. Aquat. Org. 2001, 45, 81–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Liu, Q.; Liu, S.; Yang, H.; Liu, S.; Zhu, L.; Yang, B.; Jin, J.; Ding, L.; Wang, X.; et al. A new nodavirus is associated with covert mortality disease of shrimp. J. Gen. Virol. 2014, 95, 2700–2709. [Google Scholar] [CrossRef] [Green Version]
- Kalagayan, H.; Kalagayan, H.; Godin, D.; Kanna, R.; Hagino, G.; Sweeney, J.; Wyban, J.; Brock, J. IHHN Virus as an Etiological Factor in Runt-Deformity Syndrome (RDS) of Juvenile Penaeus vannamei Cultured in Hawaii. J. World Aquac. Soc. 2010, 22, 235–243. [Google Scholar] [CrossRef]
- Mayo, M.A. Virus Taxonomy-Houston 2002. Arch. Virol. 2002, 147, 1071–1076. [Google Scholar] [PubMed]
- Tang, K.; Lightner, D. A yellow head virus gene probe: Nucleotide sequence and application for in situ hybridization. Dis. Aquat. Org. 1999, 35, 165–173. [Google Scholar] [CrossRef] [Green Version]
- Lightner, D. The penaeid shrimp viral pandemics due to IHHNV, WSSV, TSV and YHV: History in the Americas and current status. In Proceedings of the 32nd Joint UJNR Aquaculture Panel Symposium, Davis and Santa Barbara, CA, USA, January 2003; pp. 17–20. Available online: https://www.researchgate.net/publication/228580878_The_penaeid_shrimp_viral_pandemics_due_to_IHHNV_WSSV_TSV_and_YHV_history_in_the_Americas_and_current_status (accessed on 20 January 2003).
- Nunan, L.M.; Poulos, B.T.; Lightner, D.V. The detection of White Spot Syndrome Virus (WSSV) and Yellow Head Virus (YHV) in imported commodity shrimp. Aquaculture 1998, 160, 19–30. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, J.; Yuan, J.; Li, L.; Zhang, J.; Bonami, J.R.; Shi, Z. Quantitative relationship of two viruses (MrNV and XSV) in white-tail disease of Macrobrachium rosenbergii. Dis. Aquat. Org. 2006, 71, 11–17. [Google Scholar] [CrossRef] [Green Version]
- Lifeng, C. 2021 Analysis of Important Aquatic Animal Diseases in China; Agriculture Press; Fishery and Fishery Administration of the Ministry of Agriculture and Rural Affairs: Beijing, China, 2021. [Google Scholar]
- Van Hulten, M.C.; Witteveldt, J.; Peters, S.; Kloosterboer, N.; Tarchini, R.; Fiers, M.; Sandbrink, H.; Lankhorst, R.K.; Vlak, J.M. The white spot syndrome virus DNA genome sequence. Virology 2001, 286, 7–22. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; He, J.; Lin, X.; Li, Q.; Pan, D.; Zhang, X.; Xu, X. Complete genome sequence of the shrimp White spot bacilliform virus. J. Virol. 2001, 75, 11811–11820. [Google Scholar] [CrossRef] [Green Version]
- Xu, T.; Liu, S.; Li, X.; Zhang, Q. Genomic characterization of covert mortality nodavirus from farming shrimp: Evidence for a new species within the family Nodaviridae. Virus Res. 2020, 286, 198092. [Google Scholar] [CrossRef]
- Rai, P.; Safeena, M.P.; Krabsetsve, K.; La Fauce, K.; Owens, L.; Karunasagar, I. Genomics, Molecular Epidemiology and Diagnostics of Infectious hypodermal and hematopoietic necrosis virus. Indian J. Virol. Off. Organ Indian Virol. Soc. 2012, 23, 203–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mari, J.; Poulos, B.T.; Lightner, D.V.; Bonami, J.R. Shrimp Taura syndrome virus: Genomic characterization and similarity with members of the genus Cricket paralysis-like viruses. J. Gen. Virol. 2002, 83, 915–926. [Google Scholar] [CrossRef]
- Dong, X.; Liu, S.; Zhu, L.; Wan, X.; Liu, Q.; Qiu, L.; Zou, P.; Zhang, Q.; Huang, J. Complete genome sequence of an isolate of a novel genotype of yellow head virus from Fenneropenaeus chinensis indigenous in China. Arch. Virol. 2017, 162, 1149–1152. [Google Scholar] [CrossRef] [PubMed]
- Jeeva, S.; Kang, S.W.; Lee, Y.S.; Jang, I.K.; Seo, H.C.; Choi, T.J. Complete nucleotide sequence analysis of a Korean strain of hepatopancreatic parvovirus (HPV) from Fenneropenaeus chinensis. Virus Genes 2012, 44, 89–97. [Google Scholar] [CrossRef]
- Sukhumsirichart, W.; Attasart, P.; Boonsaeng, V.; Panyim, S. Complete nucleotide sequence and genomic organization of hepatopancreatic parvovirus (HPV) of Penaeus monodon. Virology 2006, 346, 266–277. [Google Scholar] [CrossRef] [Green Version]
- Tang, K.F.J.; Pantoja, C.R.; Redman, R.M.; Navarro, S.A.; Lightner, D.V. Ultrastructural and sequence characterization of Penaeus vannamei nodavirus (PvNV) from Belize. Dis. Aquat. Org. 2011, 94, 179–187. [Google Scholar] [CrossRef] [Green Version]
- Poulos, B.; Tang, K.; Pantoja, C.; Bonami, J.R.; Lightner, D. Purification and characterization of infectious myonecrosis virus of penaeid shrimp. J. Gen. Virol. 2006, 87, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Munnink, B.B.O.; Cotten, M.; Deijs, M.; Jebbink, M.F.; Bakker, M.; Farsani, S.M.J.; Canuti, M.; Kellam, P.; van der Hoek, L. A novel genus in the order Picornavirales detected in human stool. J. Gen. Virol. 2015, 96, 3440–3443. [Google Scholar] [CrossRef]
- Le Gall, O.; Christian, P.; Fauquet, C.M.; King, A.M.Q.; Knowles, N.J.; Nakashima, N.; Stanway, G.; Gorbalenya, A.E. Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T = 3 virion architecture. Arch. Virol. 2008, 153, 715. [Google Scholar] [CrossRef]
- Argos, P.; Kamer, G.; Nicklin, M.J.; Wimmer, E. Similarity in gene organization and homology between proteins of animal picornaviruses and a plant comovirus suggest common ancestry of these virus families. Nucl. Acid. Res. 1984, 12, 7251–7267. [Google Scholar] [CrossRef]
- Franssen, H.; Leunissen, J.; Goldbach, R.; Lomonossoff, G.; Zimmern, D. Homologous sequences in non-structural proteins from cowpea mosaic virus and picornaviruses. EMBO J. 1984, 3, 855–861. [Google Scholar] [CrossRef]
- Goldbach, R.W. Molecular evolution of plant rna viruses. Annu. Rev. Phytopathol. 1986, 24, 289–310. [Google Scholar] [CrossRef]
- Goldbach, R. Genome similarities between plant and animal RNA viruses. Microbiol. Sci. 1987, 4, 197–202. [Google Scholar]
- Goldbach, R.; Wellink, J. Evolution of plus-strand RNA viruses. Intervirology 1988, 29, 260–267. [Google Scholar] [CrossRef]
- Rossmann, M.G.; Johnson, J.E. Icosahedral RNA Virus Structure—Annual Review of Biochemistry. Annu. Rev. Biochem. 1989, 58, 533. [Google Scholar] [CrossRef] [PubMed]
- Liljas, L.; Tate, J.; Lin, T.; Christian, P.; Johnson, J.E. Evolutionary and taxonomic implications of conserved structural motifs between picornaviruses and insect picorna-like viruses. Arch. Virol. 2002, 147, 59–84. [Google Scholar] [CrossRef] [PubMed]
- Hasson, K.W.; Lightner, D.V.; Poulos, B.T.; Redman, R.M.; White, B.L.; Brock, J.A.; Bonami, J.R. Taura syndrome in Penaeus vannamei: Demonstration of a viral etiology. Dis. Aquat. Org. 1995, 23, 115–126. [Google Scholar] [CrossRef] [Green Version]
- Lightner, D.V. Epizootiology, distribution and the impact on international trade of two penaeid shrimp viruses in the Americas. Rev. Sci. Tech. 1996, 15, 579–601. [Google Scholar] [CrossRef]
- Overstreet, R.M.; Lightner, D.V.; Hasson, K.W.; McIlwain, S.; Lotz, J.M. Susceptibility to Taura Syndrome Virus of Some Penaeid Shrimp Species Native to the Gulf of Mexico and the Southeastern United States. J. Invertebr. Pathol. 1997, 69, 165–176. [Google Scholar] [CrossRef] [Green Version]
- Hasson, K.W.; Lightner, D.V.; Mari, J.; Bonami, J.R.; Poulos, B.T.; Mohney, L.L.; Redman, R.M.; Brock, J.A. The geographic distribution of Taura Syndrome Virus (TSV) in the Americas: Determination by histopathology and in situ hybridization using TSV-specific cDNA probes. Aquaculture 1999, 171, 13–26. [Google Scholar] [CrossRef]
- Yu, C.I.; Song, Y.L. Outbreaks of Taura Syndrome in Pacific White Shrimp Penaeus vannamei Cultured in Taiwan. Fish Pathol. 2000, 35, 21–24. [Google Scholar] [CrossRef]
- Bonami, J.R.; Shi, Z.; Qian, D.; Sri Widada, J. White tail disease of the giant freshwater prawn, Macrobrachium rosenbergii: Separation of the associated virions and characterization of MrNV as a new type of nodavirus. J. Fish Dis. 2010, 28, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Shevchenko, A.; Jensen, O.N.; Podtelejnikov, A.V.; Sagliocco, F.; Wilm, M.; Vorm, O.; Mortensen, P.; Shevchenko, A.; Boucherie, H.; Mann, M. Linking genome and proteome by mass spectrometry: Large-scale identification of yeast proteins from two dimensionalgels. Proc. Natl. Acad. Sci. USA 1996, 93, 14440–14445. [Google Scholar] [CrossRef] [Green Version]
- Calderaro, A.; Arcangeletti, M.C.; Rodighiero, I.; Buttrini, M.; Gorrini, C.; Motta, F.; Germini, D.; Medici, M.-C.; Chezzi, C.; De Conto, F. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry applied to virus identification. Sci. Rep. 2014, 4, 6803. [Google Scholar] [CrossRef] [Green Version]
- Xiu, L.; Zhang, C.; Wu, Z.; Peng, J. Establishment and Application of a Universal Coronavirus Screening Method Using MALDI-TOF Mass Spectrometry. Front. Microbiol. 2017, 8, 1510. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Zhang, Z.; Wang, M.; Sun, J.; Li, H.; Malakar, P.K.; Liu, H.; Pan, Y.; Zhao, Y. New insights into the changes of proteome and microbiome of shrimp (Litopenaeus vannamei) stored in acidic electrolyzed water ice. J. Agric. Food Chem. 2018, 66, 4966–4976. [Google Scholar] [CrossRef]
- Yong, C.Y.; Yeap, S.K.; Omar, A.R.; Tan, W.S. Advances in the study of nodavirus. PeerJ 2017, 5, e3841. [Google Scholar] [CrossRef]
- Kibenge, F.S. Emerging viruses in aquaculture. Curr. Opin. Virol. 2019, 34, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Lightner, D.V. Virus diseases of farmed shrimp in the Western Hemisphere (the Americas): A review. J. Invertebr. Pathol. 2011, 106, 110–130. [Google Scholar] [CrossRef] [PubMed]
- Carstens, B. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Lefkowitz, E., Adams, M.J., Carstens, E.B., Eds.; Elsevier: Amsterdam, The Netherlands, 2011; p. 1338. [Google Scholar]
- Bonami, J.R.; Hasson, K.W.; Mari, J.; Poulos, B.T.; Lightner, D.V. Taura syndrome of marine penaeid shrimp: Characterization of the viral agent. J. Gen. Virol. 1997, 78, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Hasson, K.W.; Lightner, D.V.; Mohney, L.L.; Redman, R.M.; Poulos, B.T.; White, B.M. Taura syndrome virus (TSV) lesion development and the disease cycle in the Pacific white shrimp Penaeus vannamei. Dis. Aquat. Org. 1999, 36, 81–93. [Google Scholar] [CrossRef] [Green Version]
- Brock, J.A.; Gose, R.; Lightner, D.V.; Hasson, K. An overview on Taura syndrome, an important disease of farmed. Swimming through troubled water. In Proceedings of the Special Session on Shrimp Farming, San Diego, CA, USA, 1–4 February 1995. [Google Scholar]
- Guo, Z.X.; He, J.G.; Xu, H.D.; Weng, S.P. Pathogenicity and complete genome sequence analysis of the mud crab dicistrovirus-1. Virus Res. 2013, 171, 8–14. [Google Scholar] [CrossRef]
- Pan, X.; Cao, Z.; Yuan, J.; Shi, Z.; Yuan, X.; Lin, L.; Xu, Y.; Yao, J.; Hao, G.; Shen, J. Isolation and Characterization of a Novel Dicistrovirus Associated with Moralities of the Great Freshwater Prawn, Macrobrachium rosenbergii. Int. J. Mol. Sci. 2016, 17, 204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, X.; Gao, Q.; Shen, J.; Xu, T. 14-3-3 is a VP3-binding protein involved in Macrobrachium rosenbergii Taihu virus infection in shrimp. Dev. Comp. Immunol. 2021, 122, 104139. [Google Scholar] [CrossRef]
- Koonin, E.V.; Wolf, Y.I.; Nagasaki, K.; Dolja, V.V. The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat. Rev. Microbiol. 2008, 6, 925–939. [Google Scholar] [CrossRef]
- Lee, T.; Pelletier, J. The biology of DHX9 and its potential as a therapeutic target. Oncotarget 2016, 7, 42716. [Google Scholar] [CrossRef] [Green Version]
- Newcomb, W.W.; Brown, J.C. Structure and capsid association of the herpesvirus large tegument protein UL36. J. Virol. 2010, 84, 9408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuchs, W.; Klupp, B.G.; Granzow, H.; Mettenleiter, T.C. Essential Function of the Pseudorabies Virus UL36 Gene Product Is Independent of Its Interaction with the UL37 Protein. J. Virol. 2004, 78, 11879–11889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Studer, M.K.; Ivanović, L.; Weber, M.E.; Marti, S.; Jonas, S. Structural basis for DEAH-helicase activation by G-patch proteins. Proc. Natl. Acad. Sci. USA 2020, 117, 7159–7170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sloan, E.K.; Bohnsack, M.T. Unravelling the Mechanisms of RNA Helicase Regulation. Trends Biochem. Sci. 2018, 43, 237–250. [Google Scholar] [CrossRef]
- Aravind, L.; Koonin, E.V. G-patch: A new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem. Ences 1999, 24, 342–344. [Google Scholar] [CrossRef]
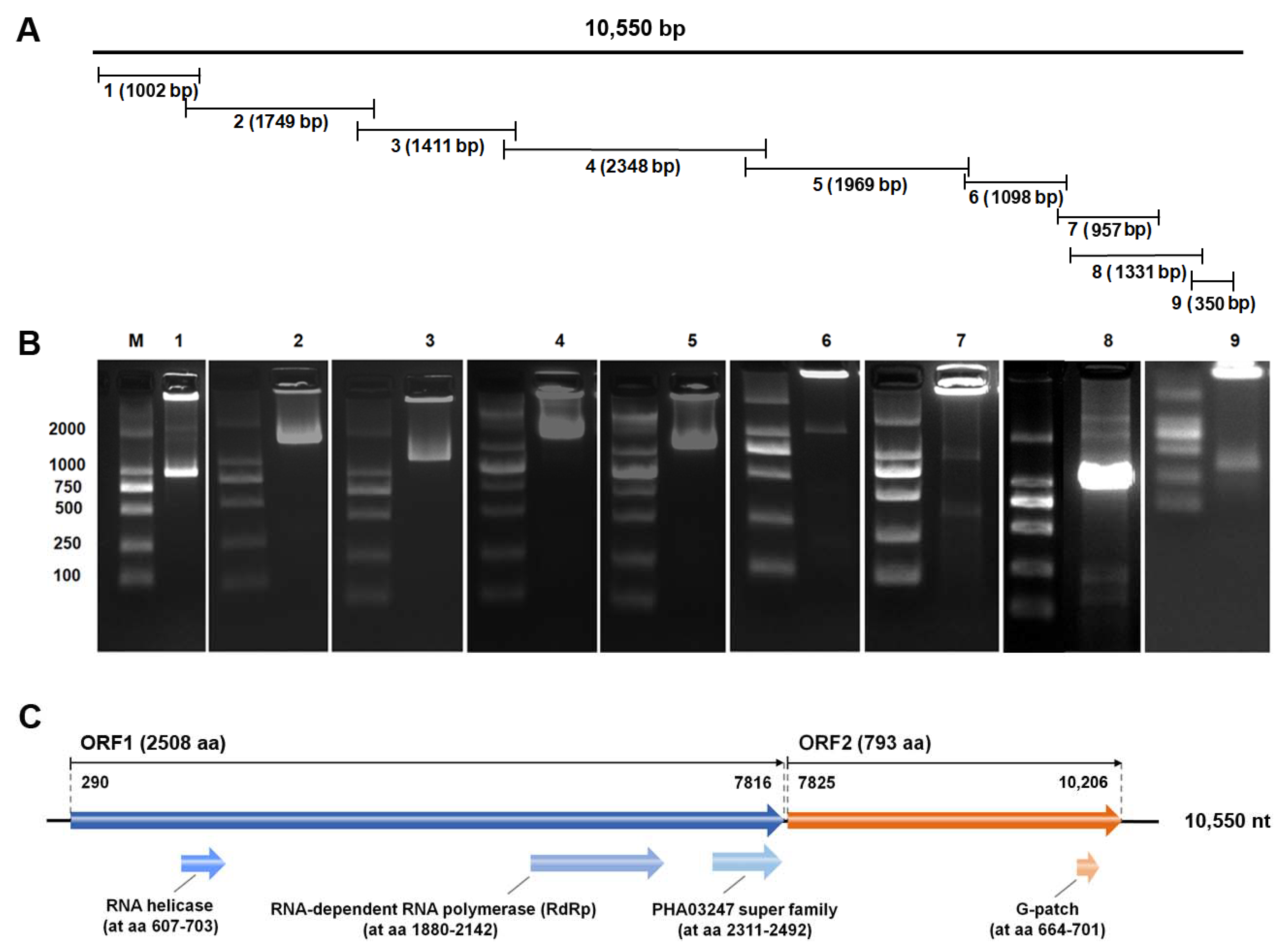
| Gene Fragments | Primer Code | PCR Annealing Temperature | Primer Sequence (5′-3′) | Size (bp) |
|---|---|---|---|---|
| 1 | RNA-F1 | 48 °C | ATCCACGGAAAGAGCC | 1002 bp |
| RNA-R1 | TAGCGGAATGCGACAA | |||
| 2 | RNA-F2 | 45 °C | CTGCCCTTTGCCGTCTTC | 1749 bp |
| RNA-R2 | CTGAGTGTCATTGTCTTGGA | |||
| 3 | RNA-F3 | 50 °C | CGTTCCCATAAGGACCCA | 1411 bp |
| RNA-R3 | ATATCGCTTTCCAGAGGC | |||
| 4 | RNA-F4 | 50 °C | CTCAGTCGTCTCCCGTGTC | 2348 bp |
| RNA-R4 | CGGTCTCAAAGTCAATCCC | |||
| 5 | RNA-F5 | 50 °C | GACGAGTTGAGCCTACAGA | 1969 bp |
| RNA-R5 | ATGCCTTGGAGGAGTGAA | |||
| 6 | RNA-F6 | 44 °C | CCCTTCACTCCTCCAA | 1098 bp |
| RNA-R6 | GAGTAATCCTGACATCCC | |||
| 7 | RNA-F7 | 40 °C | TACGACCGTAACAATG | 957 bp |
| RNA-R7 | GGCTGAGGAGGAGGAG | |||
| 8 | RNA-F8 | 39 °C | CTCTCATACTGCACCA | 1331 bp |
| RNA-R8 | AAATTGCAGGGATTAAATTG | |||
| 9 | RNA-F9 | 44 °C | ACGGTGAAGTGAACGC | 350 bp |
| RNA-R9 | TTTTCTCAAAAAGTGTGG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Xu, T.; Wang, C.; Jia, T.; Zhang, Q. A Novel Picornavirus Discovered in White Leg Shrimp Penaeus vannamei. Viruses 2021, 13, 2381. https://doi.org/10.3390/v13122381
Liu S, Xu T, Wang C, Jia T, Zhang Q. A Novel Picornavirus Discovered in White Leg Shrimp Penaeus vannamei. Viruses. 2021; 13(12):2381. https://doi.org/10.3390/v13122381
Chicago/Turabian StyleLiu, Shuang, Tingting Xu, Chong Wang, Tianchang Jia, and Qingli Zhang. 2021. "A Novel Picornavirus Discovered in White Leg Shrimp Penaeus vannamei" Viruses 13, no. 12: 2381. https://doi.org/10.3390/v13122381
APA StyleLiu, S., Xu, T., Wang, C., Jia, T., & Zhang, Q. (2021). A Novel Picornavirus Discovered in White Leg Shrimp Penaeus vannamei. Viruses, 13(12), 2381. https://doi.org/10.3390/v13122381






