How Influenza A Virus NS1 Deals with the Ubiquitin System to Evade Innate Immunity
Abstract
1. The Ubiquitin and Ubiquitin-like Systems
1.1. The Ubiquitin-Proteasome System
1.2. Ubiquitin-like Systems
1.2.1. SUMOylation
1.2.2. ISGylation
1.2.3. NEDDylation
2. Influenza A Virus and Ubiquitin
2.1. Influenza A Virus
2.2. Importance of the UPS in the IAV Life Cycle
3. IFNs and Cytokines Activation: The Innate Immune Response during IAV Infection
3.1. IFNs Activation
3.2. ISGs Activation
3.3. Inflammasome Complexes
3.4. Autophagy and Apoptosis
4. The Innate Immune Battle between UPS and IAV
4.1. Antiviral Roles of UPS Factors
4.2. IAV Hijacks UPS to Evade Innate Immune Response
5. Mechanisms Developed by NS1 to Inhibit the Innate Immune Response
5.1. IAV NS1 Protein
5.2. NS1 against Host Antiviral Response
5.2.1. IFNs, Cytokines and ISGs Inhibition by NS1
5.2.2. Host Shutoff
5.3. Ubiquitination Perturbations by NS1
5.3.1. TRIM25
5.3.2. DUBs
5.3.3. p53 and MDM2
5.3.4. SUMOylation System
5.3.5. ISG15
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Dikic, I.; Dötsch, V. Ubiquitin Linkages Make a Difference. Nat. Struct. Mol. Biol. 2009, 16, 1209–1210. [Google Scholar] [CrossRef]
- Chen, Z.J.; Sun, L.J. Nonproteolytic Functions of Ubiquitin in Cell Signaling. Mol. Cell 2009, 33, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Chau, V.; Tobias, J.W.; Bachmair, A.; Marriott, D.; Ecker, D.J.; Gonda, D.K.; Varshavsky, A. A Multiubiquitin Chain Is Confined to Specific Lysine in a Targeted Short-Lived Protein. Science 1989, 243, 1576–1583. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A. The Unravelling of the Ubiquitin System. Nat. Rev. Mol. Cell Biol. 2015, 16, 322–324. [Google Scholar] [CrossRef] [PubMed]
- Hershko, A.; Heller, H.; Elias, S.; Ciechanover, A. Components of Ubiquitin-Protein Ligase System. Resolution, Affinity Purification, and Role in Protein Breakdown. J. Biol. Chem. 1983, 258, 8206–8214. [Google Scholar] [CrossRef]
- Komander, D.; Rape, M. The Ubiquitin Code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef]
- Kulathu, Y.; Komander, D. Atypical Ubiquitylation-the Unexplored World of Polyubiquitin beyond Lys48 and Lys63 Linkages. Nat. Rev. Mol. Cell Biol. 2012, 13, 508–523. [Google Scholar] [CrossRef] [PubMed]
- Tanno, H.; Komada, M. The Ubiquitin Code and Its Decoding Machinery in the Endocytic Pathway. J. Biochem. 2013, 153, 497–504. [Google Scholar] [CrossRef]
- Tanaka, K. The Proteasome: Overview of Structure and Functions. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 12–36. [Google Scholar] [CrossRef]
- Komander, D.; Clague, M.J.; Urbé, S. Breaking the Chains: Structure and Function of the Deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563. [Google Scholar] [CrossRef]
- Clague, M.J.; Barsukov, I.; Coulson, J.M.; Liu, H.; Rigden, D.J.; Urbé, S. Deubiquitylases from Genes to Organism. Physiol. Rev. 2013, 93, 1289–1315. [Google Scholar] [CrossRef]
- Swatek, K.N.; Komander, D. Ubiquitin Modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef]
- Hao, R.; Nanduri, P.; Rao, Y.; Panichelli, R.; Ito, A.; Yoshida, M.; Yao, T. Proteasomes Activate Aggresome Disassembly and Clearance by Producing Unanchored Ubiquitin Chains. Mol. Cell 2013, 51, 819–828. [Google Scholar] [CrossRef]
- Zeng, W.; Sun, L.; Jiang, X.; Chen, X.; Hou, F.; Adhikari, A.; Xu, M.; Chen, Z.J. Reconstitution of the RIG-I Pathway Reveals a Signaling Role of Unanchored Polyubiquitin Chains in Innate Immunity. Cell 2010, 141, 315–330. [Google Scholar] [CrossRef]
- Zhou, J.; Xu, Y.; Lin, S.; Guo, Y.; Deng, W.; Zhang, Y.; Guo, A.; Xue, Y. IUUCD 2.0: An Update with Rich Annotations for Ubiquitin and Ubiquitin-like Conjugations. Nucleic Acids Res. 2018, 46, D447–D453. [Google Scholar] [CrossRef] [PubMed]
- Berndsen, C.E.; Wolberger, C. New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 2014, 21, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Shabek, N. Ubiquitin ligases: Structure, function, and regulation. Annu. Rev. Biochem. 2017, 86, 129–157. [Google Scholar] [CrossRef] [PubMed]
- Metzger, M.B.; Pruneda, J.N.; Klevit, R.E.; Weissman, A.M. RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta Mol. Cell Res. 2014, 1843, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Sluimer, J.; Distel, B. Regulating the Human HECT E3 Ligases. Cell Mol. Life Sci. 2018, 75, 3121–3141. [Google Scholar] [CrossRef]
- Metzger, M.B.; Hristova, V.A.; Weissman, A.M. HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 2012, 125, 531–537. [Google Scholar] [CrossRef]
- Bernassola, F.; Karin, M.; Ciechanover, A.; Melino, G. The HECT Family of E3 Ubiquitin Ligases: Multiple Players in Cancer Development. Cancer Cell 2008, 14, 10–21. [Google Scholar] [CrossRef]
- Bosu, D.R.; Kipreos, E.T. Cullin-RING Ubiquitin Ligases: Global Regulation and Activation Cycles. Cell Div. 2008, 3, 7. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, S. TRIM family proteins: Roles in autophagy, immunity, and carcinogenesis. Trends Biochem. Sci. 2017, 42, 297–311. [Google Scholar] [CrossRef] [PubMed]
- Morreale, F.E.; Walden, H. Types of Ubiquitin Ligases. Cell 2016, 165, 248.e1. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wang, L.; Li, S. Influenza A virus-host protein interactions control viral pathogenesis. Int. J. Mol. Sci. 2017, 18, 1673. [Google Scholar] [CrossRef] [PubMed]
- Rudnicka, A.; Yamauchi, Y. Ubiquitin in influenza virus entry and innate immunity. Viruses 2016, 8, 293. [Google Scholar] [CrossRef]
- Bedford, L.; Lowe, J.; Dick, L.R.; Mayer, R.J.; Brownell, J.E. Ubiquitin-like Protein Conjugation and the Ubiquitin-Proteasome System as Drug Targets. Nat. Rev. Drug Discov. 2011, 10, 29–46. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X. SUMO-Mediated Regulation of Nuclear Functions and Signaling Processes. Mol. Cell 2018, 71, 409–418. [Google Scholar] [CrossRef]
- Enserink, J.M. Sumo and the Cellular Stress Response. Cell Div. 2015, 10, 4. [Google Scholar] [CrossRef]
- Everett, R.D.; Boutell, C.; Hale, B.G. Interplay between Viruses and Host Sumoylation Pathways. Nat. Rev. Microbiol. 2013, 11, 400–411. [Google Scholar] [CrossRef]
- Hu, M.-M.; Liao, C.-Y.; Yang, Q.; Xie, X.-Q.; Shu, H.-B. Innate Immunity to RNA Virus Is Regulated by Temporal and Reversible Sumoylation of RIG-I and MDA5. J. Exp. Med. 2017, 214, 973–989. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.M.; Yang, Q.; Xie, X.Q.; Liao, C.Y.; Lin, H.; Liu, T.T.; Yin, L.; Shu, H. Sumoylation Promotes the Stability of the DNA Sensor CGAS and the Adaptor STING to Regulate the Kinetics of Response to DNA Virus. Immunity 2016, 45, 555–569. [Google Scholar] [CrossRef]
- Crowl, J.T.; Stetson, D.B. SUMO2 and SUMO3 Redundantly Prevent a Noncanonical Type I Interferon Response. Proc. Natl. Acad. Sci. USA 2018, 115, 6798–6803. [Google Scholar] [CrossRef]
- Decque, A.; Joffre, O.; Magalhaes, J.G.; Cossec, J.-C.; Blecher-Gonen, R.; Lapaquette, P.; Silvin, A.; Manel, N.; Joubert, P.-E.; Seeler, J.-S.; et al. Sumoylation Coordinates the Repression of Inflammatory and Anti-Viral Gene-Expression Programs during Innate Sensing. Nat. Immunol. 2016, 17, 140–149. [Google Scholar] [CrossRef]
- Zhao, C.; Denison, C.; Huibregtse, J.M.; Gygi, S.; Krug, R.M. Human ISG15 Conjugation Targets Both IFN-Induced and Constitutively Expressed Proteins Functioning in Diverse Cellular Pathways. Proc. Natl. Acad. Sci. USA 2005, 102, 10200–10205. [Google Scholar] [CrossRef] [PubMed]
- Haas, A.L.; Ahrens, P.; Bright, P.M.; Ankel, H. Interferon Induces a 15-Kilodalton Protein Exhibiting Marked Homology to Ubiquitin. J. Biol. Chem. 1987, 262, 11315–11323. [Google Scholar] [CrossRef]
- Loeb, K.R.; Haas, A.L. The Interferon-Inducible 15-KDa Ubiquitin Homolog Conjugates to Intracellular Proteins. J. Biol. Chem. 1992, 267, 7806–7813. [Google Scholar] [CrossRef]
- Narasimhan, J.; Wang, M.; Fu, Z.; Klein, J.M.; Haas, A.L.; Kim, J.-J.P. Crystal Structure of the Interferon-Induced Ubiquitin-like Protein ISG15. J. Biol. Chem. 2005, 280, 27356–27365. [Google Scholar] [CrossRef] [PubMed]
- Swaim, C.D.; Canadeo, L.A.; Monte, K.J.; Khanna, S.; Lenschow, D.J.; Huibregtse, J.M. Modulation of Extracellular ISG15 Signaling by Pathogens and Viral Effector Proteins. Cell Rep. 2020, 31, 107772. [Google Scholar] [CrossRef]
- D’Cunha, J.; Ramanujam, S.; Wagner, R.J.; Witt, P.L.; Knight, E.; Borden, E.C. In Vitro and in Vivo Secretion of Human ISG15, an IFN-Induced Immunomodulatory Cytokine. J. Immunol. 1996, 157, 4100–4108. [Google Scholar]
- Recht, M.; Borden, E.C.; Knight, E. A Human 15-KDa IFN-Induced Protein Induces the Secretion of IFN-Gamma. J. Immunol. 1991, 147, 2617–2623. [Google Scholar]
- Farrell, P.J.; Broeze, R.J.; Lengyel, P. Accumulation of an MRNA and Protein in Interferon-Treated Ehrlich Ascites Tumour Cells. Nature 1979, 279, 523–525. [Google Scholar] [CrossRef]
- Ashley, C.L.; Abendroth, A.; McSharry, B.P.; Slobedman, B. Interferon-Independent Upregulation of Interferon-Stimulated Genes during Human Cytomegalovirus Infection Is Dependent on IRF3 Expression. Viruses 2019, 11, 246. [Google Scholar] [CrossRef]
- Lertsooksawat, W.; Wongnoppavich, A.; Chairatvit, K. Up-Regulation of Interferon-Stimulated Gene 15 and Its Conjugation Machinery, UbE1L and UbcH8 Expression by Tumor Necrosis Factor-α through P38 MAPK and JNK Signaling Pathways in Human Lung Carcinoma. Mol. Cell Biochem. 2019, 462, 51–59. [Google Scholar] [CrossRef]
- Radoshevich, L.; Impens, F.; Ribet, D.; Quereda, J.J.; Nam Tham, T.; Nahori, M.-A.; Bierne, H.; Dussurget, O.; Pizarro-Cerdá, J.; Knobeloch, K.-P.; et al. ISG15 Counteracts Listeria Monocytogenes Infection. eLife 2015, 4, e06848. [Google Scholar] [CrossRef]
- Yuan, W.; Krug, R.M. Influenza B Virus NS1 Protein Inhibits Conjugation of the Interferon (IFN)-Induced Ubiquitin-like ISG15 Protein. EMBO J. 2001, 20, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.I.; Giannakopoulos, N.V.; Virgin, H.W.; Zhang, D.-E. Interferon-Inducible Ubiquitin E2, Ubc8, Is a Conjugating Enzyme for Protein ISGylation. Mol. Cell Biol. 2004, 24, 9592–9600. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Beaudenon, S.L.; Kelley, M.L.; Waddell, M.B.; Yuan, W.; Schulman, B.A.; Huibregtse, J.M.; Krug, R.M. The UbcH8 Ubiquitin E2 Enzyme Is Also the E2 Enzyme for ISG15, an IFN-α/β-Induced Ubiquitin-like Protein. Proc. Natl. Acad. Sci. USA 2004, 101, 7578–7582. [Google Scholar] [CrossRef]
- Dastur, A.; Beaudenon, S.; Kelley, M.; Krug, R.M.; Huibregtse, J.M. Herc5, an Interferon-Induced HECT E3 Enzyme, Is Required for Conjugation of ISG15 in Human Cells. J. Biol. Chem. 2006, 281, 4334–4338. [Google Scholar] [CrossRef]
- Oudshoorn, D.; van Boheemen, S.; Sánchez-Aparicio, M.T.; Rajsbaum, R.; García-Sastre, A.; Versteeg, G.A. HERC6 Is the Main E3 Ligase for Global ISG15 Conjugation in Mouse Cells. PLoS ONE 2012, 7, e29870. [Google Scholar] [CrossRef]
- Ketscher, L.; Basters, A.; Prinz, M.; Knobeloch, K.-P. MHERC6 Is the Essential ISG15 E3 Ligase in the Murine System. Biochem. Biophys. Res. Commun. 2012, 417, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Malakhov, M.P.; Malakhova, O.A.; Kim, K.I.; Ritchie, K.J.; Zhang, D.-E. UBP43 (USP18) Specifically Removes ISG15 from Conjugated Proteins. J. Biol. Chem. 2002, 277, 9976–9981. [Google Scholar] [CrossRef]
- Durfee, L.A.; Lyon, N.; Seo, K.; Huibregtse, J.M. The ISG15 Conjugation System Broadly Targets Newly Synthesized Proteins: Implications for the Antiviral Function of ISG15. Mol. Cell 2010, 38, 722–732. [Google Scholar] [CrossRef]
- Enchev, R.I.; Schulman, B.A.; Peter, M. Protein Neddylation: Beyond Cullin–RING Ligases. Nat. Rev. Mol. Cell Biol. 2015, 16, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Duda, D.M.; Borg, L.A.; Scott, D.C.; Hunt, H.W.; Hammel, M.; Schulman, B.A. Structural Insights into NEDD8 Activation of Cullin-RING Ligases: Conformational Control of Conjugation. Cell 2008, 134, 995. [Google Scholar] [CrossRef]
- Dubiel, W.; Chaithongyot, S.; Dubiel, D.; Naumann, M. The COP9 Signalosome: A Multi-DUB Complex. Biomolecules 2020, 10, 1082. [Google Scholar] [CrossRef] [PubMed]
- Flick, K.; Kaiser, P. Set Them Free: F-Box Protein Exchange by Cand1. Cell Res. 2013, 23, 870–871. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhu, H.; Liu, Y.; He, F.; Xie, P.; Zhang, L. Itch Promotes the Neddylation of JunB and Regulates JunB-Dependent Transcription. Cell. Signal. 2016, 28, 1186–1195. [Google Scholar] [CrossRef]
- Um, J.W.; Han, K.A.; Im, E.; Oh, Y.; Lee, K.; Chung, K.C. Neddylation Positively Regulates the Ubiquitin E3 Ligase Activity of Parkin. J. Neurosci. Res. 2012, 90, 1030–1042. [Google Scholar] [CrossRef]
- Xirodimas, D.P.; Saville, M.K.; Bourdon, J.-C.; Hay, R.T.; Lane, D.P. Mdm2-Mediated NEDD8 Conjugation of P53 Inhibits Its Transcriptional Activity. Cell 2004, 118, 83–97. [Google Scholar] [CrossRef]
- Batuello, C.N.; Hauck, P.M.; Gendron, J.M.; Lehman, J.A.; Mayo, L.D. Src Phosphorylation Converts Mdm2 from a Ubiquitinating to a Neddylating E3 Ligase. Proc. Natl. Acad. Sci. USA 2015, 112, 1749–1754. [Google Scholar] [CrossRef]
- Zou, T.; Zhang, J. Diverse and Pivotal Roles of Neddylation in Metabolism and Immunity. FEBS J. 2021, 288, 3884–3912. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F.; Smith, G.J.D.; Fouchier, R.A.M.; Peiris, M.; Kedzierska, K.; Doherty, P.C.; Palese, P.; Shaw, M.L.; Treanor, J.; Webster, R.G.; et al. Influenza. Nat. Rev. Dis. Primers 2018, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Vasin, A.V.; Temkina, O.A.; Egorov, V.V.; Klotchenko, S.A.; Plotnikova, M.A.; Kiselev, O.I. Molecular Mechanisms Enhancing the Proteome of Influenza A Viruses: An Overview of Recently Discovered Proteins. Virus. Res. 2014, 185, 53–63. [Google Scholar] [CrossRef]
- Subbarao, E.K.; London, W.; Murphy, B.R. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 1993, 67, 1761–1764. [Google Scholar] [CrossRef] [PubMed]
- Robb, N.C.; Smith, M.; Vreede, F.T.; Fodor, E. NS2/NEP Protein Regulates Transcription and Replication of the Influenza Virus RNA Genome. J. Gen. Virol. 2009, 90, 1398–1407. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, E.C.; Denham, E.M.; Thomas, B.; Trudgian, D.C.; Hester, S.S.; Ridlova, G.; York, A.; Turrell, L.; Fodor, E. Mapping the Phosphoproteome of Influenza A and B Viruses by Mass Spectrometry. PLoS Pathog. 2012, 8, e1002993. [Google Scholar] [CrossRef]
- Kirui, J.; Mondal, A.; Mehle, A. Ubiquitination upregulates influenza virus polymerase function. J. Virol. 2016, 90, 10906–10914. [Google Scholar] [CrossRef]
- Pal, S.; Santos, A.; Rosas, J.M.; Ortiz-Guzman, J.; Rosas-Acosta, G. Influenza A Virus Interacts Extensively with the Cellular SUMOylation System during Infection. Virus. Res. 2011, 158, 12–27. [Google Scholar] [CrossRef]
- Han, Q.; Chang, C.; Li, L.; Klenk, C.; Cheng, J.; Chen, Y.; Xia, N.; Shu, Y.; Chen, Z.; Gabriel, G.; et al. Sumoylation of Influenza A Virus Nucleoprotein Is Essential for Intracellular Trafficking and Virus Growth. J. Virol. 2014, 88, 9379–9390. [Google Scholar] [CrossRef]
- Di Pietro, A.; Kajaste-Rudnitski, A.; Oteiza, A.; Nicora, L.; Towers, G.J.; Mechti, N.; Vicenzi, E. TRIM22 Inhibits Influenza A Virus Infection by Targeting the Viral Nucleoprotein for Degradation. J. Virol. 2013, 87, 4523–4533. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Wang, L.; Ding, H.; Schwamborn, J.C.; Li, S.; Dorf, M.E. TRIM32 senses and restricts influenza A virus by ubiquitination of PB1 polymerase. PLoS Pathog. 2015, 11, e1004960. [Google Scholar] [CrossRef] [PubMed]
- Liao, T.L.; Wu, C.Y.; Su, W.C.; Jeng, K.S.; Lai, M.M.C. Ubiquitination and deubiquitination of NP protein regulates influenza A virus RNA replication. EMBO J. 2010, 29, 3879–3890. [Google Scholar] [CrossRef] [PubMed]
- Widjaja, I.; De Vries, E.; Tscherne, D.M.; García-Sastre, A.; Rottier, P.J.M.; De Haan, C.A.M. Inhibition of the Ubiquitin-Proteasome System Affects Influenza A Virus Infection at a Postfusion Step. J. Virol. 2010, 84, 9625–9631. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-; Zhou, L.; Chen, G.; Krug, R.M. Battle between Influenza A Virus and a Newly Identified Antiviral Activity of the PARP-Containing ZAPL Protein. Proc. Natl. Acad. Sci. USA 2015, 112, 14048–14053. [Google Scholar] [CrossRef]
- Chesarino, N.M.; McMichael, T.M.; Yount, J.S. E3 Ubiquitin Ligase NEDD4 Promotes Influenza Virus Infection by Decreasing Levels of the Antiviral Protein IFITM3. PLoS Pathog. 2015, 11, e1005095. [Google Scholar] [CrossRef]
- Su, W.C.; Chen, Y.C.; Tseng, C.H.; Hsu, P.W.C.; Tung, K.F.; Jeng, K.S.; Lai, M.M.C. Pooled RNAi Screen Identifies Ubiquitin Ligase Itch as Crucial for Influenza A Virus Release from the Endosome during Virus Entry. Proc. Natl. Acad. Sci. USA 2013, 110, 17516–17521. [Google Scholar] [CrossRef]
- Watanabe, T.; Watanabe, S.; Kawaoka, Y. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe 2010, 7, 427–439. [Google Scholar] [CrossRef]
- Khor, R.; McElroy, L.J.; Whittaker, G.R. The Ubiquitin-Vacuolar Protein Sorting System Is Selectively Required during Entry of Influenza Virus into Host Cells. Traffic 2003, 4, 857–868. [Google Scholar] [CrossRef]
- Calistri, A.; Munegato, D.; Carli, I.; Parolin, C.; Palu, G. The Ubiquitin-Conjugating System: Multiple Roles in Viral Replication and Infection. Cells 2014, 3, 386–417. [Google Scholar] [CrossRef]
- Wimmer, P.; Schreiner, S. Viral mimicry to usurp ubiquitin and sumo host pathways. Viruses 2015, 7, 4854–4877. [Google Scholar] [CrossRef]
- Smith, M.C.; Boutell, C.; Davido, D.J. HSV-1 ICP0: Paving the Way for Viral Replication. Future Virol. 2011, 6, 421–429. [Google Scholar] [CrossRef]
- Kroeker, A.L.; Ezzati, P.; Coombs, K.M.; Halayko, A.J. Influenza A Infection of Primary Human Airway Epithelial Cells Up-Regulates Proteins Related to Purine Metabolism and Ubiquitin-Related Signaling. J. Proteome Res. 2013, 12, 3139–3151. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Ito, T.; Suzuki, T.; Holland, R.E., Jr.; Chambers, T.M.; Kiso, M.; Ishida, H.; Kawaoka, Y. Sialic acid species as a determinant of the host range of influenza A viruses. J. Virol. 2000, 74, 11825–11831. [Google Scholar] [CrossRef] [PubMed]
- Pinto, L.H.; Holsinger, L.J.; Lamb, R.A. Influenza virus M2 protein has ion channel activity. Cell 1992, 69, 517–528. [Google Scholar] [CrossRef]
- Stauffer, S.; Feng, Y.; Nebioglu, F.; Heilig, R.; Picotti, P.; Helenius, A. Stepwise priming by acidic pH and a high K+ concentration is required for efficient uncoating of influenza a virus cores after penetration. J. Virol. 2014, 88, 13029–13046. [Google Scholar] [CrossRef]
- White, J.; Kartenbeck, J.; Helenius, A. Membrane fusion activity of influenza virus. EMBO J. 1982, 1, 217–222. [Google Scholar] [CrossRef]
- Huotari, J.; Meyer-Schaller, N.; Hubner, M.; Stauffer, S.; Katheder, N.; Horvath, P.; Mancini, R.; Helenius, A.; Peter, M. Cullin-3 Regulates Late Endosome Maturation. Proc. Natl. Acad. Sci. USA 2012, 109, 823–828. [Google Scholar] [CrossRef]
- Hubner, M.; Peter, M. Cullin-3 and the endocytic system: New functions of ubiquitination for endosome maturation. Cell Logist 2012, 2, 166–168. [Google Scholar] [CrossRef]
- Cros, J.F.; García-Sastre, A.; Palese, P. An unconventional NLS is critical for the nuclear import of the influenza A virus nucleoprotein and ribonucleoprotein. Traffic 2005, 6, 205–213. [Google Scholar] [CrossRef]
- Eisfeld, A.J.; Neumann, G.; Kawaoka, Y. At the centre: Influenza A virus ribonucleoproteins. Nat. Rev. Microbiol. 2015, 13, 28–41. [Google Scholar] [CrossRef]
- Wu, W.W.H.; Sun, Y.-B.; Panté, N. Nuclear Import of Influenza A Viral Ribonucleoprotein Complexes Is Mediated by Two Nuclear Localization Sequences on Viral Nucleoprotein. Virol. J. 2007, 4, 49. [Google Scholar] [CrossRef]
- Ko, H.S.; Lee, Y.; Shin, J.-H.; Karuppagounder, S.S.; Gadad, B.S.; Koleske, A.J.; Pletnikova, O.; Troncoso, J.C.; Dawson, V.L.; Dawson, T.M. Phosphorylation by the C-Abl Protein Tyrosine Kinase Inhibits Parkin’s Ubiquitination and Protective Function. Proc. Natl. Acad. Sci. USA 2010, 107, 16691–16696. [Google Scholar] [CrossRef]
- Lee, Y.; Karuppagounder, S.S.; Shin, J.-H.; Lee, Y.-I.; Ko, H.S.; Swing, D.; Jiang, H.; Kang, S.-U.; Lee, B.D.; Kang, H.C.; et al. Parthanatos Mediates AIMP2 Activated Age Dependent Dopaminergic Neuronal Loss. Nat. Neurosci. 2013, 16, 1392–1400. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Wu, J.; Liu, R.; Li, J.; Song, L.; Teng, Y.; Sheng, C.; Liu, D.; Yao, C.; Chen, H.; et al. Interaction of NS2 with AIMP2 Facilitates the Switch from Ubiquitination to SUMOylation of M1 in Influenza a Virus-Infected Cells. J. Virol. 2015, 89, 300–311. [Google Scholar] [CrossRef]
- Wu, C.-; Jeng, K.; Lai, M.M. The SUMOylation of Matrix Protein M1 Modulates the Assembly and Morphogenesis of Influenza A Virus. J. Virol. 2011, 85, 6618–6628. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A.; Pillai, P.S. Innate Immunity to Influenza Virus Infection. Nat. Rev. Immunol. 2014, 14, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.E.; Gack, M.U. Ubiquitination in the Antiviral Immune Response. Virology 2015, 479, 52–65. [Google Scholar] [CrossRef]
- Feng, W.; Sun, X.; Shi, N.; Zhang, M.; Guan, Z.; Duan, M. Influenza a Virus NS1 Protein Induced A20 Contributes to Viral Replication by Suppressing Interferon-Induced Antiviral Response. Biochem. Biophys. Res. Commun. 2017, 482, 1107–1113. [Google Scholar] [CrossRef]
- Davidson, S.; Crotta, S.; McCabe, T.M.; Wack, A. Pathogenic Potential of Interferon Aβ in Acute Influenza Infection. Nat. Commun. 2014, 5, 3864. [Google Scholar] [CrossRef]
- Davidson, S.; McCabe, T.M.; Crotta, S.; Gad, H.H.; Hessel, E.M.; Beinke, S.; Hartmann, R.; Wack, A. IFNλ Is a Potent Anti-influenza Therapeutic without the Inflammatory Side Effects of IFNα Treatment. EMBO Mol. Med. 2016, 8, 1099–1112. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Lou, D.I.; Sawyer, S.L. Identification of a Genomic Reservoir for New TRIM Genes in Primate Genomes. PLoS Genet. 2011, 7, e1002388. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Li, N.L.; Wang, J.; Shi, P.-Y.; Wang, T.; Miller, M.A.; Li, K. Overlapping and Distinct Molecular Determinants Dictating the Antiviral Activities of TRIM56 against Flaviviruses and Coronavirus. J. Virol. 2014, 88, 13821–13835. [Google Scholar] [CrossRef]
- Rajsbaum, R.; García-Sastre, A.; Versteeg, G.A. TRIMmunity: The Roles of the TRIM E3-Ubiquitin Ligase Family in Innate Antiviral Immunity. J. Mol. Biol. 2014, 426, 1265–1284. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-R.; Lee, M.K.; Kim, C.W.; Kim, M. TRIM Proteins and Their Roles in the Influenza Virus Life Cycle. Microorganisms 2020, 8, 1424. [Google Scholar] [CrossRef]
- Gack, M.U.; Shin, Y.C.; Joo, C.-; Urano, T.; Liang, C.; Sun, L.; Takeuchi, O.; Akira, S.; Chen, Z.; Inoue, S.; et al. TRIM25 RING-Finger E3 Ubiquitin Ligase Is Essential for RIG-I-Mediated Antiviral Activity. Nature 2007, 446, 916–920. [Google Scholar] [CrossRef]
- Gack, M.U.; Albrecht, R.A.; Urano, T.; Inn, K.-; Huang, I.-; Carnero, E.; Farzan, M.; Inoue, S.; Jung, J.U.; García-Sastre, A. Influenza A Virus NS1 Targets the Ubiquitin Ligase TRIM25 to Evade Recognition by the Host Viral RNA Sensor RIG-I. Cell Host Microbe 2009, 5, 439–449. [Google Scholar] [CrossRef]
- Pauli, E.-; Chan, Y.K.; Davis, M.E.; Gableske, S.; Wang, M.K.; Feister, K.F.; Gack, M.U. The Ubiquitin-Specific Protease USP15 Promotes RIG-I-Mediated Antiviral Signaling by Deubiquitylating TRIM25. Sci. Signal. 2014, 7, ra3. [Google Scholar] [CrossRef]
- Marín, I. Origin and diversification of TRIM ubiquitin ligases. PLoS ONE 2012, 7, e50030. [Google Scholar] [CrossRef]
- Esposito, D.; Koliopoulos, M.G.; Rittinger, K. Structural Determinants of TRIM Protein Function. Biochem. Soc. Trans. 2017, 45, 183–191. [Google Scholar] [CrossRef]
- Reymond, A.; Meroni, G.; Fantozzi, A.; Merla, G.; Cairo, S.; Luzi, L.; Riganelli, D.; Zanaria, E.; Messali, S.; Cainarca, S.; et al. The Tripartite Motif Family Identifies Cell Compartments. EMBO J. 2001, 20, 2140–2151. [Google Scholar] [CrossRef]
- Scherer, M.; Klingl, S.; Sevvana, M.; Otto, V.; Schilling, E.-; Stump, J.D.; Müller, R.; Reuter, N.; Sticht, H.; Muller, Y.A.; et al. Crystal Structure of Cytomegalovirus IE1 Protein Reveals Targeting of TRIM Family Member PML via Coiled-Coil Interactions. PLoS Pathog. 2014, 10, e1004512. [Google Scholar] [CrossRef]
- Schilling, E.-M.; Scherer, M.; Reuter, N.; Schweininger, J.; Muller, Y.A.; Stamminger, T. The Human Cytomegalovirus IE1 Protein Antagonizes PML Nuclear Body-Mediated Intrinsic Immunity via the Inhibition of PML De Novo SUMOylation. J. Virol. 2017, 91, e02049-16. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.M.; Gao, J.; Wang, J.; Yang, M.; Potts, P.R. MAGE-RING Protein Complexes Comprise a Family of E3 Ubiquitin Ligases. Mol. Cell 2010, 39, 963–974. [Google Scholar] [CrossRef] [PubMed]
- Pineda, C.T.; Ramanathan, S.; Fon Tacer, K.; Weon, J.L.; Potts, M.B.; Ou, Y.-; White, M.A.; Potts, P.R. Degradation of AMPK by a Cancer-Specific Ubiquitin Ligase. Cell 2015, 160, 715–728. [Google Scholar] [CrossRef]
- Van Gent, M.; Sparrer, K.M.J.; Gack, M.U. TRIM Proteins and Their Roles in Antiviral Host Defenses. Annu. Rev. Virol. 2018, 5, 385–405. [Google Scholar] [CrossRef]
- Cambiaghi, V.; Giuliani, V.; Lombardi, S.; Marinelli, C.; Toffalorio, F.; Pelicci, P.G. TRIM Proteins in Cancer. Adv. Exp. Med. Biol. 2012, 770, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Koliopoulos, M.G.; Lethier, M.; Van Der Veen, A.G.; Haubrich, K.; Hennig, J.; Kowalinski, E.; Stevens, R.V.; Martin, S.R.; Reis e Sousa, C.; Cusack, S.; et al. Molecular mechanism of influenza A NS1-mediated TRIM25 recognition and inhibition. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Yoneyama, M.; Fujita, T. Structural Mechanism of RNA Recognition by the RIG-I-like Receptors. Immunity 2008, 29, 178–181. [Google Scholar] [CrossRef]
- Yoneyama, M.; Kikuchi, M.; Natsukawa, T.; Shinobu, N.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Akira, S.; Fujita, T. The RNA Helicase RIG-I Has an Essential Function in Double-Stranded RNA-Induced Innate Antiviral Responses. Nat. Immunol. 2004, 5, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Baum, A.; Sachidanandam, R.; García-Sastre, A. Preference of RIG-I for Short Viral RNA Molecules in Infected Cells Revealed by next-Generation Sequencing. Proc. Natl. Acad. Sci. USA 2010, 107, 16303–16308. [Google Scholar] [CrossRef]
- Versteeg, G.A.; García-Sastre, A. Viral Tricks to Grid-Lock the Type I Interferon System. Curr. Opin. Microbiol. 2010, 13, 508–516. [Google Scholar] [CrossRef]
- Hornung, V.; Ellegast, J.; Kim, S.; Brzózka, K.; Jung, A.; Kato, H.; Poeck, H.; Akira, S.; Conzelmann, K.; Schlee, M.; et al. 5′Triphosphate RNA Is the Ligand for RIG-I. Science 2006, 314, 994–997. [Google Scholar] [CrossRef]
- Weber-Gerlach, M.; Weber, F. Standing on Three Legs: Antiviral Activities of RIG-I against Influenza Viruses. Curr. Opin. Immunol. 2016, 42, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Pichlmair, A.; Schulz, O.; Tan, C.P.; Näslund, T.I.; Liljeström, P.; Weber, F.; Sousa, C.R.E. RIG-I-Mediated Antiviral Responses to Single-Stranded RNA Bearing 5′-Phosphates. Science 2006, 314, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Rehwinkel, J.; Tan, C.P.; Goubau, D.; Schulz, O.; Pichlmair, A.; Bier, K.; Robb, N.; Vreede, F.; Barclay, W.; Fodor, E.; et al. RIG-I Detects Viral Genomic RNA during Negative-Strand RNA Virus Infection. Cell 2010, 140, 397–408. [Google Scholar] [CrossRef]
- Weber, M.; Sediri, H.; Felgenhauer, U.; Binzen, I.; Bänfer, S.; Jacob, R.; Brunotte, L.; García-Sastre, A.; Schmid-Burgk, J.L.; Schmidt, T.; et al. Influenza Virus Adaptation PB2-627K Modulates Nucleocapsid Inhibition by the Pathogen Sensor RIG-I. Cell Host Microbe 2015, 17, 309–319. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, W.; Duggan, E.S.; Booth, J.L.; Zou, M.-H.; Metcalf, J.P. RIG-I and TLR3 Are Both Required for Maximum Interferon Induction by Influenza Virus in Human Lung Alveolar Epithelial Cells. Virology 2015, 482, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Kolakofsky, D.; Kowalinski, E.; Cusack, S. A Structure-Based Model of RIG-I Activation. RNA 2012, 18, 2118–2127. [Google Scholar] [CrossRef] [PubMed]
- Reikine, S.; Nguyen, J.B.; Modis, Y. Pattern Recognition and Signaling Mechanisms of RIG-I and MDA5. Front. Immunol. 2014, 5, 342. [Google Scholar] [CrossRef]
- Patel, J.R.; Jain, A.; Chou, Y.; Baum, A.; Ha, T.; García-Sastre, A. ATPase-Driven Oligomerization of RIG-I on RNA Allows Optimal Activation of Type-I Interferon. EMBO Rep. 2013, 14, 780–787. [Google Scholar] [CrossRef] [PubMed]
- Ferrage, F.; Dutta, K.; Nistal-Villán, E.; Patel, J.R.; Sánchez-Aparicio, M.T.; De Ioannes, P.; Buku, A.; Aseguinolaza, G.G.; García-Sastre, A.; Aggarwal, A.K. Structure and Dynamics of the Second CARD of Human RIG-I Provide Mechanistic Insights into Regulation of RIG-I Activation. Structure 2012, 20, 2048–2061. [Google Scholar] [CrossRef]
- Kowalinski, E.; Lunardi, T.; McCarthy, A.A.; Louber, J.; Brunel, J.; Grigorov, B.; Gerlier, D.; Cusack, S. Structural Basis for the Activation of Innate Immune Pattern-Recognition Receptor RIG-I by Viral RNA. Cell 2011, 147, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Oshiumi, H.; Miyashita, M.; Inoue, N.; Okabe, M.; Matsumoto, M.; Seya, T. The ubiquitin ligase riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe 2010, 8, 496–509. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Yang, Y.-K.; Wang, R.-P.; Zhou, X.; Diao, F.-C.; Li, M.-D.; Zhai, Z.-H.; Jiang, Z.-F.; Chen, D.-Y. REUL Is a Novel E3 Ubiquitin Ligase and Stimulator of Retinoic-Acid-Inducible Gene-I. PLoS ONE 2009, 4, e5760. [Google Scholar] [CrossRef]
- Oshiumi, H.; Matsumoto, M.; Hatakeyama, S.; Seya, T. Riplet/RNF135, a RING Finger Protein, Ubiquitinates RIG-I to Promote Interferon-Beta Induction during the Early Phase of Viral Infection. J. Biol. Chem. 2009, 284, 807–817. [Google Scholar] [CrossRef]
- Sun, X.; Xian, H.; Tian, S.; Sun, T.; Qin, Y.; Zhang, S.; Cui, J. A hierarchical mechanism of RIG-I ubiquitination provides sensitivity, robustness and synergy in antiviral immune responses. Sci. Rep. 2016, 6, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Cadena, C.; Ahmad, S.; Xavier, A.; Willemsen, J.; Park, S.; Park, J.W.; Oh, S.-W.; Fujita, T.; Hou, F.; Binder, M.; et al. Ubiquitin-Dependent and -Independent Roles of E3 Ligase RIPLET in Innate Immunity. Cell 2019, 177, 1187–1200.e16. [Google Scholar] [CrossRef]
- Sanchez, J.G.; Chiang, J.J.; Sparrer, K.M.J.; Alam, S.L.; Chi, M.; Roganowicz, M.D.; Sankaran, B.; Gack, M.U.; Pornillos, O. Mechanism of TRIM25 Catalytic Activation in the Antiviral RIG-I Pathway. Cell Rep. 2016, 16, 1315–1325. [Google Scholar] [CrossRef]
- D’Cruz, A.A.; Kershaw, N.J.; Chiang, J.J.; Wang, M.K.; Nicola, N.A.; Babon, J.J.; Gack, M.U.; Nicholson, S.E. Crystal Structure of the TRIM25 B30.2 (PRYSPRY) Domain: A Key Component of Antiviral Signalling. Biochem. J. 2013, 456, 231–240. [Google Scholar] [CrossRef]
- Peisley, A.; Wu, B.; Xu, H.; Chen, Z.J.; Hur, S. Structural Basis for Ubiquitin-Mediated Antiviral Signal Activation by RIG-I. Nature 2014, 508, 110–114. [Google Scholar] [CrossRef]
- Okamoto, M.; Kouwaki, T.; Fukushima, Y.; Oshiumi, H. Regulation of RIG-I activation by K63-linked polyubiquitination. Front. Immunol. 2018, 8, 1942. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Chen, Z.J. Structural Insights into the Activation of RIG-I, a Nanosensor for Viral RNAs. EMBO Rep. 2012, 13, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Aparicio, M.T.; Ayllón, J.; Leo-Macias, A.; Wolff, T.; García-Sastre, A. Subcellular Localizations of RIG-I, TRIM25, and MAVS Complexes. J. Virol. 2017, 91, e01155-16. [Google Scholar] [CrossRef] [PubMed]
- Seth, R.B.; Sun, L.; Ea, C.-K.; Chen, Z.J. Identification and Characterization of MAVS, a Mitochondrial Antiviral Signaling Protein That Activates NF-ΚB and IRF3. Cell 2005, 122, 669–682. [Google Scholar] [CrossRef]
- Kawai, T.; Takahashi, K.; Sato, S.; Coban, C.; Kumar, H.; Kato, H.; Ishii, K.J.; Takeuchi, O.; Akira, S. IPS-1, an Adaptor Triggering RIG-I- and Mda5-Mediated Type I Interferon Induction. Nat. Immunol. 2005, 6, 981–988. [Google Scholar] [CrossRef]
- Wu, B.; Peisley, A.; Tetrault, D.; Li, Z.; Egelman, E.H.; Magor, K.E.; Walz, T.; Penczek, P.A.; Hur, S. Molecular Imprinting as a Signal-Activation Mechanism of the Viral RNA Sensor RIG-I. Mol. Cell 2014, 55, 511–523. [Google Scholar] [CrossRef]
- Xu, H.; He, X.; Zheng, H.; Huang, L.J.; Hou, F.; Yu, Z.; de la Cruz, M.J.; Borkowski, B.; Zhang, X.; Chen, Z.J.; et al. Correction: Structural Basis for the Prion-like MAVS Filaments in Antiviral Innate Immunity. eLife 2015, 4, e07546. [Google Scholar] [CrossRef]
- Li, X.-D.; Sun, L.; Seth, R.B.; Pineda, G.; Chen, Z.J. Hepatitis C Virus Protease NS3/4A Cleaves Mitochondrial Antiviral Signaling Protein off the Mitochondria to Evade Innate Immunity. Proc. Natl. Acad. Sci. USA 2005, 102, 17717–17722. [Google Scholar] [CrossRef]
- Hou, F.; Sun, L.; Zheng, H.; Skaug, B.; Jiang, Q.-X.; Chen, Z.J. MAVS Forms Functional Prion-like Aggregates to Activate and Propagate Antiviral Innate Immune Response. Cell 2011, 146, 448–461. [Google Scholar] [CrossRef]
- Xu, L.-G.; Wang, Y.-Y.; Han, K.-J.; Li, L.-Y.; Zhai, Z.; Shu, H.-B. VISA Is an Adapter Protein Required for Virus-Triggered IFN-Beta Signaling. Mol. Cell 2005, 19, 727–740. [Google Scholar] [CrossRef] [PubMed]
- Gack, M.U. Mechanisms of RIG-I-Like Receptor Activation and Manipulation by Viral Pathogens. J. Virol. 2014, 88, 5213–5216. [Google Scholar] [CrossRef]
- Jensen, S.; Thomsen, A.R. Sensing of RNA Viruses: A Review of Innate Immune Receptors Involved in Recognizing RNA Virus Invasion. J. Virol. 2012, 86, 2900–2910. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, O.; Akira, S. Innate Immunity to Virus Infection. Immunol. Rev. 2009, 227, 75–86. [Google Scholar] [CrossRef]
- Wolff, T.; Ludwig, S. Influenza Viruses Control the Vertebrate Type I Interferon System: Factors, Mechanisms, and Consequences. J. Interferon Cytokine Res. 2009, 29, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Hermant, P.; Michiels, T. Interferon-λ in the Context of Viral Infections: Production, Response and Therapeutic Implications. J. Innate. Immun. 2014, 6, 563–574. [Google Scholar] [CrossRef]
- Orian, A.; Gonen, H.; Bercovich, B.; Fajerman, I.; Eytan, E.; Israël, A.; Mercurio, F.; Iwai, K.; Schwartz, A.L.; Ciechanover, A. SCFβ-TrCP Ubiquitin Ligase-Mediated Processing of NF-ΚB P105 Requires Phosphorylation of Its C-Terminus by IκB Kinase. EMBO J. 2000, 19, 2580–2591. [Google Scholar] [CrossRef]
- Mogensen, T.H.; Paludan, S.R. Molecular Pathways in Virus-Induced Cytokine Production. Microbiol. Mol. Biol. Rev. 2001, 65, 131–150. [Google Scholar] [CrossRef]
- Balachandran, S.; Beg, A.A. Defining Emerging Roles for NF-ΚB in Antivirus Responses: Revisiting the Interferon-β Enhanceosome Paradigm. PLoS Pathog. 2011, 7, e1002165. [Google Scholar] [CrossRef]
- Adhikari, A.; Xu, M.; Chen, Z.J. Ubiquitin-Mediated Activation of TAK1 and IKK. Oncogene 2007, 26, 3214–3226. [Google Scholar] [CrossRef]
- Fang, R.; Jiang, Q.; Zhou, X.; Wang, C.; Guan, Y.; Tao, J.; Xi, J.; Feng, J.-M.; Jiang, Z. MAVS Activates TBK1 and IKKε through TRAFs in NEMO Dependent and Independent Manner. PLoS Pathog. 2017, 13, e1006720. [Google Scholar] [CrossRef] [PubMed]
- Andreakos, E.; Salagianni, M.; Galani, I.E.; Koltsida, O. Interferon-Λs: Front-Line Guardians of Immunity and Homeostasis in the Respiratory Tract. Front. Immunol. 2017, 8, 1232. [Google Scholar] [CrossRef]
- Galani, I.E.; Triantafyllia, V.; Eleminiadou, E.-E.; Koltsida, O.; Stavropoulos, A.; Manioudaki, M.; Thanos, D.; Doyle, S.E.; Kotenko, S.V.; Thanopoulou, K.; et al. Interferon-λ Mediates Non-Redundant Front-Line Antiviral Protection against Influenza Virus Infection without Compromising Host Fitness. Immunity 2017, 46, 875–890.e6. [Google Scholar] [CrossRef] [PubMed]
- Levy, D.E.; Marié, I.J.; Durbin, J.E. Induction and Function of Type I and III Interferon in Response to Viral Infection. Curr. Opin. Virol. 2011, 1, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Schindler, C.; Levy, D.E.; Decker, T. JAK-STAT Signaling: From Interferons to Cytokines. J. Biol. Chem. 2007, 282, 20059–20063. [Google Scholar] [CrossRef]
- Hoffmann, H.-H.; Schneider, W.M.; Rice, C.M. Interferons and Viruses: An Evolutionary Arms Race of Molecular Interactions. Trends Immunol. 2015, 36, 124–138. [Google Scholar] [CrossRef]
- Villalón-Letelier, F.; Brooks, A.G.; Saunders, P.M.; Londrigan, S.L.; Reading, P.C. Host Cell Restriction Factors That Limit Influenza A Infection. Viruses 2017, 9, 376. [Google Scholar] [CrossRef]
- Qu, H.; Li, J.; Yang, L.; Sun, L.; Liu, W.; He, H. Influenza A Virus-Induced Expression of ISG20 Inhibits Viral Replication by Interacting with Nucleoprotein. Virus Genes 2016, 52, 759–767. [Google Scholar] [CrossRef]
- Desai, T.M.; Marin, M.; Chin, C.R.; Savidis, G.; Brass, A.L.; Melikyan, G.B. IFITM3 Restricts Influenza A Virus Entry by Blocking the Formation of Fusion Pores Following Virus-Endosome Hemifusion. PLoS Pathog. 2014, 10, e1004048. [Google Scholar] [CrossRef]
- Turan, K.; Mibayashi, M.; Sugiyama, K.; Saito, S.; Numajiri, A.; Nagata, K. Nuclear MxA Proteins Form a Complex with Influenza Virus NP and Inhibit the Transcription of the Engineered Influenza Virus Genome. Nucleic Acids Res. 2004, 32, 643–652. [Google Scholar] [CrossRef]
- Pavlovic, J.; Arzet, H.A.; Hefti, H.P.; Frese, M.; Rost, D.; Ernst, B.; Kolb, E.; Staeheli, P.; Haller, O. Enhanced Virus Resistance of Transgenic Mice Expressing the Human MxA Protein. J. Virol. 1995, 69, 4506–4510. [Google Scholar] [CrossRef]
- Haller, O.; Staeheli, P.; Schwemmle, M.; Kochs, G. Mx GTPases: Dynamin-like Antiviral Machines of Innate Immunity. Trends Microbiol. 2015, 23, 154–163. [Google Scholar] [CrossRef]
- Feeley, E.M.; Sims, J.S.; John, S.P.; Chin, C.R.; Pertel, T.; Chen, L.M.; Gaiha, G.D.; Ryan, B.J.; Donis, R.O.; Elledge, S.J.; et al. IFITM3 Inhibits Influenza a Virus Infection by Preventing Cytosolic Entry. PLoS Pathog. 2011, 7, e1002337. [Google Scholar] [CrossRef]
- Brass, A.L.; Huang, I.-C.; Benita, Y.; John, S.P.; Krishnan, M.N.; Feeley, E.M.; Ryan, B.J.; Weyer, J.L.; van der Weyden, L.; Fikrig, E.; et al. The IFITM Proteins Mediate Cellular Resistance to Influenza A H1N1 Virus, West Nile Virus, and Dengue Virus. Cell 2009, 139, 1243–1254. [Google Scholar] [CrossRef]
- Bailey, C.C.; Zhong, G.; Huang, I.-C.; Farzan, M. IFITM-Family Proteins: The Cell’s First Line of Antiviral Defense. Annu. Rev. Virol. 2014, 1, 261–283. [Google Scholar] [CrossRef]
- Clemens, M.J.; Williams, B.R.G. Inhibition of Cell-Free Protein Synthesis by PppA2′ P5′ A2′ P5′ A: A Novel Oligonucleotide Synthesized by Interferon-Treated L Cell Extracts. Cell 1978, 13, 565–572. [Google Scholar] [CrossRef]
- Silverman, R.H.; Weiss, S.R. Viral Phosphodiesterases That Antagonize Double-Stranded RNA Signaling to RNase L by Degrading 2–5A. J. Interferon Cytokine Res. 2014, 34, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Malathi, K.; Dong, B.; Gale, M.; Silverman, R.H. Small Self-RNA Generated by RNase L Amplifies Antiviral Innate Immunity. Nature 2007, 448, 816–819. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.C.; Sen, G.C. PACT, a Protein Activator of the Interferon-Induced Protein Kinase, PKR. EMBO J. 1998, 17, 4379–4390. [Google Scholar] [CrossRef]
- Kumar, A.; Haque, J.; Lacoste, J.; Hiscott, J.; Williams, B.R. Double-Stranded RNA-Dependent Protein Kinase Activates Transcription Factor NF-Kappa B by Phosphorylating I Kappa, B. Proc. Natl. Acad. Sci. USA 1994, 91, 6288–6292. [Google Scholar] [CrossRef]
- Zhang, P.; Samuel, C.E. Induction of Protein Kinase PKR-Dependent Activation of Interferon Regulatory Factor 3 by Vaccinia Virus Occurs through Adapter IPS-1 Signaling. J. Biol. Chem. 2008, 283, 34580–34587. [Google Scholar] [CrossRef]
- Schulz, O.; Pichlmair, A.; Rehwinkel, J.; Rogers, N.C.; Scheuner, D.; Kato, H.; Takeuchi, O.; Akira, S.; Kaufman, R.J.; Reis e Sousa, C. Protein Kinase R Contributes to Immunity against Specific Viruses by Regulating Interferon MRNA Integrity. Cell Host Microbe 2010, 7, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.-Y.; Ichinohe, T. Response of Host Inflammasomes to Viral Infection. Trends Microbiol. 2015, 23, 55–63. [Google Scholar] [CrossRef]
- Groslambert, M.; Py, B.F. Spotlight on the NLRP3 Inflammasome Pathway. J. Inflamm. Res. 2018, 11, 359–374. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, M.; Chen, I.-Y.; Kawaguchi, A.; Koshiba, T.; Nagata, K.; Takeyama, H.; Hasegawa, H.; Ichinohe, T. The RNA- and TRIM25-Binding Domains of Influenza Virus NS1 Protein Are Essential for Suppression of NLRP3 Inflammasome-Mediated Interleukin-1β Secretion. J. Virol. 2016, 90, 4105–4114. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jiang, W.; Yan, Y.; Gong, T.; Han, J.; Tian, Z.; Zhou, R. RNA Viruses Promote Activation of the NLRP3 Inflammasome through a RIP1-RIP3-DRP1 Signaling Pathway. Nat. Immunol. 2014, 15, 1126–1133. [Google Scholar] [CrossRef]
- Chen, W.; Xu, Y.; Li, H.; Tao, W.; Xiang, Y.; Huang, B.; Niu, J.; Zhong, J.; Meng, G. HCV Genomic RNA Activates the NLRP3 Inflammasome in Human Myeloid Cells. PLoS ONE 2014, 9, e84953. [Google Scholar] [CrossRef]
- Pothlichet, J.; Meunier, I.; Davis, B.K.; Ting, J.P.-Y.; Skamene, E.; von Messling, V.; Vidal, S.M. Type I IFN Triggers RIG-I/TLR3/NLRP3-Dependent Inflammasome Activation in Influenza A Virus Infected Cells. PLoS Pathog. 2013, 9, e1003256. [Google Scholar] [CrossRef]
- Lu, B.; Nakamura, T.; Inouye, K.; Li, J.; Tang, Y.; Lundbäck, P.; Valdes-Ferrer, S.I.; Olofsson, P.S.; Kalb, T.; Roth, J.; et al. Novel Role of PKR in Inflammasome Activation and HMGB1 Release. Nature 2012, 488, 670–674. [Google Scholar] [CrossRef]
- Ichinohe, T.; Pang, I.K.; Iwasaki, A. Influenza Virus Activates Inflammasomes via Its Intracellular M2 Ion Channel. Nat. Immunol. 2010, 11, 404–410. [Google Scholar] [CrossRef]
- McAuley, J.L.; Tate, M.D.; MacKenzie-Kludas, C.J.; Pinar, A.; Zeng, W.; Stutz, A.; Latz, E.; Brown, L.E.; Mansell, A. Activation of the NLRP3 Inflammasome by IAV Virulence Protein PB1-F2 Contributes to Severe Pathophysiology and Disease. PLoS Pathog. 2013, 9, e1003392. [Google Scholar] [CrossRef]
- Schroder, K.; Tschopp, J. The Inflammasomes. Cell 2010, 140, 821–832. [Google Scholar] [CrossRef]
- García-Sastre, A. Induction and Evasion of Type I Interferon Responses by Influenza Viruses. Virus Res. 2011, 162, 12–18. [Google Scholar] [CrossRef]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of Cells and Tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef]
- Meyerson, N.R.; Zhou, L.; Guo, Y.R.; Zhao, C.; Tao, Y.J.; Krug, R.M.; Sawyer, S.L. Nuclear TRIM25 Specifically Targets Influenza Virus Ribonucleoproteins to Block the Onset of RNA Chain Elongation. Cell Host Microbe 2017, 22, 627–638. [Google Scholar] [CrossRef]
- Liu, B.; Li, N.L.; Shen, Y.; Bao, X.; Fabrizio, T.; Elbahesh, H.; Webby, R.J.; Li, K. The C-Terminal Tail of TRIM56 Dictates Antiviral Restriction of Influenza A and B Viruses by Impeding Viral RNA Synthesis. J. Virol. 2016, 90, 4369–4382. [Google Scholar] [CrossRef]
- Ma, J.; Sun, Q.; Mi, R.; Zhang, H. Avian Influenza A Virus H5N1 Causes Autophagy-Mediated Cell Death through Suppression of MTOR Signaling. J. Genet. Genom. 2011, 38, 533–537. [Google Scholar] [CrossRef] [PubMed]
- Beale, R.; Wise, H.; Stuart, A.; Ravenhill, B.J.; Digard, P.; Randow, F. A LC3-Interacting Motif in the Influenza A Virus M2 Protein Is Required to Subvert Autophagy and Maintain Virion Stability. Cell Host Microbe 2014, 15, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Gannagé, M.; Dormann, D.; Albrecht, R.; Dengjel, J.; Torossi, T.; Rämer, P.C.; Lee, M.; Strowig, T.; Arrey, F.; Conenello, G.; et al. Matrix Protein 2 of Influenza A Virus Blocks Autophagosome Fusion with Lysosomes. Cell Host Microbe 2009, 6, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Elmore, S. Apoptosis: A Review of Programmed Cell Death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Galluzzi, L.; Brenner, C.; Morselli, E.; Touat, Z.; Kroemer, G. Viral Control of Mitochondrial Apoptosis. PLoS Pathog. 2008, 4, e1000018. [Google Scholar] [CrossRef] [PubMed]
- Tallóczy, Z.; Jiang, W.; Virgin, H.W.; Leib, D.A.; Scheuner, D.; Kaufman, R.J.; Eskelinen, E.-L.; Levine, B. Regulation of Starvation- and Virus-Induced Autophagy by the EIF2alpha Kinase Signaling Pathway. Proc. Natl. Acad. Sci. USA 2002, 99, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, M.; Cheng, A.; Yang, Q.; Wu, Y.; Jia, R.; Liu, M.; Zhu, D.; Chen, S.; Zhang, S.; et al. The Role of Host EIF2α in Viral Infection. Virol. J. 2020, 17, 112. [Google Scholar] [CrossRef]
- Wu, X.; Wang, J.; Wang, S.; Wu, F.; Chen, Z.; Li, C.; Cheng, G.; Qin, F.X. Inhibition of Influenza A Virus Replication by TRIM14 via Its Multifaceted Protein-Protein Interaction with NP. Front. Microbiol. 2019, 10, 344. [Google Scholar] [CrossRef]
- Patil, G.; Zhao, M.; Song, K.; Hao, W.; Bouchereau, D.; Wang, L.; Li, S. TRIM41- Mediated Ubiquitination of Nucleoprotein Limits Influenza A Virus Infection. J. Virol. 2018, 92, e00905-18. [Google Scholar] [CrossRef]
- Pagani, I.; Di Pietro, A.; Oteiz, A.; Ghitti, M.; Mechti, N.; Naffakh, N.; Vicenzi, E. Mutations Conferring Increased Sensitivity to Tripartite Motif 22 Restriction Accumulated Progressively in the Nucleoprotein of Seasonal Influenza A (H1N1) Viruses between 1918 and 2009. MSphere 2018, 3, e00110-18. [Google Scholar] [CrossRef] [PubMed]
- Di Rienzo, M.; Romagnoli, A.; Antonioli, M.; Piacentini, M.; Fimia, G.M. TRIM Proteins in Autophagy: Selective Sensors in Cell Damage and Innate Immune Responses. Cell Death Differ. 2020, 27, 887–902. [Google Scholar] [CrossRef]
- Yang, B.; Wang, J.; Wang, Y.; Zhou, H.; Wu, X.; Tian, Z.; Sun, B. Novel Function of Trim44 Promotes an Antiviral Response by Stabilizing VISA. J. Immunol. 2013, 190, 3613–3619. [Google Scholar] [CrossRef]
- Krischuns, T.; Günl, F.; Henschel, L.; Binder, M.; Willemsen, J.; Schloer, S.; Rescher, U.; Gerlt, V.; Zimmer, G.; Nordhoff, C.; et al. Phosphorylation of TRIM28 Enhances the Expression of IFN-β and Proinflammatory Cytokines During HPAIV Infection of Human Lung Epithelial Cells. Front. Immunol. 2018, 9, 2229. [Google Scholar] [CrossRef]
- Fan, Y.; Mao, R.; Yu, Y.; Liu, S.; Shi, Z.; Cheng, J.; Zhang, H.; An, L.; Zhao, Y.; Xu, X.; et al. USP21 Negatively Regulates Antiviral Response by Acting as a RIG-I Deubiquitinase. J. Exp. Med. 2014, 211, 313–328. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhao, W.; Zhang, M.; Wang, P.; Zhao, K.; Zhao, X.; Yang, S.; Gao, C. USP4 Positively Regulates RIG-I-Mediated Antiviral Response through Deubiquitination and Stabilization of RIG-I. J. Virol. 2013, 87, 4507–4515. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, J.; Qian, L.; Feng, Q.; Wang, X.; Yuan, Y.; Zuo, Y.; Cheng, Q.; Miao, Y.; Guo, T.; et al. Induction of OTUD1 by RNA Viruses Potently Inhibits Innate Immune Responses by Promoting Degradation of the MAVS/TRAF3/TRAF6 Signalosome. PLoS Pathog. 2018, 14, e1007067. [Google Scholar] [CrossRef]
- Jahan, A.S.; Biquand, E.; Muñoz-Moreno, R.; Le Quang, A.; Mok, C.K.-P.; Wong, H.H.; Teo, Q.W.; Valkenburg, S.A.; Chin, A.W.H.; Man Poon, L.L.; et al. OTUB1 Is a Key Regulator of RIG-I-Dependent Immune Signaling and Is Targeted for Proteasomal Degradation by Influenza A NS1. Cell Rep. 2020, 30, 1570–1584.e6. [Google Scholar] [CrossRef] [PubMed]
- Herhaus, L.; Al-Salihi, M.; Macartney, T.; Weidlich, S.; Sapkota, G.P. OTUB1 Enhances TGFβ Signalling by Inhibiting the Ubiquitylation and Degradation of Active SMAD2/3. Nat. Commun. 2013, 4, 2519. [Google Scholar] [CrossRef]
- Sun, X.-X.; Challagundla, K.B.; Dai, M.-S. Positive Regulation of P53 Stability and Activity by the Deubiquitinating Enzyme Otubain 1. EMBO J. 2012, 31, 576–592. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, M.A.; Bowman, J.W.; Fujita, H.; Orazio, N.; Shi, M.; Liang, Q.; Amatya, R.; Kelly, T.J.; Iwai, K.; Ting, J.; et al. The Linear Ubiquitin Assembly Complex (LUBAC) Is Essential for NLRP3 Inflammasome Activation. J. Exp. Med. 2014, 211, 1333–1347. [Google Scholar] [CrossRef]
- Weng, L.; Mitoma, H.; Trichot, C.; Tricot, C.; Bao, M.; Liu, Y.; Zhang, Z.; Liu, Y.-J. The E3 Ubiquitin Ligase Tripartite Motif 33 Is Essential for Cytosolic RNA-Induced NLRP3 Inflammasome Activation. J. Immunol. 2014, 193, 3676–3682. [Google Scholar] [CrossRef]
- Py, B.F.; Kim, M.-S.; Vakifahmetoglu-Norberg, H.; Yuan, J. Deubiquitination of NLRP3 by BRCC3 Critically Regulates Inflammasome Activity. Mol. Cell 2013, 49, 331–338. [Google Scholar] [CrossRef]
- Schmidt, N.; Domingues, P.; Golebiowski, F.; Patzina, C.; Tatham, M.H.; Hay, R.T.; Hale, B.G. An Influenza Virus-Triggered SUMO Switch Orchestrates Co-Opted Endogenous Retroviruses to Stimulate Host Antiviral Immunity. Proc. Natl. Acad. Sci. USA 2019, 116, 17399–17408. [Google Scholar] [CrossRef]
- Morales, D.J.; Lenschow, D.J. The Antiviral Activities of ISG15. J. Mol. Biol. 2013, 425, 4995–5008. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chai, W.; Min, J.; Ye, Z.; Tong, X.; Qi, D.; Liu, W.; Luo, E.; Li, J.; Ye, X. Neddylation of M1 Negatively Regulates the Replication of Influenza A Virus. J. Gen. Virol. 2020, 101, 1242–1250. [Google Scholar] [CrossRef]
- Rowe, H.M.; Jakobsson, J.; Mesnard, D.; Rougemont, J.; Reynard, S.; Aktas, T.; Maillard, P.V.; Layard-Liesching, H.; Verp, S.; Marquis, J.; et al. KAP1 Controls Endogenous Retroviruses in Embryonic Stem Cells. Nature 2010, 463, 237–240. [Google Scholar] [CrossRef]
- Turelli, P.; Castro-Diaz, N.; Marzetta, F.; Kapopoulou, A.; Raclot, C.; Duc, J.; Tieng, V.; Quenneville, S.; Trono, D. Interplay of TRIM28 and DNA Methylation in Controlling Human Endogenous Retroelements. Genome Res. 2014, 24, 1260–1270. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Leung, D.; Miyashita, H.; Maksakova, I.A.; Miyachi, H.; Kimura, H.; Tachibana, M.; Lorincz, M.C.; Shinkai, Y. Proviral Silencing in Embryonic Stem Cells Requires the Histone Methyltransferase ESET. Nature 2010, 464, 927–931. [Google Scholar] [CrossRef] [PubMed]
- Schultz, D.C.; Friedman, J.R.; Rauscher, F.J. Targeting Histone Deacetylase Complexes via KRAB-Zinc Finger Proteins: The PHD and Bromodomains of KAP-1 Form a Cooperative Unit That Recruits a Novel Isoform of the Mi-2α Subunit of NuRD. Genes Dev. 2001, 15, 428–443. [Google Scholar] [CrossRef]
- Ivanov, A.V.; Peng, H.; Yurchenko, V.; Yap, K.L.; Negorev, D.G.; Schultz, D.C.; Psulkowski, E.; Fredericks, W.J.; White, D.E.; Maul, G.G.; et al. PHD Domain-Mediated E3 Ligase Activity Directs Intramolecular Sumoylation of an Adjacent Bromodomain Which Is Required for Gene Silencing. Mol. Cell 2007, 28, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Mascle, X.H.; Germain-Desprez, D.; Huynh, P.; Estephan, P.; Aubry, M. Sumoylation of the Transcriptional Intermediary Factor 1beta (TIF1beta), the Co-Repressor of the KRAB Multifinger Proteins, Is Required for Its Transcriptional Activity and Is Modulated by the KRAB Domain. J. Biol. Chem. 2007, 282, 10190–10202. [Google Scholar] [CrossRef]
- Roulois, D.; Yau, H.L.; Singhania, R.; Wang, Y.; Danesh, A.; Shen, S.Y.; Han, H.; Liang, G.; Pugh, T.J.; Jones, P.A.; et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell 2015, 162, 961–973. [Google Scholar] [CrossRef]
- Cuellar, T.L.; Herzner, A.-M.; Zhang, X.; Goyal, Y.; Watanabe, C.; Friedman, B.A.; Janakiraman, V.; Durinck, S.; Stinson, J.; Arnott, D.; et al. Silencing of Retrotransposons by SETDB1 Inhibits the Interferon Response in Acute Myeloid Leukemia. J. Cell Biol. 2017, 216, 3535–3549. [Google Scholar] [CrossRef]
- Chiappinelli, K.B.; Strissel, P.L.; Desrichard, A.; Li, H.; Henke, C.; Akman, B.; Hein, A.; Rote, N.S.; Cope, L.M.; Snyder, A.; et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via DsRNA Including Endogenous Retroviruses. Cell 2015, 162, 974–986. [Google Scholar] [CrossRef]
- Rajagopalan, D.; Tirado-Magallanes, R.; Bhatia, S.S.; Teo, W.S.; Sian, S.; Hora, S.; Lee, K.K.; Zhang, Y.; Jadhav, S.P.; Wu, Y.; et al. TIP60 Represses Activation of Endogenous Retroviral Elements. Nucleic. Acids. Res. 2018, 46, 9456–9470. [Google Scholar] [CrossRef] [PubMed]
- Tie, C.H.; Fernandes, L.; Conde, L.; Robbez-Masson, L.; Sumner, R.P.; Peacock, T.; Rodriguez-Plata, M.T.; Mickute, G.; Gifford, R.; Towers, G.J.; et al. KAP1 Regulates Endogenous Retroviruses in Adult Human Cells and Contributes to Innate Immune Control. EMBO Rep. 2018, 19, e45000. [Google Scholar] [CrossRef]
- Liu, M.; Ohtani, H.; Zhou, W.; Ørskov, A.D.; Charlet, J.; Zhang, Y.W.; Shen, H.; Baylin, S.B.; Liang, G.; Grønbæk, K.; et al. Vitamin C Increases Viral Mimicry Induced by 5-Aza-2′-Deoxycytidine. Proc. Natl. Acad. Sci. USA 2016, 113, 10238–10244. [Google Scholar] [CrossRef]
- Zhao, C.; Sridharan, H.; Chen, R.; Baker, D.P.; Wang, S.; Krug, R.M. Influenza B Virus Non-Structural Protein 1 Counteracts ISG15 Antiviral Activity by Sequestering ISGylated Viral Proteins. Nat. Commun. 2016, 7, 12754. [Google Scholar] [CrossRef]
- Hsiang, T.-Y.; Zhao, C.; Krug, R.M. Interferon-Induced ISG15 Conjugation Inhibits Influenza A Virus Gene Expression and Replication in Human Cells. J. Virol. 2009, 83, 5971–5977. [Google Scholar] [CrossRef]
- Zhang, T.; Ye, Z.; Yang, X.; Qin, Y.; Hu, Y.; Tong, X.; Lai, W.; Ye, X. NEDDylation of PB2 Reduces Its Stability and Blocks the Replication of Influenza A Virus. Sci. Rep. 2017, 7, 43691. [Google Scholar] [CrossRef]
- Sun, H.; Yao, W.; Wang, K.; Qian, Y.; Chen, H.; Jung, Y.-S. Inhibition of Neddylation Pathway Represses Influenza Virus Replication and Pro-Inflammatory Responses. Virology 2018, 514, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhao, Z.; Xu, C.; Sun, L.; Chen, J.; Zhang, L.; Liu, W. Cyclophilin a Restricts Influenza a Virus Replication through Degradation of the M1 Protein. PLoS ONE 2012, 7, e31063. [Google Scholar] [CrossRef] [PubMed]
- Zinzula, L.; Tramontano, E. Strategies of Highly Pathogenic RNA Viruses to Block DsRNA Detection by RIG-I-like Receptors: Hide, Mask, Hit. Antiviral Res. 2013, 100, 615–635. [Google Scholar] [CrossRef]
- Liu, Y.; Olagnier, D.; Lin, R. Host and Viral Modulation of RIG-I-Mediated Antiviral Immunity. Front. Immunol. 2017, 7, 662. [Google Scholar] [CrossRef]
- Varga, Z.T.; Grant, A.; Manicassamy, B.; Palese, P. Influenza Virus Protein PB1-F2 Inhibits the Induction of Type I Interferon by Binding to MAVS and Decreasing Mitochondrial Membrane Potential. J. Virol. 2012, 86, 8359–8366. [Google Scholar] [CrossRef] [PubMed]
- Graef, K.M.; Vreede, F.T.; Lau, Y.-F.; McCall, A.W.; Carr, S.M.; Subbarao, K.; Fodor, E. The PB2 Subunit of the Influenza Virus RNA Polymerase Affects Virulence by Interacting with the Mitochondrial Antiviral Signaling Protein and Inhibiting Expression of Beta Interferon. J. Virol. 2010, 84, 8433–8445. [Google Scholar] [CrossRef]
- Sharma, K.; Tripathi, S.; Ranjan, P.; Kumar, P.; Garten, R.; Deyde, V.; Katz, J.M.; Cox, N.J.; Lal, R.B.; Sambhara, S.; et al. Influenza A Virus Nucleoprotein Exploits Hsp40 to Inhibit PKR Activation. PLoS ONE 2011, 6, e20215. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Vijayan, M.; Pritzl, C.J.; Fuchs, S.Y.; McDermott, A.B.; Hahm, B. Hemagglutinin of influenza A virus antagonizes type I interferon (IFN) responses by inducing degradation of type I IFN receptor 1. J. Virol. 2016, 90, 2403–2417. [Google Scholar] [CrossRef] [PubMed]
- Su, W.-C.; Yu, W.-Y.; Huang, S.-H.; Lai, M.M.C. Ubiquitination of the Cytoplasmic Domain of Influenza A Virus M2 Protein Is Crucial for Production of Infectious Virus Particles. J. Virol. 2018, 92, e01972-17. [Google Scholar] [CrossRef]
- Talon, J.; Horvath, C.M.; Polley, R.; Basler, C.F.; Muster, T.; Palese, P.; Garcia-Sastre, A. Activation of Interferon Regulatory Factor 3 Is Inhibited by the Influenza a Virus NS1 Protein. J. Virol. 2000, 74, 7989–7996. [Google Scholar] [CrossRef]
- Rückle, A.; Haasbach, E.; Julkunen, I.; Planz, O.; Ehrhardt, C.; Ludwig, S. The NS1 Protein of Influenza A Virus Blocks RIG-I-Mediated Activation of the Noncanonical NF-ΚB Pathway and P52/RelB-Dependent Gene Expression in Lung Epithelial Cells. J. Virol. 2012, 86, 10211–10217. [Google Scholar] [CrossRef]
- Wang, X.; Li, M.; Zheng, H.; Muster, T.; Palese, P.; Beg, A.A.; García-Sastre, A. Influenza A Virus NS1 Protein Prevents Activation of NF-ΚB and Induction of Alpha/Beta Interferon. J. Virol. 2000, 74, 11566. [Google Scholar] [CrossRef]
- Min, J.-Y.; Li, S.; Sen, G.C.; Krug, R.M. A Site on the Influenza A Virus NS1 Protein Mediates Both Inhibition of PKR Activation and Temporal Regulation of Viral RNA Synthesis. Virology 2007, 363, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Min, J.-Y.; Krug, R.M. The Primary Function of RNA Binding by the Influenza A Virus NS1 Protein in Infected Cells: Inhibiting the 2’-5’ Oligo (A) Synthetase/RNase L Pathway. Proc. Natl. Acad. Sci. USA 2006, 103, 7100–7105. [Google Scholar] [CrossRef] [PubMed]
- Marc, D. Influenza virus non-structural protein NS1: Interferon antagonism and beyond. J. Gen. Virol. 2014, 95, 2594–2611. [Google Scholar] [CrossRef]
- Carrillo, B.; Choi, J.-; Bornholdt, Z.A.; Sankaran, B.; Rice, A.P.; Venkataram Prasad, B.V. The influenza A virus protein NS1 displays structural polymorphism. J. Virol. 2014, 88, 4113–4122. [Google Scholar] [CrossRef]
- Ludwig, S.; Wang, X.; Ehrhardt, C.; Zheng, H.; Donelan, N.; Planz, O.; Pleschka, S.; García-Sastre, A.; Heins, G.; Wolff, T. The Influenza A Virus NS1 Protein Inhibits Activation of Jun N-Terminal Kinase and AP-1 Transcription Factors. J. Virol. 2002, 76, 11166–11171. [Google Scholar] [CrossRef] [PubMed]
- Mibayashi, M.; Martínez-Sobrido, L.; Loo, Y.-M.; Cárdenas, W.B.; Gale, M.; García-Sastre, A. Inhibition of Retinoic Acid-Inducible Gene I-Mediated Induction of Beta Interferon by the NS1 Protein of Influenza A Virus. J. Virol. 2007, 81, 514–524. [Google Scholar] [CrossRef] [PubMed]
- Lamb, R.A.; Lai, C.-J. Sequence of Interrupted and Uninterrupted MRNAs and Cloned DNA Coding for the Two Overlapping Nonstructural Proteins of Influenza Virus. Cell 1980, 21, 475–485. [Google Scholar] [CrossRef]
- Robb, N.C.; Jackson, D.; Vreede, F.T.; Fodor, E. Splicing of Influenza A Virus NS1 MRNA Is Independent of the Viral NS1 Protein. J. Gen. Virol. 2010, 91, 2331–2340. [Google Scholar] [CrossRef] [PubMed]
- Dubois, J.; Terrier, O.; Rosa-Calatrava, M. Influenza Viruses and MRNA Splicing: Doing More with Less. MBio 2014, 5, e00070-14. [Google Scholar] [CrossRef] [PubMed]
- Lohrmann, F.; Dijkman, R.; Stertz, S.; Thiel, V.; Haller, O.; Staeheli, P.; Kochs, G. Emergence of a C-Terminal Seven-Amino-Acid Elongation of NS1 in Around 1950 Conferred a Minor Growth Advantage to Former Seasonal Influenza A Viruses. J. Virol. 2013, 87, 11300–11303. [Google Scholar] [CrossRef]
- Chauché, C.; Nogales, A.; Zhu, H.; Goldfarb, D.; Ahmad Shanizza, A.I.; Gu, Q.; Parrish, C.R.; Martínez-Sobrido, L.; Marshall, J.F.; Murcia, P.R. Mammalian Adaptation of an Avian Influenza A Virus Involves Stepwise Changes in NS1. J. Virol. 2018, 92, e01875-17. [Google Scholar] [CrossRef]
- Chien, C.Y.; Tejero, R.; Huang, Y.; Zimmerman, D.E.; Ríos, C.B.; Krug, R.M.; Montelione, G.T. A Novel RNA-Binding Motif in Influenza A Virus Non-Structural Protein 1. Nat. Struct. Biol. 1997, 4, 891–895. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lynch, P.A.; Chien, C.-; Montelione, G.T.; Krug, R.M.; Berman, H.M. Crystal Structure of the Unique RNA-Binding Domain of the Influenza Virus NS1 Protein. Nat. Struct. Biol. 1997, 4, 896–899. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Riedel, K.; Lynch, P.; Chien, C.; Montelione, G.T.; Krug, R.M. RNA Binding by the Novel Helical Domain of the Influenza Virus NS1 Protein Requires Its Dimer Structure and a Small Number of Specific Basic Amino Acids. RNA 1999, 5, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Donelan, N.R.; Basler, C.F.; García-Sastre, A. A Recombinant Influenza A Virus Expressing an RNA-Binding-Defective NS1 Protein Induces High Levels of Beta Interferon and Is Attenuated in Mice. J. Virol. 2003, 77, 13257–13266. [Google Scholar] [CrossRef]
- Kerry, P.S.; Ayllon, J.; Taylor, M.A.; Hass, C.; Lewis, A.; García-Sastre, A.; Randall, R.E.; Hale, B.G.; Russell, R.J. A Transient Homotypic Interaction Model for the Influenza A Virus NS1 Protein Effector Domain. PLoS ONE 2011, 6, e17946. [Google Scholar] [CrossRef]
- Aramini, J.M.; Hamilton, K.; Ma, L.-; Swapna, G.V.T.; Leonard, P.G.; Ladbury, J.E.; Krug, R.M.; Montelione, G.T. F NMR Reveals Multiple Conformations at the Dimer Interface of the Nonstructural Protein 1 Effector Domain from Influenza A Virus. Structure 2014, 22, 515–525. [Google Scholar] [CrossRef]
- Aramini, J.M.; Ma, L.C.; Zhou, L.; Schauder, C.M.; Hamilton, K.; Amer, B.R.; Mack, T.R.; Lee, H.W.; Ciccosanti, C.T.; Zhao, L.; et al. Dimer Interface of the Effector Domain of Non-Structural Protein 1 from Influenza A Virus: An Interface with Multiple Functions. J. Biol. Chem. 2011, 286, 26050–26060. [Google Scholar] [CrossRef]
- Cheng, A.; Wong, S.M.; Yuan, Y.A. Structural basis for dsRNA recognition by NS1 protein of influenza A virus. Cell Res. 2009, 19, 187–195. [Google Scholar] [CrossRef]
- Greenspan, D.; Palese, P.; Krystal, M. Two Nuclear Location Signals in the Influenza Virus NS1 Nonstructural Protein. J. Virol. 1988, 62, 3020–3026. [Google Scholar] [CrossRef]
- Melén, K.; Kinnunen, L.; Fagerlund, R.; Ikonen, N.; Twu, K.Y.; Krug, R.M.; Julkunen, I. Nuclear and Nucleolar Targeting of Influenza A Virus NS1 Protein: Striking Differences between Different Virus Subtypes. J. Virol. 2007, 81, 5995–6006. [Google Scholar] [CrossRef]
- Li, Y.; Yamakita, Y.; Krug, R.M. Regulation of a Nuclear Export Signal by an Adjacent Inhibitory Sequence: The Effector Domain of the Influenza Virus NS1 Protein. Proc. Natl. Acad. Sci. USA 1998, 95, 4864–4869. [Google Scholar] [CrossRef]
- Hale, B.G.; Randall, R.E.; Ortín, J.; Jackson, D. The Multifunctional NS1 Protein of Influenza A Viruses. J. Gen. Virol. 2008, 89, 2359–2376. [Google Scholar] [CrossRef]
- Hale, B.G. Conformational Plasticity of the Influenza A Virus NS1 Protein. J. Gen. Virol. 2014, 95, 2099–2105. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.; Hossain, M.J.; Hickman, D.; Perez, D.R.; Lamb, R.A. A New Influenza Virus Virulence Determinant: The NS1 Protein Four C-Terminal Residues Modulate Pathogenicity. Proc. Natl. Acad. Sci. USA 2008, 105, 4381–4386. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Sebastiano, V.; Moshkina, N.; Mena, N.; Hultquist, J.; Jimenez-Morales, D.; Ma, Y.; Rialdi, A.; Albrecht, R.; Fenouil, R.; et al. Influenza Virus Infection Causes Global RNAPII Termination Defects. Nat. Struct. Mol. Biol. 2018, 25, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Klemm, C.; Boergeling, Y.; Ludwig, S.; Ehrhardt, C. Immunomodulatory Nonstructural Proteins of Influenza A Viruses. Trends Microbiol. 2018, 26, 624–636. [Google Scholar] [CrossRef] [PubMed]
- García-Sastre, A.; Egorov, A.; Matassov, D.; Brandt, S.; Levy, D.E.; Durbin, J.E.; Palese, P.; Muster, T. Influenza A Virus Lacking the NS1 Gene Replicates in Interferon-Deficient Systems. Virology 1998, 252, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Kochs, G.; Koerner, I.; Thiel, L.; Kothlow, S.; Kaspers, B.; Ruggli, N.; Summerfield, A.; Pavlovic, J.; Stech, J.; Staeheli, P. Properties of H7N7 Influenza A Virus Strain SC35M Lacking Interferon Antagonist NS1 in Mice and Chickens. J. Gen. Virol. 2007, 88, 1403–1409. [Google Scholar] [CrossRef]
- Nogales, A.; Chauché, C.; DeDiego, M.L.; Topham, D.J.; Parrish, C.R.; Murcia, P.R.; Martínez-Sobrido, L. The K186E Amino Acid Substitution in the Canine Influenza Virus H3N8 NS1 Protein Restores Its Ability To Inhibit Host Gene Expression. J. Virol. 2017, 91, e00877-17. [Google Scholar] [CrossRef]
- Nogales, A.; Baker, S.F.; Ortiz-Riaño, E.; Dewhurst, S.; Topham, D.J.; Martínez-Sobrido, L. Influenza A Virus Attenuation by Codon Deoptimization of the NS Gene for Vaccine Development. J. Virol. 2014, 88, 10525–10540. [Google Scholar] [CrossRef]
- DeDiego, M.L.; Nogales, A.; Lambert-Emo, K.; Martinez-Sobrido, L.; Topham, D.J. NS1 Protein Mutation I64T Affects Interferon Responses and Virulence of Circulating H3N2 Human Influenza A Viruses. J. Virol. 2016, 90, 9693–9711. [Google Scholar] [CrossRef]
- Nogales, A.; Martinez-Sobrido, L.; Topham, D.J.; DeDiego, M.L. NS1 Protein Amino Acid Changes D189N and V194I Affect Interferon Responses, Thermosensitivity, and Virulence of Circulating H3N2 Human Influenza A Viruses. J. Virol. 2017, 91, e01930-16. [Google Scholar] [CrossRef]
- Rajsbaum, R.; Albrecht, R.A.; Wang, M.K.; Maharaj, N.P.; Versteeg, G.A.; Nistal-Villán, E.; García-Sastre, A.; Gack, M.U. Species-Specific Inhibition of RIG-I Ubiquitination and IFN Induction by the Influenza A Virus NS1 Protein. PLoS Pathog. 2012, 8, e1003059. [Google Scholar] [CrossRef]
- Qian, W.; Wei, X.; Guo, K.; Li, Y.; Lin, X.; Zou, Z.; Zhou, H.; Jin, M. The C-Terminal Effector Domain of Non-Structural Protein 1 of Influenza A Virus Blocks IFN-β Production by Targeting TNF Receptor-Associated Factor 3. Front. Immunol. 2017, 8, 779. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Song, L.; Li, J.; Zhang, Z.; Peng, H.; Jiang, W.; Wang, Q.; Kang, T.; Chen, S.; Huang, W. Influenza A Virus-Encoded NS1 Virulence Factor Protein Inhibits Innate Immune Response by Targeting IKK. Cell. Microbiol. 2012, 14, 1849–1866. [Google Scholar] [CrossRef]
- Newby, C.M.; Sabin, L.; Pekosz, A. The RNA Binding Domain of Influenza A Virus NS1 Protein Affects Secretion of Tumor Necrosis Factor Alpha, Interleukin-6, and Interferon in Primary Murine Tracheal Epithelial Cells. J. Virol. 2007, 81, 9469–9480. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, L.; Xu, G.; Cao, Z.; Wang, Q.; Li, S.; Peng, N.; Yin, J.; Yu, H.; Li, M.; et al. Epigenetic Modification Is Regulated by the Interaction of Influenza A Virus Nonstructural Protein 1 with the De Novo DNA Methyltransferase DNMT3B and Subsequent Transport to the Cytoplasm for K48-Linked Polyubiquitination. J. Virol. 2019, 93, e01587-18. [Google Scholar] [CrossRef]
- Nogales, A.; Martinez-Sobrido, L.; Topham, D.; DeDiego, M. Modulation of Innate Immune Responses by the Influenza A NS1 and PA-X Proteins. Viruses 2018, 10, 708. [Google Scholar] [CrossRef]
- Wang, B.X.; Fish, E.N. Interactions Between NS1 of Influenza A Viruses and Interferon-α/β: Determinants for Vaccine Development. J. Interferon Cytokine Res. 2017, 37, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Krug, R.M. Functions of the Influenza A Virus NS1 Protein In Antiviral Defense. Curr. Opin. Virol. 2015, 12, 1–6. [Google Scholar] [CrossRef]
- Cheong, W.-C.; Kang, H.-R.; Yoon, H.; Kang, S.-J.; Ting, J.P.-Y.; Song, M.J. Influenza A Virus NS1 Protein Inhibits the NLRP3 Inflammasome. PLoS ONE 2015, 10, e0126456. [Google Scholar] [CrossRef]
- Park, H.-S.; Liu, G.; Thulasi Raman, S.N.; Landreth, S.L.; Liu, Q.; Zhou, Y. NS1 Protein of 2009 Pandemic Influenza A Virus Inhibits Porcine NLRP3 Inflammasome-Mediated Interleukin-1 Beta Production by Suppressing ASC Ubiquitination. J. Virol. 2018, 92, e00022-18. [Google Scholar] [CrossRef] [PubMed]
- Ehrhardt, C.; Ludwig, S. A New Player in a Deadly Game: Influenza Viruses and the PI3K/Akt Signalling Pathway. Cell Microbiol. 2009, 11, 863–871. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.-K.; Liu, Q.; Tikoo, S.K.; Babiuk, L.A.; Zhou, Y. Influenza A Virus NS1 Protein Activates the Phosphatidylinositol 3-Kinase (PI3K)/Akt Pathway by Direct Interaction with the P85 Subunit of PI3K. J. Gen. Virol. 2007, 88, 13–18. [Google Scholar] [CrossRef]
- Ponnuswamy, A.; Hupp, T.; Fåhraeus, R. Concepts in MDM2 Signaling. Genes Cancer 2012, 3, 291–297. [Google Scholar] [CrossRef][Green Version]
- Hale, B.G.; Jackson, D.; Chen, Y.-H.; Lamb, R.A.; Randall, R.E. Influenza A Virus NS1 Protein Binds P85β and Activates Phosphatidylinositol-3-Kinase Signaling. Proc. Natl. Acad. Sci. USA 2006, 103, 14194–14199. [Google Scholar] [CrossRef]
- Hale, B.G.; Kerry, P.S.; Jackson, D.; Precious, B.L.; Gray, A.; Killip, M.J.; Randall, R.E.; Russell, R.J. Structural Insights into Phosphoinositide 3-Kinase Activation by the Influenza A Virus NS1 Protein. Proc. Natl. Acad. Sci. USA 2010, 107, 1954–1959. [Google Scholar] [CrossRef] [PubMed]
- Lopes, A.M.; Domingues, P.; Zell, R.; Hale, B.G. Structure-Guided Functional Annotation of the Influenza A Virus NS1 Protein Reveals Dynamic Evolution of the P85β-Binding Site during Circulation in Humans. J. Virol. 2017, 91, e01081-17. [Google Scholar] [CrossRef]
- Ehrhardt, C.; Wolff, T.; Pleschka, S.; Planz, O.; Beermann, W.; Bode, J.G.; Schmolke, M.; Ludwig, S. Influenza A Virus NS1 Protein Activates the PI3K/Akt Pathway To Mediate Antiapoptotic Signaling Responses. J. Virol. 2007, 81, 3058–3067. [Google Scholar] [CrossRef] [PubMed]
- Hrincius, E.R.; Dierkes, R.; Anhlan, D.; Wixler, V.; Ludwig, S.; Ehrhardt, C. Phosphatidylinositol-3-Kinase (PI3K) Is Activated by Influenza Virus VRNA via the Pathogen Pattern Receptor Rig-I to Promote Efficient Type I Interferon Production. Cell. Microbiol. 2011, 13, 1907–1919. [Google Scholar] [CrossRef]
- Lu, X.; Masic, A.; Liu, Q.; Zhou, Y. Regulation of Influenza A Virus Induced CXCL-10 Gene Expression Requires PI3K/Akt Pathway and IRF3 Transcription Factor. Mol. Immunol. 2011, 48, 1417–1423. [Google Scholar] [CrossRef]
- Ampomah, P.B.; Lim, L.H.K. Influenza A Virus-Induced Apoptosis and Virus Propagation. Apoptosis 2020, 25, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wurzer, W.J.; Planz, O.; Ehrhardt, C.; Giner, M.; Silberzahn, T.; Pleschka, S.; Ludwig, S. Caspase 3 Activation Is Essential for Efficient Influenza Virus Propagation. EMBO J. 2003, 22, 2717–2728. [Google Scholar] [CrossRef]
- Das, K.; Ma, L.-C.; Xiao, R.; Radvansky, B.; Aramini, J.; Zhao, L.; Marklund, J.; Kuo, R.-L.; Twu, K.Y.; Arnold, E.; et al. Structural Basis for Suppression of a Host Antiviral Response by Influenza A Virus. Proc. Natl. Acad. Sci. USA 2008, 105, 13093–13098. [Google Scholar] [CrossRef]
- Noah, D.L.; Twu, K.Y.; Krug, R.M. Cellular Antiviral Responses against Influenza A Virus Are Countered at the Posttranscriptional Level by the Viral NS1A Protein via Its Binding to a Cellular Protein Required for the 3′ End Processing of Cellular Pre-MRNAS. Virology 2003, 307, 386–395. [Google Scholar] [CrossRef]
- Nemeroff, M.E.; Barabino, S.M.L.; Li, Y.; Keller, W.; Krug, R.M. Influenza Virus NS1 Protein Interacts with the Cellular 30 KDa Subunit of CPSF and Inhibits 3′ End Formation of Cellular Pre-MRNAs. Mol. Cell 1998, 1, 991–1000. [Google Scholar] [CrossRef]
- Levene, R.E.; Gaglia, M.M. Host Shutoff in Influenza A Virus: Many Means to an End. Viruses 2018, 10, 475. [Google Scholar] [CrossRef]
- Chan, S.; Choi, E.-A.; Shi, Y. Pre-MRNA 3′-End Processing Complex Assembly and Function. WIREs RNA 2011, 2, 321–335. [Google Scholar] [CrossRef]
- Chen, Z. Influenza A Virus NS1 Protein Targets Poly(A)-Binding Protein II of the Cellular 3′-End Processing Machinery. EMBO J. 1999, 18, 2273–2283. [Google Scholar] [CrossRef] [PubMed]
- Anastasina, M.; Le May, N.; Bugai, A.; Fu, Y.; Söderholm, S.; Gaelings, L.; Ohman, T.; Tynell, J.; Kyttänen, S.; Barboric, M.; et al. Influenza Virus NS1 Protein Binds Cellular DNA to Block Transcription of Antiviral Genes. Biochim. Biophys. Acta Gene Regul. Mech. 2016, 1859, 1440–1448. [Google Scholar] [CrossRef] [PubMed]
- Satterly, N.; Tsai, P.-L.; van Deursen, J.; Nussenzveig, D.R.; Wang, Y.; Faria, P.A.; Levay, A.; Levy, D.E.; Fontoura, B.M.A. Influenza Virus Targets the MRNA Export Machinery and the Nuclear Pore Complex. Proc. Natl. Acad. Sci. USA 2007, 104, 1853–1858. [Google Scholar] [CrossRef] [PubMed]
- Aragón, T. Eukaryotic Translation Initiation Factor 4GI Is a Cellular Target for NS1 Protein, a Translational Activator of Influenza Virus. Mol. Cell. Biol. 2000, 20, 10. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Chen, L.; Zeng, H.; Gomez, J.A.; Plowden, J.; Fujita, T.; Katz, J.M.; Donis, R.O.; Sambhara, S. NS1 Protein of Influenza A Virus Inhibits the Function of Intracytoplasmic Pathogen Sensor, RIG-I. Am. J. Respir Cell Mol. Biol. 2007, 36, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Opitz, B.; Rejaibi, A.; Dauber, B.; Eckhard, J.; Vinzing, M.; Schmeck, B.; Hippenstiel, S.; Suttorp, N.; Wolff, T. IFNβ Induction by Influenza A Virus Is Mediated by RIG-I Which Is Regulated by the Viral NS1 Protein. Cell. Microbiol. 2007, 9, 930–938. [Google Scholar] [CrossRef]
- Jureka, A.S.; Kleinpeter, A.B.; Tipper, J.L.; Harrod, K.S.; Petit, C.M. The Influenza NS1 Protein Modulates RIG-I Activation via a Strain-Specific Direct Interaction with the Second CARD of RIG-I. J. Biol. Chem. 2020, 295, 1153–1164. [Google Scholar] [CrossRef]
- Jureka, A.S.; Kleinpeter, A.B.; Cornilescu, G.; Cornilescu, C.C.; Petit, C.M. Structural Basis for a Novel Interaction between the NS1 Protein Derived from the 1918 Influenza Virus and RIG-I. Structure 2015, 23, 2001–2010. [Google Scholar] [CrossRef]
- Kuo, R.-L.; Li, L.-H.; Lin, S.-J.; Li, Z.-H.; Chen, G.-W.; Chang, C.-K.; Wang, Y.-R.; Tam, E.-H.; Gong, Y.-N.; Krug, R.M.; et al. Role of N Terminus-Truncated NS1 Proteins of Influenza A Virus in Inhibiting IRF3 Activation. J. Virol. 2016, 90, 4696–4705. [Google Scholar] [CrossRef]
- Kuo, R.-L.; Zhao, C.; Malur, M.; Krug, R.M. Influenza A Virus Strains That Circulate in Humans Differ in the Ability of Their NS1 Proteins to Block the Activation of IRF3 and Interferon-β Transcription. Virology 2010, 408, 146–158. [Google Scholar] [CrossRef]
- Marcos-Villar, L.; Nistal-Villan, E.; Zamarreño, N.; Garaigorta, U.; Gastaminza, P.; Nieto, A. Interferon-β Stimulation Elicited by the Influenza Virus Is Regulated by the Histone Methylase Dot1L through the RIG-I-TRIM25 Signaling Axis. Cells 2020, 9, 732. [Google Scholar] [CrossRef]
- Catrysse, L.; Vereecke, L.; Beyaert, R.; van Loo, G. A20 in Inflammation and Autoimmunity. Trends Immunol. 2014, 35, 22–31. [Google Scholar] [CrossRef]
- Maelfait, J.; Roose, K.; Vereecke, L.; Guire, C.M.; Sze, M.; Schuijs, M.J.; Willart, M.; Ibañez, L.I.; Hammad, H.; Lambrecht, B.N.; et al. A20 Deficiency in Lung Epithelial Cells Protects against Influenza A Virus Infection. PLoS Pathog. 2016, 12, e1005410. [Google Scholar] [CrossRef]
- Arguello, M.; Paz, S.; Ferran, C.; Moll, H.P.; Hiscott, J. Anti-Viral Tetris: Modulation of the Innate Anti-Viral Immune Response by A20. Adv. Exp. Med. Biol. 2014, 809, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Wertz, I.E.; O’Rourke, K.M.; Zhou, H.; Eby, M.; Aravind, L.; Seshagiri, S.; Wu, P.; Wiesmann, C.; Baker, R.; Boone, D.L.; et al. De-Ubiquitination and Ubiquitin Ligase Domains of A20 Downregulate NF-KappaB Signalling. Nature 2004, 430, 694–699. [Google Scholar] [CrossRef]
- Wang, Y.-Y.; Li, L.; Han, K.-J.; Zhai, Z.; Shu, H.-B. A20 Is a Potent Inhibitor of TLR3- and Sendai Virus-Induced Activation of NF-ΚB and ISRE and IFN-β Promoter. FEBS Lett. 2004, 576, 86–90. [Google Scholar] [CrossRef]
- Lane, D.; Levine, A. P53 Research: The Past Thirty Years and the Next Thirty Years. Cold Spring Harb. Perspect. Biol. 2010, 2, a000893. [Google Scholar] [CrossRef]
- Bieging, K.T.; Mello, S.S.; Attardi, L.D. Unravelling Mechanisms of P53-Mediated Tumour Suppression. Nat. Rev. Cancer 2014, 14, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Turpin, E.; Luke, K.; Jones, J.; Tumpey, T.; Konan, K.; Schultz-Cherry, S. Influenza Virus Infection Increases P53 Activity: Role of P53 in Cell Death and Viral Replication. J. Virol. 2005, 79, 8802–8811. [Google Scholar] [CrossRef]
- Zhirnov, O.P.; Klenk, H.-D. Control of Apoptosis in Influenza Virus-Infected Cells by up-Regulation of Akt and P53 Signaling. Apoptosis 2007, 12, 1419–1432. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Wang, X.; Guo, L.; Qiu, Y.; Li, X.; Yu, H.; Xiang, H.; Tong, G.; Ma, Z. Influenza A Virus Induces P53 Accumulation in a Biphasic Pattern. Biochem. Biophys Res. Commun. 2009, 382, 331–335. [Google Scholar] [CrossRef]
- Terrier, O.; Josset, L.; Textoris, J.; Marcel, V.; Cartet, G.; Ferraris, O.; N’Guyen, C.; Lina, B.; Diaz, J.-J.; Bourdon, J.-C.; et al. Cellular Transcriptional Profiling in Human Lung Epithelial Cells Infected by Different Subtypes of Influenza A Viruses Reveals an Overall Down-Regulation of the Host P53 Pathway. Virol. J. 2011, 8, 285. [Google Scholar] [CrossRef]
- Muñoz-Fontela, C.; Mandinova, A.; Aaronson, S.A.; Lee, S.W. Emerging Roles of P53 and Other Tumour-Suppressor Genes in Immune Regulation. Nat. Rev. Immunol. 2016, 16, 741–750. [Google Scholar] [CrossRef]
- Lazo, P.A.; Santos, C.R. Interference with P53 Functions in Human Viral Infections, a Target for Novel Antiviral Strategies? Rev. Med. Virol. 2011, 21, 285–300. [Google Scholar] [CrossRef] [PubMed]
- Terrier, O.; Diederichs, A.; Dubois, J.; Cartet, G.; Lina, B.; Bourdon, J.-C.; Rosa-Calatrava, M. Influenza NS1 Interacts with P53 and Alters Its Binding to P53-Responsive Genes, in a Promoter-Dependent Manner. FEBS Lett. 2013, 587, 2965–2971. [Google Scholar] [CrossRef]
- Terrier, O.; Marcel, V.; Cartet, G.; Lane, D.P.; Lina, B.; Rosa-Calatrava, M.; Bourdon, J.-C. Influenza A Viruses Control Expression of Proviral Human P53 Isoforms P53β and Δ133p53α. J. Virol. 2012, 86, 8452–8460. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Deng, X.; Yan, W.; Zhu, Z.; Shen, Y.; Qiu, Y.; Shi, Z.; Shao, D.; Wei, J.; Xia, X.; et al. Stabilization of P53 in Influenza A Virus-Infected Cells Is Associated with Compromised MDM2-Mediated Ubiquitination of P53. J. Biol. Chem. 2012, 287, 18366–18375. [Google Scholar] [CrossRef] [PubMed]
- Pizzorno, A.; Dubois, J.; Machado, D.; Cartet, G.; Traversier, A.; Julien, T.; Lina, B.; Bourdon, J.-C.; Rosa-Calatrava, M.; Terrier, O. Influenza A Viruses Alter the Stability and Antiviral Contribution of Host E3-Ubiquitin Ligase Mdm2 during the Time-Course of Infection. Sci. Rep. 2018, 8, 3746. [Google Scholar] [CrossRef]
- Bohlman, S.; Manfredi, J.J. P53-Independent Effects of Mdm2. Subcell Biochem. 2014, 85, 235–246. [Google Scholar] [CrossRef]
- Thomasova, D.; Mulay, S.R.; Bruns, H.; Anders, H.-J. P53-Independent Roles of MDM2 in NF-ΚB Signaling: Implications for Cancer Therapy, Wound Healing, and Autoimmune Diseases. Neoplasia 2012, 14, 1097–1101. [Google Scholar] [CrossRef]
- Xu, K.; Klenk, C.; Liu, B.; Keiner, B.; Cheng, J.; Zheng, B.-J.; Li, L.; Han, Q.; Wang, C.; Li, T.; et al. Modification of Nonstructural Protein 1 of Influenza A Virus by SUMO1. J. Virol. 2011, 85, 1086–1098. [Google Scholar] [CrossRef]
- Santos, A.; Pal, S.; Chacon, J.; Meraz, K.; Gonzalez, J.; Prieto, K.; Rosas-Acosta, G. SUMOylation Affects the Interferon Blocking Activity of the Influenza A Nonstructural Protein NS1 without Affecting Its Stability or Cellular Localization. J. Virol. 2013, 87, 5602–5620. [Google Scholar] [CrossRef][Green Version]
- Way, G.; Xiong, Z.; Wang, G.; Dai, H.; Zheng, S.; García-Sastre, A.; Liao, J. A Novel SUMOylation Site in the Influenza a Virus NS1 Protein Identified with a Highly Sensitive FRET Assay. J. Biotechnol. 2020, 323, 121–127. [Google Scholar] [CrossRef]
- Jiang, J.; Li, J.; Fan, W.; Zheng, W.; Yu, M.; Chen, C.; Sun, L.; Bi, Y.; Ding, C.; Gao, G.F.; et al. Robust Lys63-Linked Ubiquitination of RIG-I Promotes Cytokine Eruption in Early Influenza B Virus Infection. J. Virol. 2016, 90, 6263–6275. [Google Scholar] [CrossRef] [PubMed]
- Versteeg, G.A.; Hale, B.G.; van Boheemen, S.; Wolff, T.; Lenschow, D.J.; García-Sastre, A. Species-Specific Antagonism of Host ISGylation by the Influenza B Virus NS1 Protein. J. Virol. 2010, 84, 5423–5430. [Google Scholar] [CrossRef] [PubMed]
- Guan, R.; Ma, L.-C.; Leonard, P.G.; Amer, B.R.; Sridharan, H.; Zhao, C.; Krug, R.M.; Montelione, G.T. Structural Basis for the Sequence-Specific Recognition of Human ISG15 by the NS1 Protein of Influenza B Virus. Proc. Natl. Acad. Sci. USA 2011, 108, 13468–13473. [Google Scholar] [CrossRef] [PubMed]
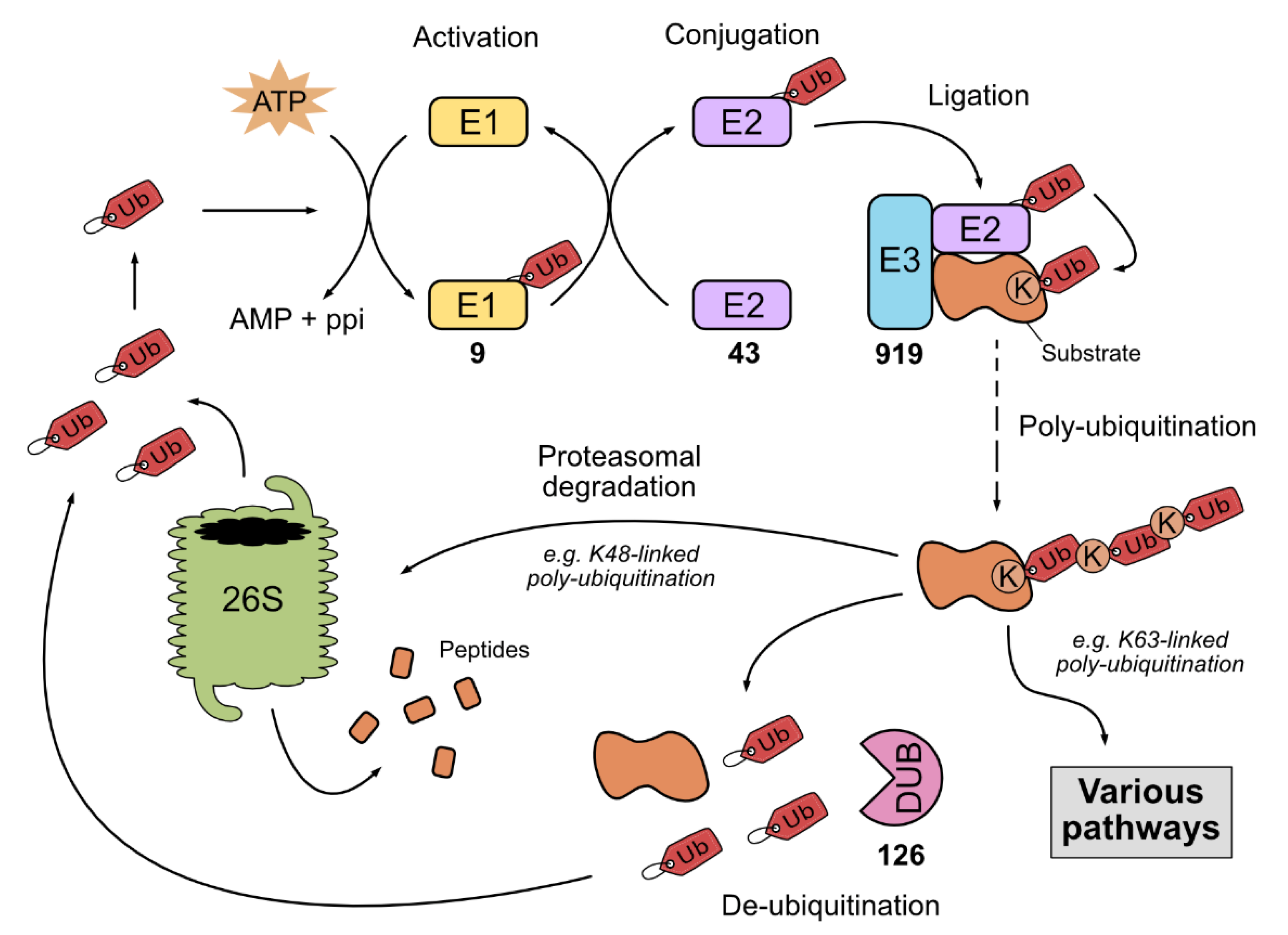


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lamotte, L.-A.; Tafforeau, L. How Influenza A Virus NS1 Deals with the Ubiquitin System to Evade Innate Immunity. Viruses 2021, 13, 2309. https://doi.org/10.3390/v13112309
Lamotte L-A, Tafforeau L. How Influenza A Virus NS1 Deals with the Ubiquitin System to Evade Innate Immunity. Viruses. 2021; 13(11):2309. https://doi.org/10.3390/v13112309
Chicago/Turabian StyleLamotte, Laurie-Anne, and Lionel Tafforeau. 2021. "How Influenza A Virus NS1 Deals with the Ubiquitin System to Evade Innate Immunity" Viruses 13, no. 11: 2309. https://doi.org/10.3390/v13112309
APA StyleLamotte, L.-A., & Tafforeau, L. (2021). How Influenza A Virus NS1 Deals with the Ubiquitin System to Evade Innate Immunity. Viruses, 13(11), 2309. https://doi.org/10.3390/v13112309






