Isolation of the Novel Phage PHB09 and Its Potential Use against the Plant Pathogen Pseudomonas syringae pv. actinidiae
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Phage Isolation and Purification
2.3. Transmission Electron Microscopy
2.4. Determination of the Optimal Multiplicity of Infection and One-Step Growth Curve
2.5. Phage Host Range
2.6. Stability of the Phage under Various Thermal, Ultraviolet, and pH Conditions
2.7. In Vitro and In Vivo Phage Efficacy in Psa Control
2.8. DNA Extraction
2.9. Genome Sequencing and Bioinformatics Analysis
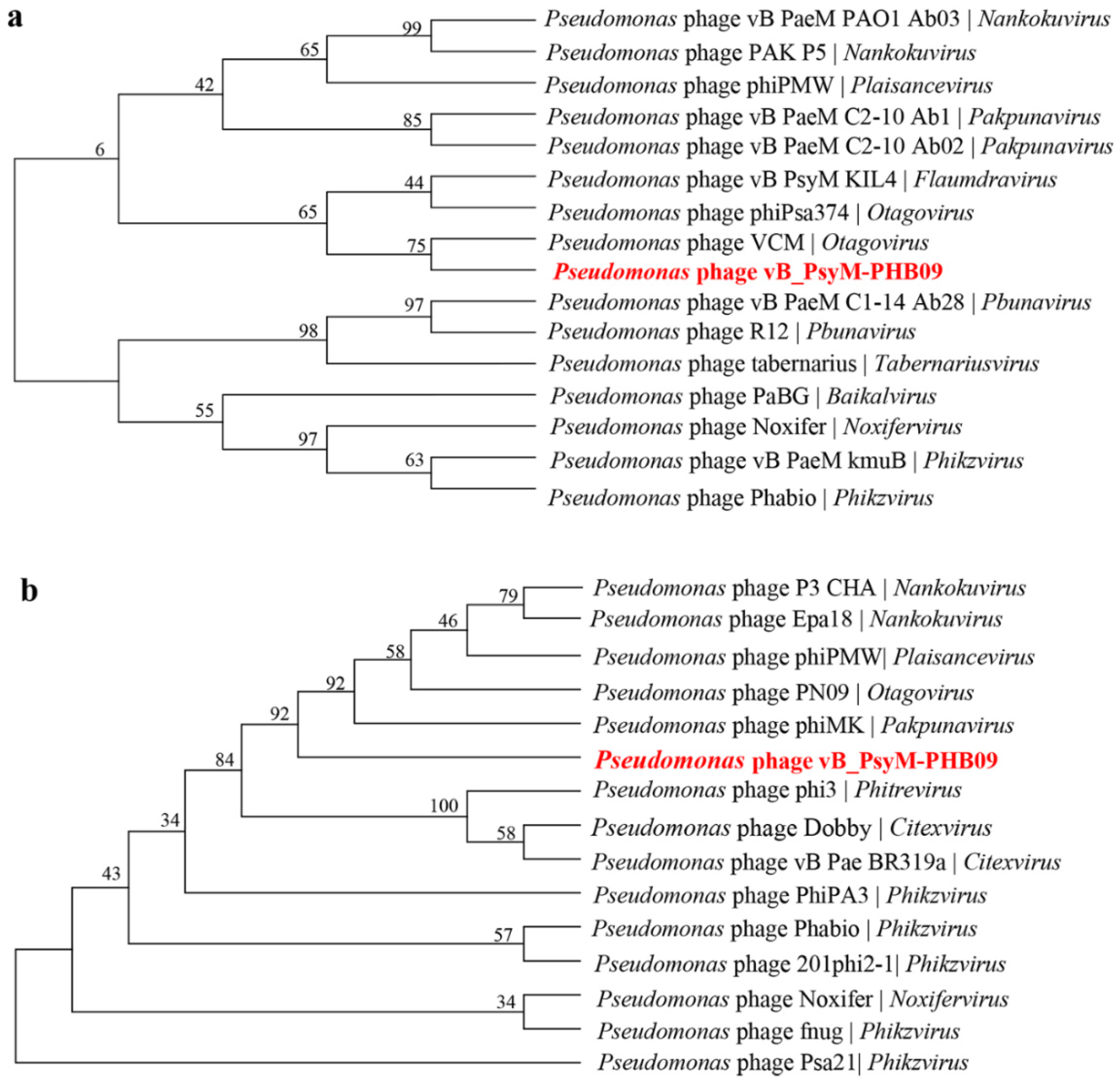
2.10. Phylogenetic Analysis
2.11. Statistical Analysis
3. Results and Discussion
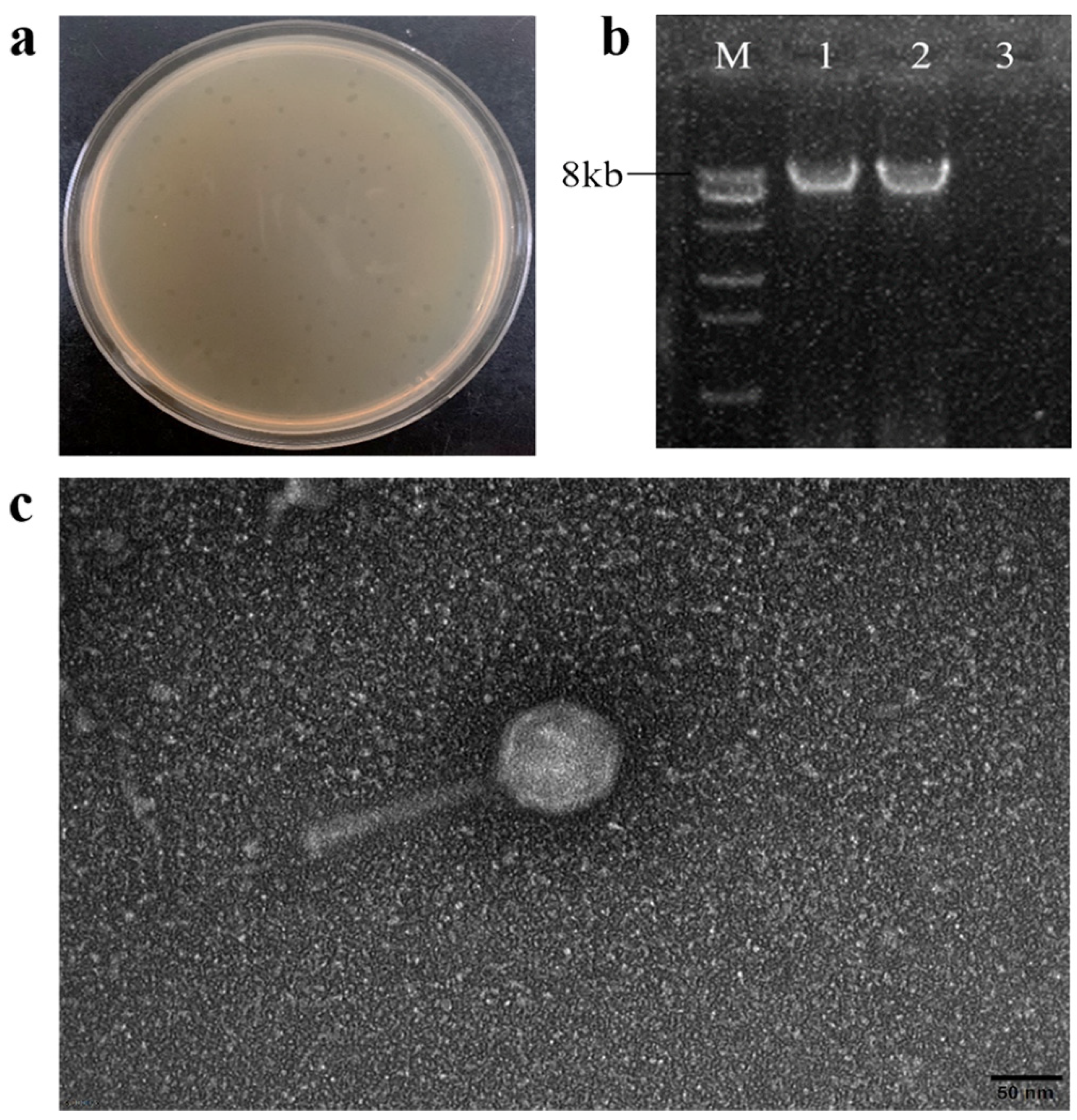
3.1. Isolation and Morphology of Phage PHB09
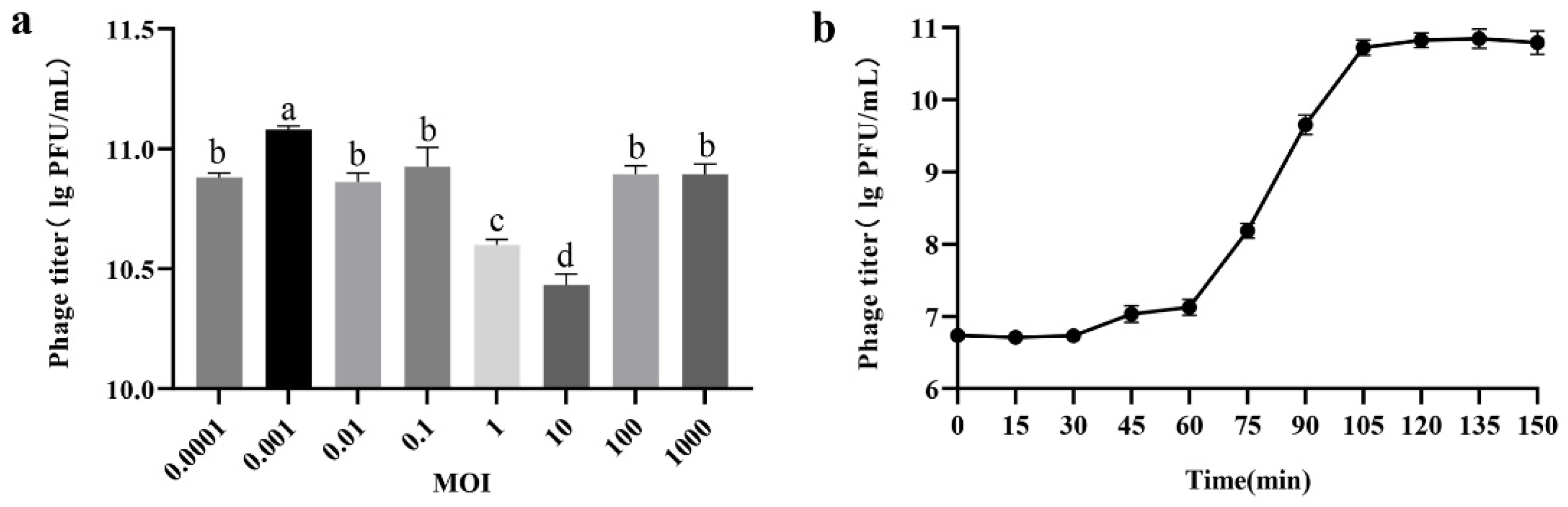
3.2. Biological Characterization of Phage PHB09
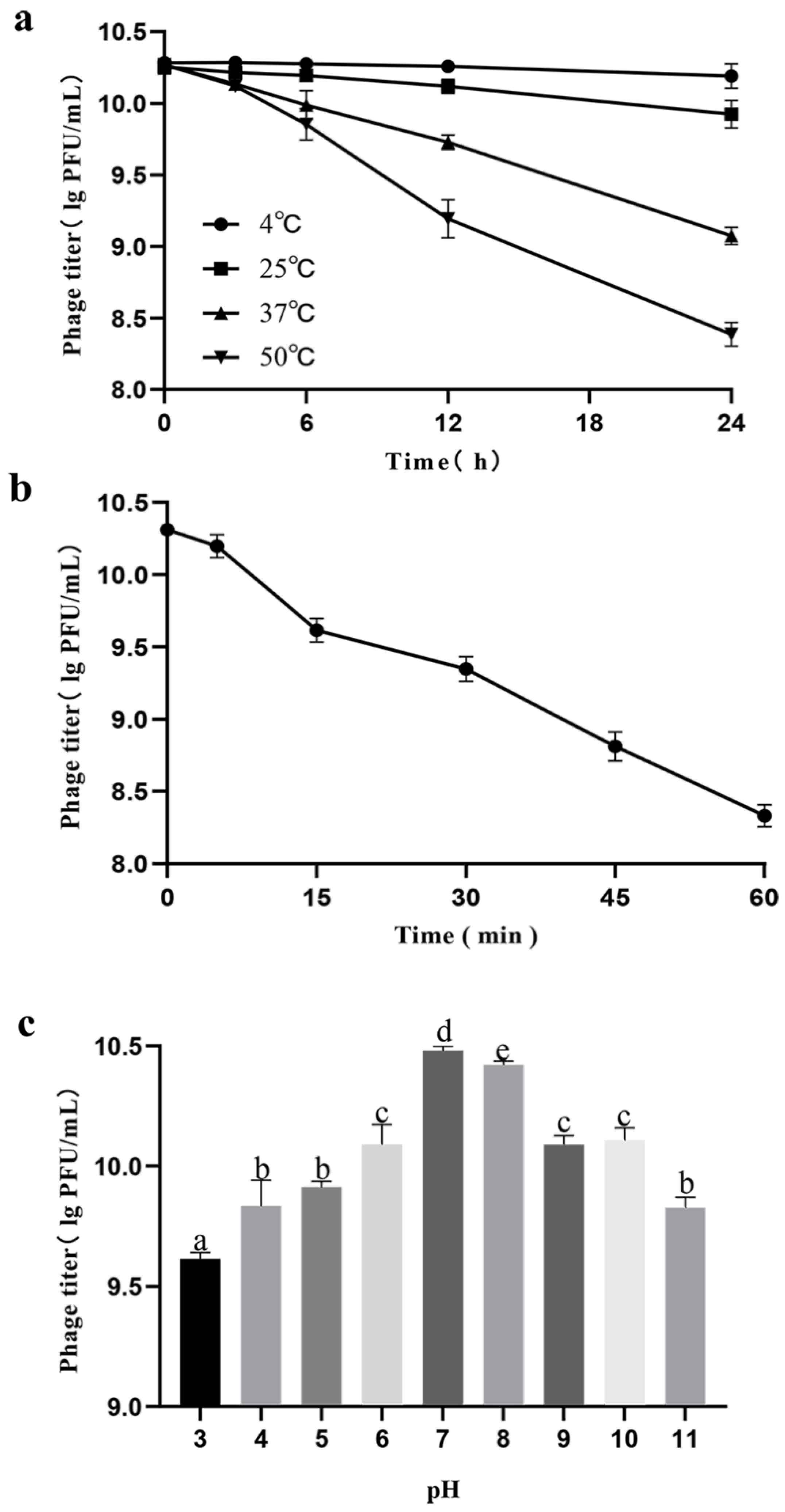
3.3. Thermal, Ultraviolet, and pH Stability Tests
3.4. Host Range of Phage PHB09
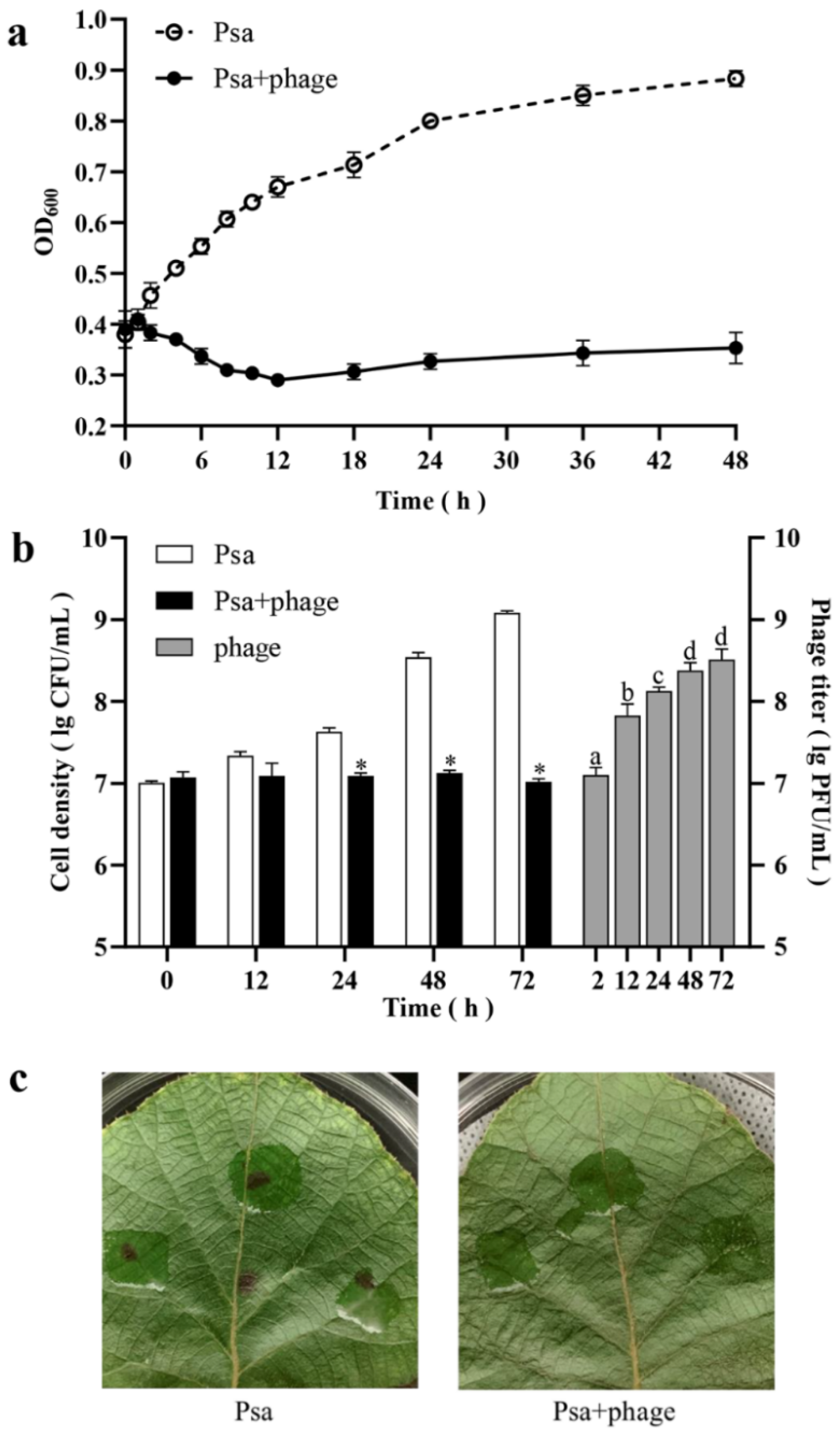
3.5. In Vitro and In Vivo Efficacy of Phage PHB09
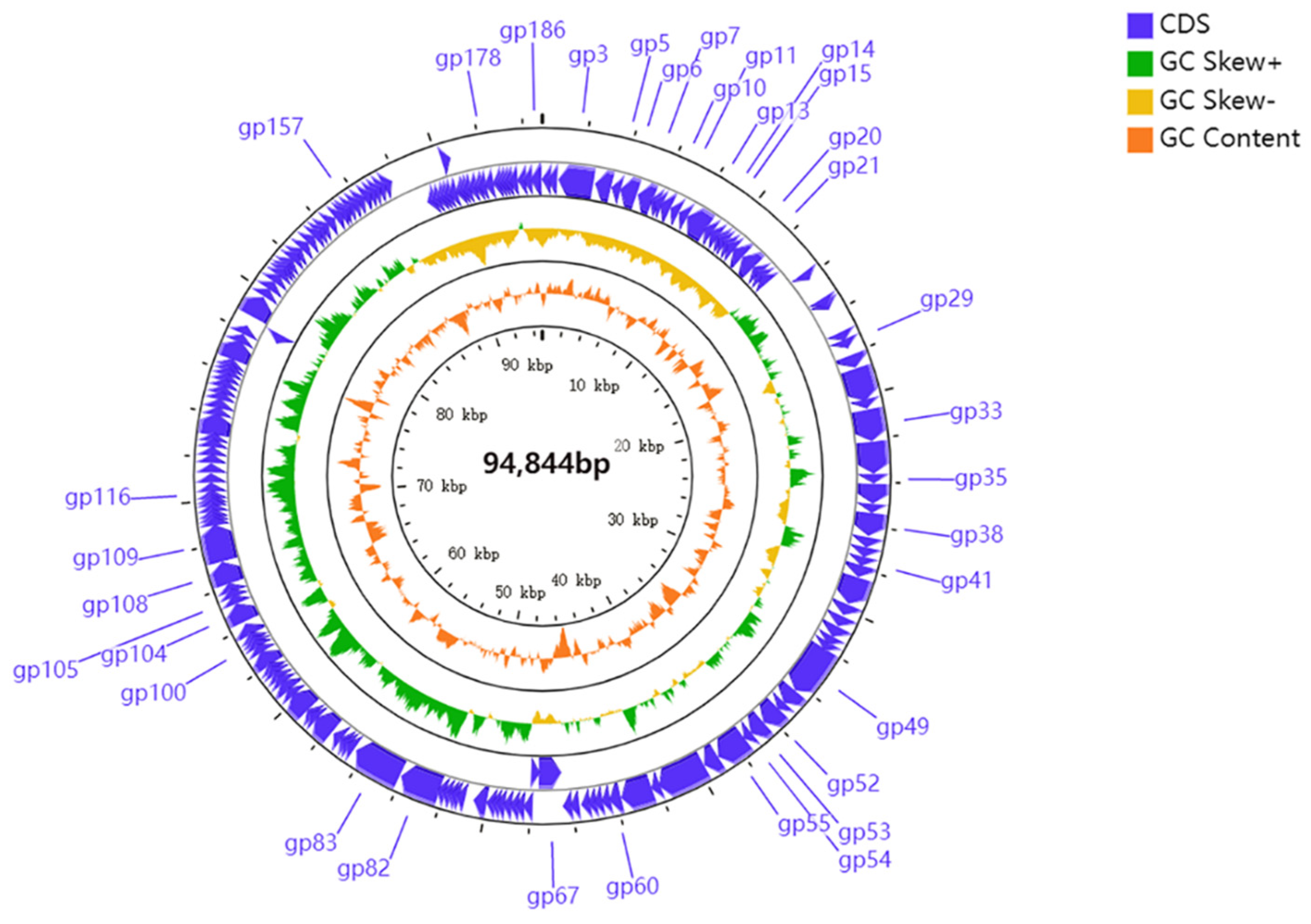
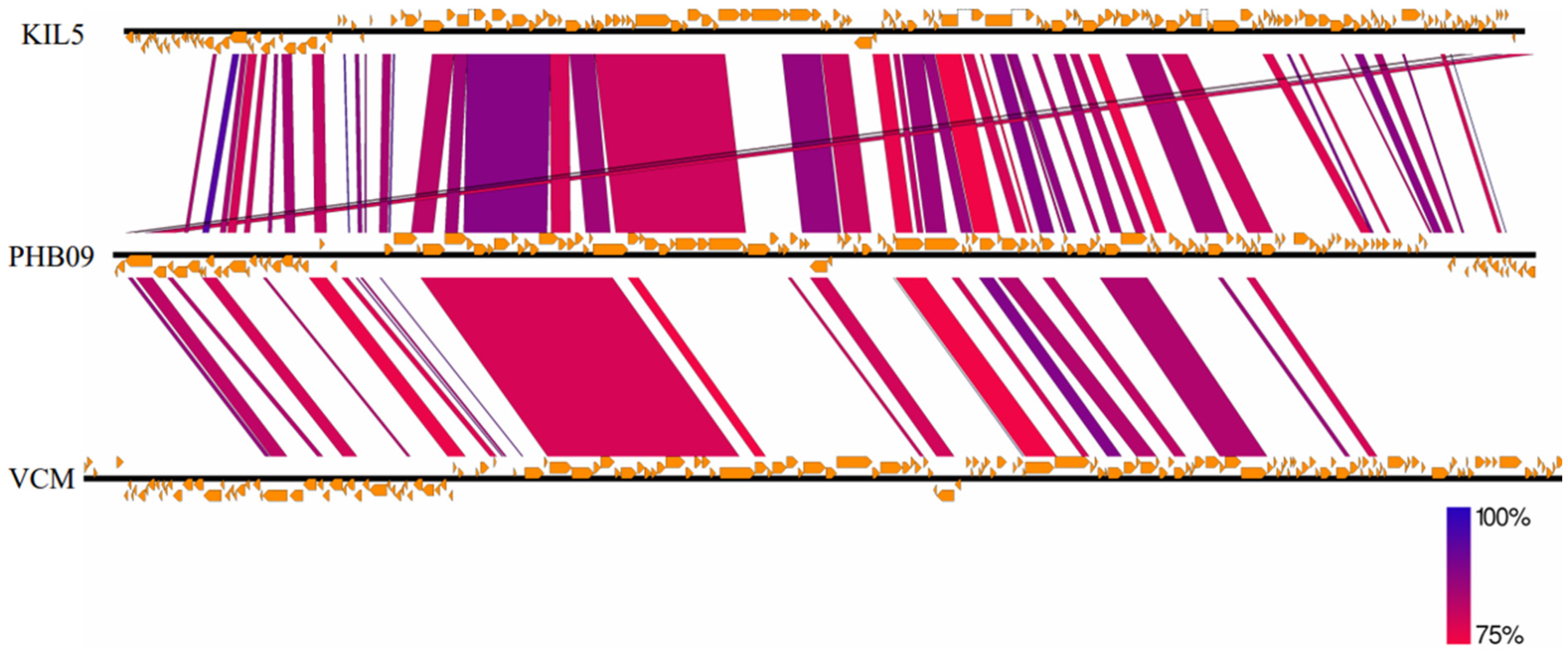
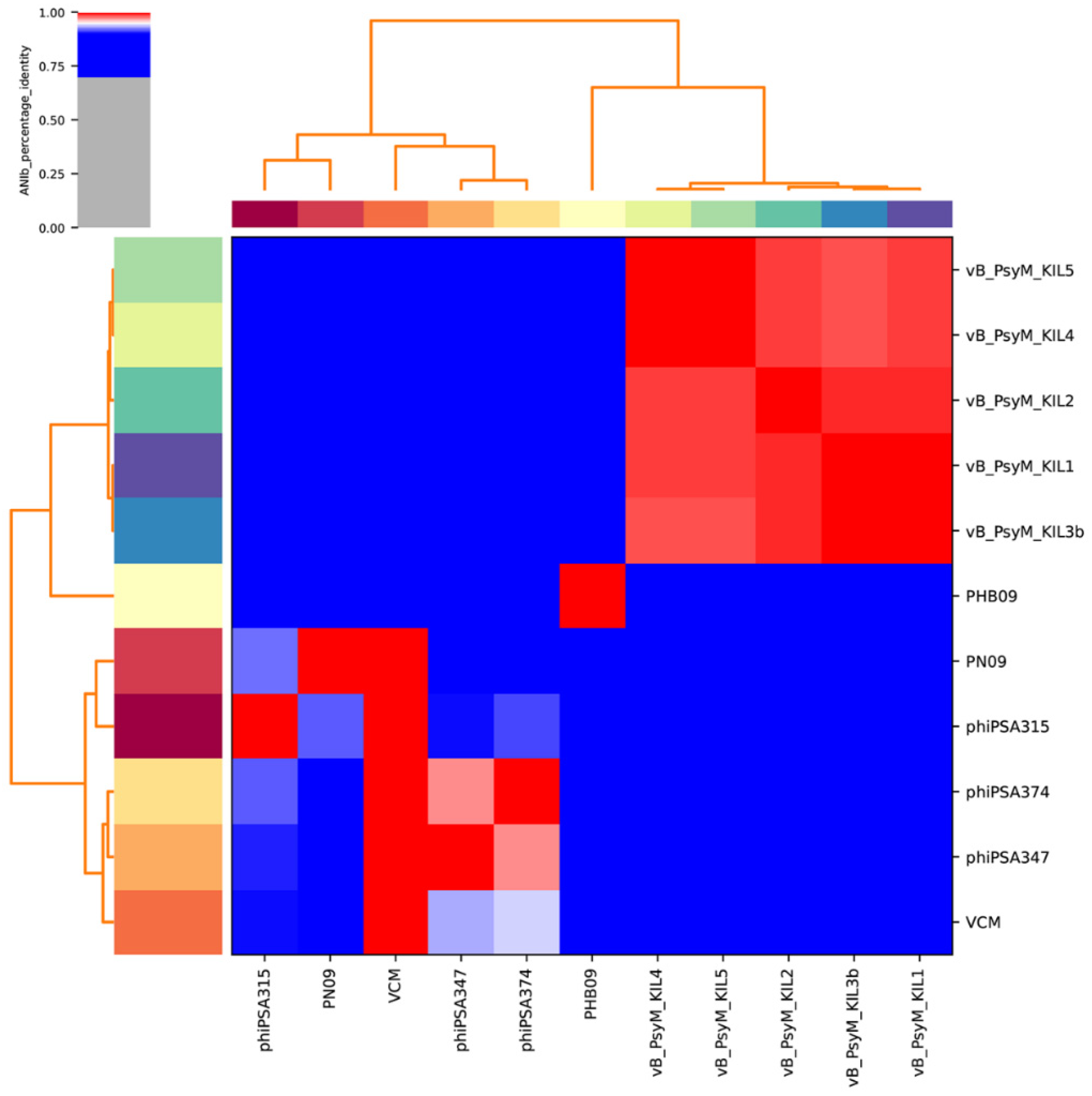
3.6. Genome Characterization and Comparative Genomic Analysis
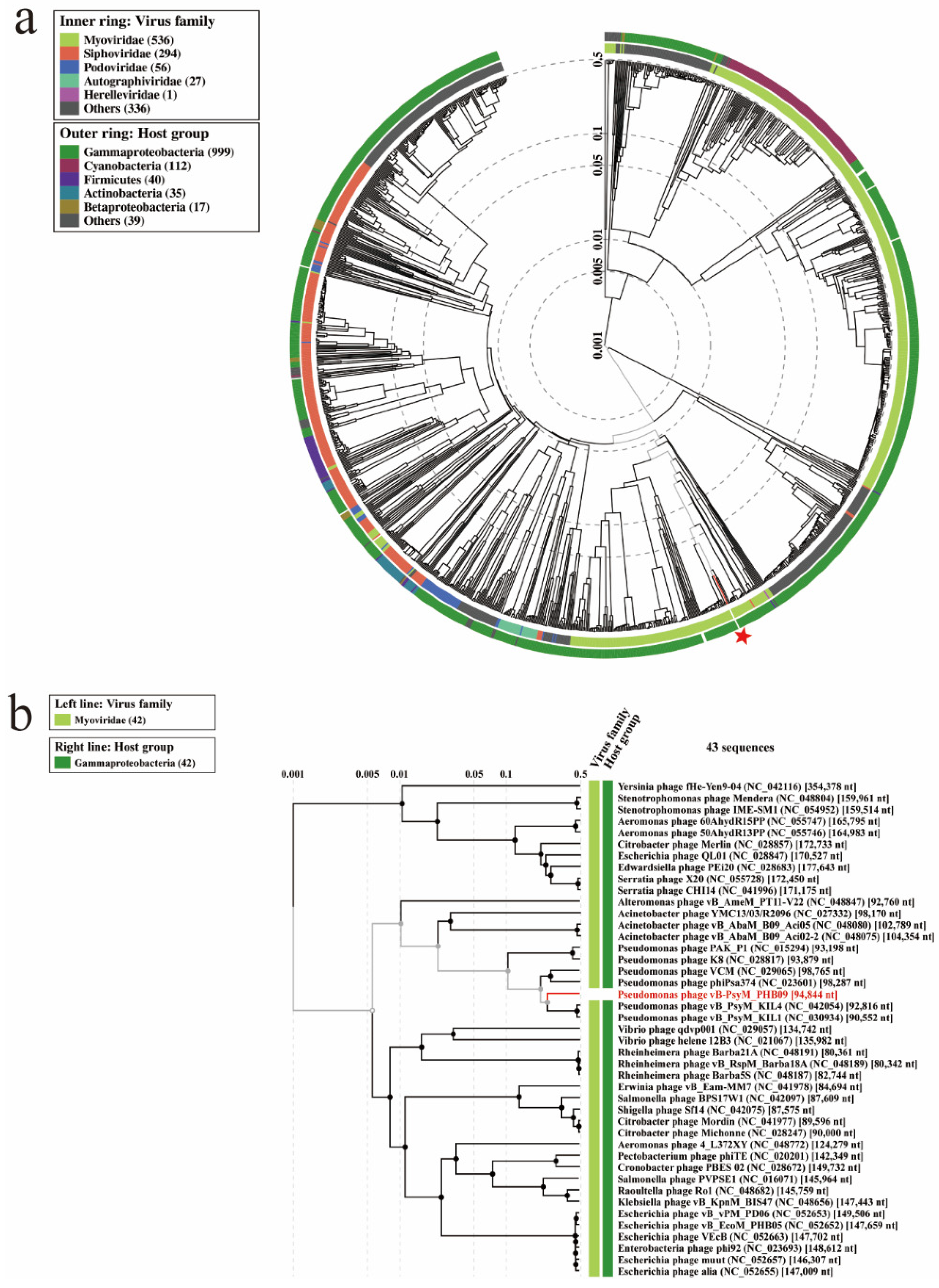
3.7. Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pinheiro, L.A.M.; Pereira, C.; Frazão, C.; Balcão, V.M.; Almeida, A. Efficiency of Phage Φ6 for Biocontrol of Pseudomonas syringae pv. syringae: An in Vitro Preliminary Study. Microorganisms 2019, 7, 286. [Google Scholar] [CrossRef]
- Xin, X.-F.; Kvitko, B.; He, S.Y. Pseudomonas syringae: What It Takes to Be a Pathogen. Nat. Rev. Microbiol. 2018, 16, 316–328. [Google Scholar] [CrossRef]
- Kim, G.H.; Kim, K.-H.; Son, K.I.; Choi, E.D.; Lee, Y.S.; Jung, J.S.; Koh, Y.J. Outbreak and Spread of Bacterial Canker of Kiwifruit Caused by Pseudomonas syringae pv. actinidiae Biovar 3 in Korea. Plant Pathol. J. 2016, 32, 545–551. [Google Scholar] [CrossRef]
- Koh, Y.J.; Kim, G.H.; Koh, H.S.; Lee, Y.S.; Kim, S.-C.; Jung, J.S. Occurrence of a New Type of Pseudomonas syringae pv. actinidiae Strain of Bacterial Canker on Kiwifruit in Korea. Plant Pathol. J. 2012, 28, 423–427. [Google Scholar] [CrossRef][Green Version]
- Poulter, R.T.M.; Ho, J.; Handley, T.; Taiaroa, G.; Butler, M.I. Comparison between Complete Genomes of an Isolate of Pseudomonas syringae pv. actinidiae from Japan and a New Zealand Isolate of the Pandemic Lineage. Sci. Rep. 2018, 8, 10915. [Google Scholar] [CrossRef] [PubMed]
- Maleki, S.; Maleki-Zanjani, B.; Gallego, P.P. Kiwifruit Status in Iran: Management and Production. Acta Hortic. 2018, 39–44. [Google Scholar] [CrossRef]
- Figueira, D.; Garcia, E.; Ares, A.; Tiago, I.; Veríssimo, A.; Costa, J. Genetic Diversity of Pseudomonas syringae pv. actinidiae: Seasonal and Spatial Population Dynamics. Microorganisms 2020, 8, 931. [Google Scholar] [CrossRef] [PubMed]
- Marcelletti, S.; Ferrante, P.; Petriccione, M.; Firrao, G.; Scortichini, M. Pseudomonas syringae pv. actinidiae Draft Genomes Comparison Reveal Strain-Specific Features Involved in Adaptation and Virulence to Actinidia Species. PLoS ONE 2011, 6, e27297. [Google Scholar] [CrossRef]
- Chapman, J.R.; Taylor, R.K.; Weir, B.S.; Romberg, M.K.; Vanneste, J.L.; Luck, J.; Alexander, B.J.R. Phylogenetic Relationships among Global Populations of Pseudomonas syringae pv. actinidiae. Phytopathology 2012, 102, 1034–1044. [Google Scholar] [CrossRef]
- McCann, H.C.; Li, L.; Liu, Y.; Li, D.; Pan, H.; Zhong, C.; Rikkerink, E.H.A.; Templeton, M.D.; Straub, C.; Colombi, E.; et al. Origin and Evolution of the Kiwifruit Canker Pandemic. Genome Biol. Evol. 2017, 9, 932–944. [Google Scholar] [CrossRef]
- Yu, J.-G.; Lim, J.-A.; Song, Y.-R.; Heu, S.; Kim, G.H.; Koh, Y.J.; Oh, C.-S. Isolation and Characterization of Bacteriophages Against Pseudomonas syringae pv. actinidiae Causing Bacterial Canker Disease in Kiwifruit. J. Microbiol. Biotechnol. 2016, 26, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Fujikawa, T.; Sawada, H. Genome Analysis of Pseudomonas syringae pv. actinidiae Biovar 6, Which Produces the Phytotoxins, Phaseolotoxin and Coronatine. Sci. Rep. 2019, 9, 3836. [Google Scholar] [CrossRef] [PubMed]
- Fujikawa, T.; Sawada, H. Genome Analysis of the Kiwifruit Canker Pathogen Pseudomonas syringae pv. actinidiae Biovar 5. Sci. Rep. 2016, 6, 21399. [Google Scholar] [CrossRef]
- Frampton, R.A.; Taylor, C.; Holguín Moreno, A.V.; Visnovsky, S.B.; Petty, N.K.; Pitman, A.R.; Fineran, P.C. Identification of Bacteriophages for Biocontrol of the Kiwifruit Canker Phytopathogen Pseudomonas syringae pv. actinidiae. Appl. Environ. Microbiol. 2014, 80, 2216–2228. [Google Scholar] [CrossRef]
- Altimira, F.; Yáñez, C.; Bravo, G.; González, M.; Rojas, L.A.; Seeger, M. Characterization of Copper-Resistant Bacteria and Bacterial Communities from Copper-Polluted Agricultural Soils of Central Chile. BMC Microbiol. 2012, 12, 193. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, Z.; Cao, Z.; Wang, L.; Li, X.; Li, S.; Xu, Y. Bacteriophages as Antimicrobial Agents against Major Pathogens in Swine: A Review. J. Anim. Sci. Biotechnol. 2015, 6, 39. [Google Scholar] [CrossRef] [PubMed]
- Harada, L.K.; Silva, E.C.; Campos, W.F.; Del Fiol, F.S.; Vila, M.; Dąbrowska, K.; Krylov, V.N.; Balcão, V.M. Biotechnological Applications of Bacteriophages: State of the Art. Microbiol. Res. 2018, 212–213, 38–58. [Google Scholar] [CrossRef] [PubMed]
- Frampton, R.A.; Pitman, A.R.; Fineran, P.C. Advances in Bacteriophage-Mediated Control of Plant Pathogens. Int. J. Microbiol. 2012, 2012, 1–11. [Google Scholar] [CrossRef]
- Czajkowski, R.; Ozymko, Z.; Lojkowska, E. Isolation and Characterization of Novel Soilborne Lytic Bacteriophages Infecting Dickeya spp. Biovar 3 (‘D. Solani’). Plant Pathol. 2014, 63, 758–772. [Google Scholar] [CrossRef]
- Hirata, H.; Kashihara, M.; Horiike, T.; Suzuki, T.; Dohra, H.; Netsu, O.; Tsuyumu, S. Genome Sequence of Pectobacterium carotovorum Phage PPWS1, Isolated from Japanese Horseradish [Eutrema japonicum (Miq.) Koidz] Showing Soft-Rot Symptoms. Genome Announc. 2016, 4. [Google Scholar] [CrossRef]
- Bae, J.Y. Biocontrol Potential of a Lytic Bacteriophage PE204 against Bacterial Wilt of Tomato. J. Microbiol. Biotechnol. 2012, 22, 1613–1620. [Google Scholar] [CrossRef] [PubMed]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage Applications for Food Production and Processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, L.A.M.; Pereira, C.; Barreal, M.E.; Gallego, P.P.; Balcão, V.M.; Almeida, A. Use of Phage Φ6 to Inactivate Pseudomonas syringae pv. actinidiae in Kiwifruit Plants: In Vitro and Ex Vivo Experiments. Appl. Microbiol. Biotechnol. 2020, 104, 1319–1330. [Google Scholar] [CrossRef] [PubMed]
- Costa, P.; Pereira, C.; Gomes, A.; Almeida, A. Efficiency of Single Phage Suspensions and Phage Cocktail in the Inactivation of Escherichia coli and Salmonella typhimurium: An In Vitro Preliminary Study. Microorganisms 2019, 7, 94. [Google Scholar] [CrossRef]
- Flores, O.; Retamales, J.; Núñez, M.; León, M.; Salinas, P.; Besoain, X.; Yañez, C.; Bastías, R. Characterization of Bacteriophages against Pseudomonas syringae pv. actinidiae with Potential Use as Natural Antimicrobials in Kiwifruit Plants. Microorganisms 2020, 8, 974. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.-R.; Vu, N.T.; Park, J.; Hwang, I.S.; Jeong, H.-J.; Cho, Y.-S.; Oh, C.-S. Phage PPPL-1, A New Biological Agent to Control Bacterial Canker Caused by Pseudomonas syringae pv. actinidiae in Kiwifruit. Antibiotics 2021, 10, 554. [Google Scholar] [CrossRef]
- Maniloff, J.; Ackermann, H.-W. Taxonomy of Bacterial Viruses: Establishment of Tailed Virus Genera and the Other Caudovirales. Arch. Virol. 1998, 143, 2051–2063. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lim, J.-A.; Yu, J.-G.; Oh, C.-S. Genomic Features and Lytic Activity of the Bacteriophage PPPL-1 Effective against Pseudomonas syringae pv. actinidiae, a Cause of Bacterial Canker in Kiwifruit. J. Microbiol. Biotechnol. 2018, 28, 1542–1546. [Google Scholar] [CrossRef]
- Doss, J.; Culbertson, K.; Hahn, D.; Camacho, J.; Barekzi, N. A Review of Phage Therapy against Bacterial Pathogens of Aquatic and Terrestrial Organisms. Viruses 2017, 9, 50. [Google Scholar] [CrossRef]
- Yang, Y.; Lu, S.; Shen, W.; Zhao, X.; Shen, M.; Tan, Y.; Li, G.; Li, M.; Wang, J.; Hu, F.; et al. Characterization of the First Double-Stranded RNA Bacteriophage Infecting Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 38795. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Peng, Y.; Leung, H.C.M.; Yiu, S.M.; Chin, F.Y.L. IDBA-UD: A de Novo Assembler for Single-Cell and Metagenomic Sequencing Data with Highly Uneven Depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef]
- Olsson, M.; Pauliny, A.; Wapstra, E.; Uller, T.; Schwartz, T.; Blomqvist, D. Sex Differences in Sand Lizard Telomere Inheritance: Paternal Epigenetic Effects Increases Telomere Heritability and Offspring Survival. PLoS ONE 2011, 6, e17473. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Chan, P.P. TRNAscan-SE On-Line: Integrating Search and Context for Analysis of Transfer RNA Genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Liang, Y.; Huang, S.; Zhang, J.; Wang, J.; Chen, H.; Ye, Y.; Gao, X.; Wu, Q.; Tan, Z. Isolation and Characterization of the Novel Phages VB_VpS_BA3 and VB_VpS_CA8 for Lysing Vibrio parahaemolyticus. Front. Microbiol. 2020, 11, 259. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Li, C.; Wu, Q.; Ding, Y.; Yang, M.; Hu, Y.; Zeng, H.; Zhang, J. Isolation and Characterization of New Phage VB_CtuP_A24 and Application to Control Cronobacter spp. in Infant Milk Formula and Lettuce. Food Res. Int. 2021, 141, 110109. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A Genome Comparison Visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. ViPTree: The Viral Proteomic Tree Server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef]
- Lu, S.; Le, S.; Tan, Y.; Zhu, J.; Li, M.; Rao, X.; Zou, L.; Li, S.; Wang, J.; Jin, X.; et al. Genomic and Proteomic Analyses of the Terminally Redundant Genome of the Pseudomonas aeruginosa Phage PaP1: Establishment of Genus PaP1-Like Phages. PLoS ONE 2013, 8, e62933. [Google Scholar] [CrossRef]
- Chen, Y.; Sun, E.; Song, J.; Yang, L.; Wu, B. Complete Genome Sequence of a Novel T7-Like Bacteriophage from a Pasteurella multocida Capsular Type A Isolate. Curr. Microbiol. 2018, 75, 574–579. [Google Scholar] [CrossRef]
- Ceyssens, P.-J.; Lavigne, R. Bacteriophages of Pseudomonas. Future Microbiol. 2010, 5, 1041–1055. [Google Scholar] [CrossRef]
- Rombouts, S.; Volckaert, A.; Venneman, S.; Declercq, B.; Vandenheuvel, D.; Allonsius, C.N.; Van Malderghem, C.; Jang, H.B.; Briers, Y.; Noben, J.P.; et al. Characterization of Novel Bacteriophages for Biocontrol of Bacterial Blight in Leek Caused by Pseudomonas syringae pv. porri. Front. Microbiol. 2016, 7, 279. [Google Scholar] [CrossRef]
- Di Lallo, G.; Evangelisti, M.; Mancuso, F.; Ferrante, P.; Marcelletti, S.; Tinari, A.; Superti, F.; Migliore, L.; D’Addabbo, P.; Frezza, D.; et al. Isolation and Partial Characterization of Bacteriophages Infecting Pseudomonas syringae pv. actinidiae, Causal Agent of Kiwifruit Bacterial Canker. J. Basic Microbiol. 2014, 54, 1210–1221. [Google Scholar] [CrossRef] [PubMed]
- Jończyk, E.; Kłak, M.; Międzybrodzki, R.; Górski, A. The Influence of External Factors on Bacteriophages—Review. Folia Microbiol. 2011, 56, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Das, Q.; Islam, M.R.; Marcone, M.F.; Warriner, K.; Diarra, M.S. Potential of Berry Extracts to Control Foodborne Pathogens. Food Control 2017, 73, 650–662. [Google Scholar] [CrossRef]
- Mojica, K.D.A.; Brussaard, C.P.D. Factors Affecting Virus Dynamics and Microbial Host—Virus Interactions in Marine Environments. FEMS Microbiol. Ecol. 2014, 89, 495–515. [Google Scholar] [CrossRef] [PubMed]
- Duarte, J.; Pereira, C.; Moreirinha, C.; Salvio, R.; Lopes, A.; Wang, D.; Almeida, A. New Insights on Phage Efficacy to Control Aeromonas Salmonicida in Aquaculture Systems: An in Vitro Preliminary Study. Aquaculture 2018, 495, 970–982. [Google Scholar] [CrossRef]
- Reimer, J.D.; Sinniger, F.; Nonaka, M.; Uchida, S. Non-Invasive Internal Morphological Examination of Epizoic Zoanthids Utilizing CT Scanners. Coral Reefs 2009, 28, 621. [Google Scholar] [CrossRef]
- Fujiwara, A.; Fujisawa, M.; Hamasaki, R.; Kawasaki, T.; Fujie, M.; Yamada, T. Biocontrol of Ralstonia solanacearum by Treatment with Lytic Bacteriophages. Appl. Environ. Microbiol. 2011, 77, 4155–4162. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Ryu, S. Characterization of a T5-like Coliphage, SPC35, and Differential Development of Resistance to SPC35 in Salmonella enterica Serovar Typhimurium and Escherichia coli. Appl. Environ. Microbiol. 2011, 77, 2042–2050. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Zhang, J.; Niu, Y.D.; Cui, N.; Ma, Y.; Cao, F.; Jin, L.; Li, Z.; Xu, Y. Isolation and Characterization of a “PhiKMV-like” Bacteriophage and Its Therapeutic Effect on Mink Hemorrhagic Pneumonia. PLoS ONE 2015, 10, e0116571. [Google Scholar] [CrossRef]
- Adriaenssens, E.; Brister, J.R. How to Name and Classify Your Phage: An Informal Guide. Viruses 2017, 9, 70. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.; Kropinski, A.M.; Adriaenssens, E.M. A Roadmap for Genome-Based Phage Taxonomy. Viruses 2021, 13, 506. [Google Scholar] [CrossRef] [PubMed]
| Accession No. | Protein | Organism | Genus |
|---|---|---|---|
| HE983845 | Terminase large subunit | Pseudomonas phage vB_PaeM_C2-10_Ab1 | Pakpunavirus |
| CEF88981 | Terminase large subunit | Pseudomonas phage vB_PaeM_C2-10_Ab02 | Pakpunavirus |
| YP_009616728 | Terminase large subunit | Pseudomonas phage vB_PsyM_KIL4 | Flaumdravirus |
| NC_041994 | Terminase large subunit | Pseudomonas phage Noxifer | Noxifervirus |
| YP_009124453 | Terminase large subunit | Pseudomonas phage vB_PaeM_PAO1_Ab03 | Nankokuvirus |
| YP_008857039 | Terminase large subunit | Pseudomonas phage PAK_P5 | Nankokuvirus |
| YP_008433440 | Terminase large subunit | Pseudomonas phage PaBG | Baikalvirus |
| QPB10483 | Terminase large subunit | Pseudomonas phage VCM | Otagovirus |
| QNO00383 | Terminase large subunit | Pseudomonas phage phiPsa374 | Otagovirus |
| YP_009966987 | Terminase large subunit | Pseudomonas phage vB_PaeM_C1-14_Ab28 | Pbunavirus |
| YP_009914209 | Terminase large subunit | Pseudomonas phage R12 | Pbunavirus |
| YP_001956731 | Terminase large subunit | Pseudomonas phage vB_PaeM_kmuB | Phikzvirus |
| ARV76634 | Terminase large subunit | Pseudomonas phage Phabio | Phikzvirus |
| YP_009595351 | Terminase large subunit | Pseudomonas phage phiPMW | Plaisancevirus |
| YP_009620857 | Terminase large subunit | Pseudomonas phage tabernarius | Tabernariusvirus |
| YP_009291185 | Major capsid protein | Pseudomonas phage phiMK | Pakpunavirus |
| ALY08259 | Major capsid protein | Pseudomonas phage phi3 | Phitrevirus |
| YP_009595355 | Major capsid protein | Pseudomonas phage phiPMW | Plaisancevirus |
| ADX32002 | Major capsid protein | Pseudomonas phage P3_CHA | Nankokuvirus |
| YP_009609037 | Major capsid protein | Pseudomonas phage Noxifer | Noxifervirus |
| AZF87846 | Major capsid protein | Pseudomonas phage Dobby | Citexvirus |
| QJB22791 | Major capsid protein | Pseudomonas phage fnug | Phikzvirus |
| QVJ13065 | Major capsid protein | Pseudomonas phage Psa21 | Phikzvirus |
| AEH03559 | Major capsid protein | Pseudomonas phage PhiPA3 | Phikzvirus |
| ARV76818 | Major capsid protein | Pseudomonas phage Phabio | Phikzvirus |
| QPB10488 | Major capsid protein | Pseudomonas phage PN09 | Otagovirus |
| YP_001956923 | Major capsid protein | Pseudomonas phage 201phi2-1 | Phikzvirus |
| Bacteria Strain | Accession Number | Source | Plaque Formation a |
|---|---|---|---|
| Psa BJ530 (biovar 3) | 1.19157 | CGMCC | + |
| Psa BJ9 (biovar 3) | Isolate from kiwi orchard | + | |
| Psa BST (biovar 3) | Isolate from kiwi orchard | + | |
| Psa (biovar 2) | 10584 | KACC | + |
| Psa (biovar 2) | 10592 | KACC | + |
| Psa (biovar 2) | 10587 | KACC | + |
| Pseudomonas oryzihabitans | 1.3158 | CGMCC | − |
| Pseudomonas oryzihabitans | 1.2879 | CGMCC | − |
| Pseudomonas oryzihabitans | 1.15148 | CGMCC | − |
| Pseudomonas fluorescens | 1.7375 | CGMCC | − |
| Pseudomonas fluorescens | 1.7373 | CGMCC | − |
| Pseudomonas fluorescens | 1.7306 | CGMCC | − |
| Pseudomonas putida | 1.8829 | CGMCC | − |
| Pseudomonas putida | 1.7662 | CGMCC | − |
| Pseudomonas gessardii | 1.6429 | CGMCC | − |
| Pseudomonas gessardii | 1.874 | CGMCC | − |
| Pseudomonas fragi | 1.7759 | CGMCC | − |
| Pseudomonas fragi | 1.7757 | CGMCC | − |
| Pseudomonas fragi | 1.7752 | CGMCC | − |
| Enterobacter hormaechei | 1.10608 | CGMCC | − |
| Enterobacter cloacae | 1.8726 | CGMCC | − |
| Escherichia coli | 1.12883 | CGMCC | − |
| Gene Product | Genebank ID | Identity (%) | e-Value | Putative Function | Organism | Functional Categories |
|---|---|---|---|---|---|---|
| Gp3 | UAV89895.1 | 81.28 | 0 | nicotinamide phosphoribosyltransferase | Pseudomonas phage REC | auxiliary metabolism |
| Gp5 | UAV89899.1 | 90 | 3.53 × 10−84 | polynucleotide kinase | Pseudomonas phage REC | nucleotides metabolism |
| Gp6 | QNO00057.1 | 76.28 | 6.22 × 10−163 | RNA ligase | Pseudomonas phage phiPsa300 | transcription |
| Gp7 | WP_198844968.1 | 71.79 | 6.1 × 10−139 | SPFH domain-containing protein | Pseudomonas sp. MF7453 | structure |
| Gp10 | UAV89634.1 | 87.30 | 2.25 × 10−123 | phosphohydrolase | Pseudomonas phage M5.1 | nucleotides metabolism |
| Gp11 | UAV89635.1 | 65.60 | 5.35 × 10−70 | hydrolase | Pseudomonas phage M5.1 | auxiliary metabolism |
| Gp13 | AMR57593.1 | 79.14 | 0 | putative DNA ligase | Pseudomonas phage vB_PsyM_KIL3 | DNA replication and repair |
| Gp14 | ATN92916.1 | 55.12 | 1.17 × 10−17 | putative DNA ligase | Pseudomonas phage PPSC2 | DNA replication and repair |
| Gp15 | YP_009616712.1 | 81.59 | 9.59 × 10−98 | putative deoxycytidylate deaminase | Pseudomonas phage vB_PsyM_KIL4 | auxiliary metabolism |
| Gp20 | AMR57443.1 | 95.61 | 1.5 × 10−142 | putative serine protease | Pseudomonas phage vB_PsyM_KIL2 | auxiliary metabolism |
| Gp21 | AMR57444.1 | 97.67 | 0 | putative phosphate starvation protein | Pseudomonas phage vB_PsyM_KIL2 | nucleotides metabolism |
| Gp29 | AMR57452.1 | 83.33 | 3.56 × 10−81 | putative HNH endonuclease | Pseudomonas phage vB_PsyM_KIL2 | DNA replication and repair |
| Gp33 | YP_009222712.1 | 94.27 | 0 | terminase-like family protein | Pseudomonas phage VCM | packing |
| Gp35 | YP_009275981.1 | 91.94 | 6.26 × 10−95 | putative methyltransferase | Pseudomonas phage vB_PsyM_KIL1 | nucleotides metabolism |
| Gp38 | YP_009616734.1 | 97.73 | 0 | major capsid protein | Pseudomonas phage vB_PsyM_KIL4 | structure |
| Gp41 | UAV89665.1 | 81.96 | 9.54 × 10−70 | head-to-tail stopper | Pseudomonas phage M5.1 | structure |
| Gp49 | YP_009616750.1 | 85.96 | 0 | putative tape measure protein | Pseudomonas phage vB_PsyM_KIL4 | structure |
| Gp52 | YP_009276001.1 | 93.48 | 0 | putative structural protein | Pseudomonas phage vB_PsyM_KIL1 | structure |
| Gp53 | YP_009276002.1 | 88.53 | 9.3 × 10−169 | putative baseplate protein | Pseudomonas phage vB_PsyM_KIL1 | structure |
| Gp54 | YP_009276003.1 | 90.08 | 8.85 × 10−76 | putative tail lysozyme | Pseudomonas phage vB_PsyM_KIL1 | lysis |
| Gp55 | YP_009276004.1 | 91.34 | 0 | putative baseplate component | Pseudomonas phage vB_PsyM_KIL1 | structure |
| Gp60 | YP_009276009.1 | 94.62 | 5.6 × 10−129 | putative endolysin | Pseudomonas phage vB_PsyM_KIL1 | lysis |
| Gp67 | AMR57493.1 | 85.26 | 0 | putative RNA ligase | Pseudomonas phage vB_PsyM_KIL2 | transcription |
| Gp82 | YP_009276030.1 | 93.82 | 0 | putative DNA primase/helicase | Pseudomonas phage vB_PsyM_KIL1 | DNA replication and repair |
| Gp83 | UAV89975.1 | 80.31 | 0 | DNA polymerase | Pseudomonas phage REC | DNA replication and repair |
| Gp100 | YP_009276002.1 | 88.53 | 9.3 × 10−169 | putative baseplate protein | Pseudomonas phage vB_PsyM_KIL1 | structure |
| Gp104 | UAV89994.1 | 76.03 | 1.3 × 10−173 | flavin-dependent thymidylate synthase | Pseudomonas phage REC | nucleotides metabolism |
| Gp105 | UAV84600.1 | 100 | 4.3 × 10−105 | HNH homing endonuclease | Pseudomonas phage PHB09 | auxiliary metabolism |
| Gp108 | YP_009222647.1 | 90.46 | 0 | ribonucleotide-diphosphate reductase subunit beta | Pseudomonas phage VCM | DNA replication and repair |
| Gp109 | YP_009222646.1 | 95.48 | 0 | ribonucleoside-diphosphate reductase NrdZ | Pseudomonas phage VCM | nucleotides metabolism |
| Gp116 | YP_009276062.1 | 87.83 | 2.22 × 10−40 | putative glutaredoxin | Pseudomonas phage vB_PsyM_KIL1 | nucleotides metabolism |
| Gp157 | AXF53051.1 | 43.37 | 2.94 × 10−15 | NADH oxidase H2O-forming | Siphoviridae sp. | auxiliary metabolism |
| Gp178 | UAV89606.1 | 74.21 | 4.59 × 10−74 | peptide chain release factor | Pseudomonas phage M5.1 | auxiliary metabolism |
| Gp186 | YP_009222746.1 | 82.19 | 3.4 × 10−114 | DNA recombination-mediator protein A | Pseudomonas phage VCM | DNA replication and repair |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Liu, M.; Hu, R.; Bai, J.; He, X.; Jin, Y. Isolation of the Novel Phage PHB09 and Its Potential Use against the Plant Pathogen Pseudomonas syringae pv. actinidiae. Viruses 2021, 13, 2275. https://doi.org/10.3390/v13112275
Liu Y, Liu M, Hu R, Bai J, He X, Jin Y. Isolation of the Novel Phage PHB09 and Its Potential Use against the Plant Pathogen Pseudomonas syringae pv. actinidiae. Viruses. 2021; 13(11):2275. https://doi.org/10.3390/v13112275
Chicago/Turabian StyleLiu, Yanxi, Mengjiao Liu, Ran Hu, Jun Bai, Xiaoqing He, and Yi Jin. 2021. "Isolation of the Novel Phage PHB09 and Its Potential Use against the Plant Pathogen Pseudomonas syringae pv. actinidiae" Viruses 13, no. 11: 2275. https://doi.org/10.3390/v13112275
APA StyleLiu, Y., Liu, M., Hu, R., Bai, J., He, X., & Jin, Y. (2021). Isolation of the Novel Phage PHB09 and Its Potential Use against the Plant Pathogen Pseudomonas syringae pv. actinidiae. Viruses, 13(11), 2275. https://doi.org/10.3390/v13112275







