Genetic Characterization of Novel H7Nx Low Pathogenic Avian Influenza Viruses from Wild Birds in South Korea during the Winter of 2020–2021
Abstract
:1. Introduction
2. Materials and Methods
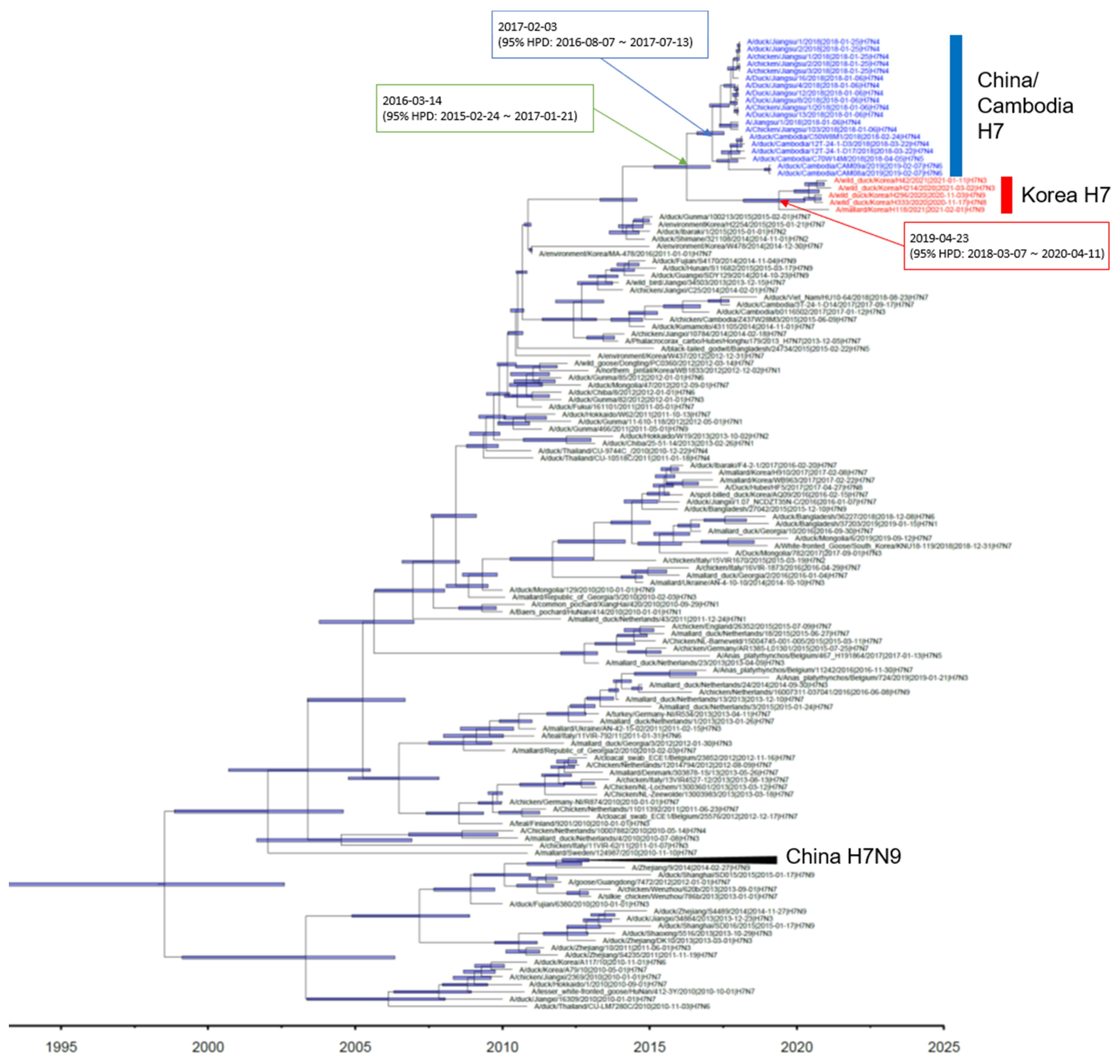
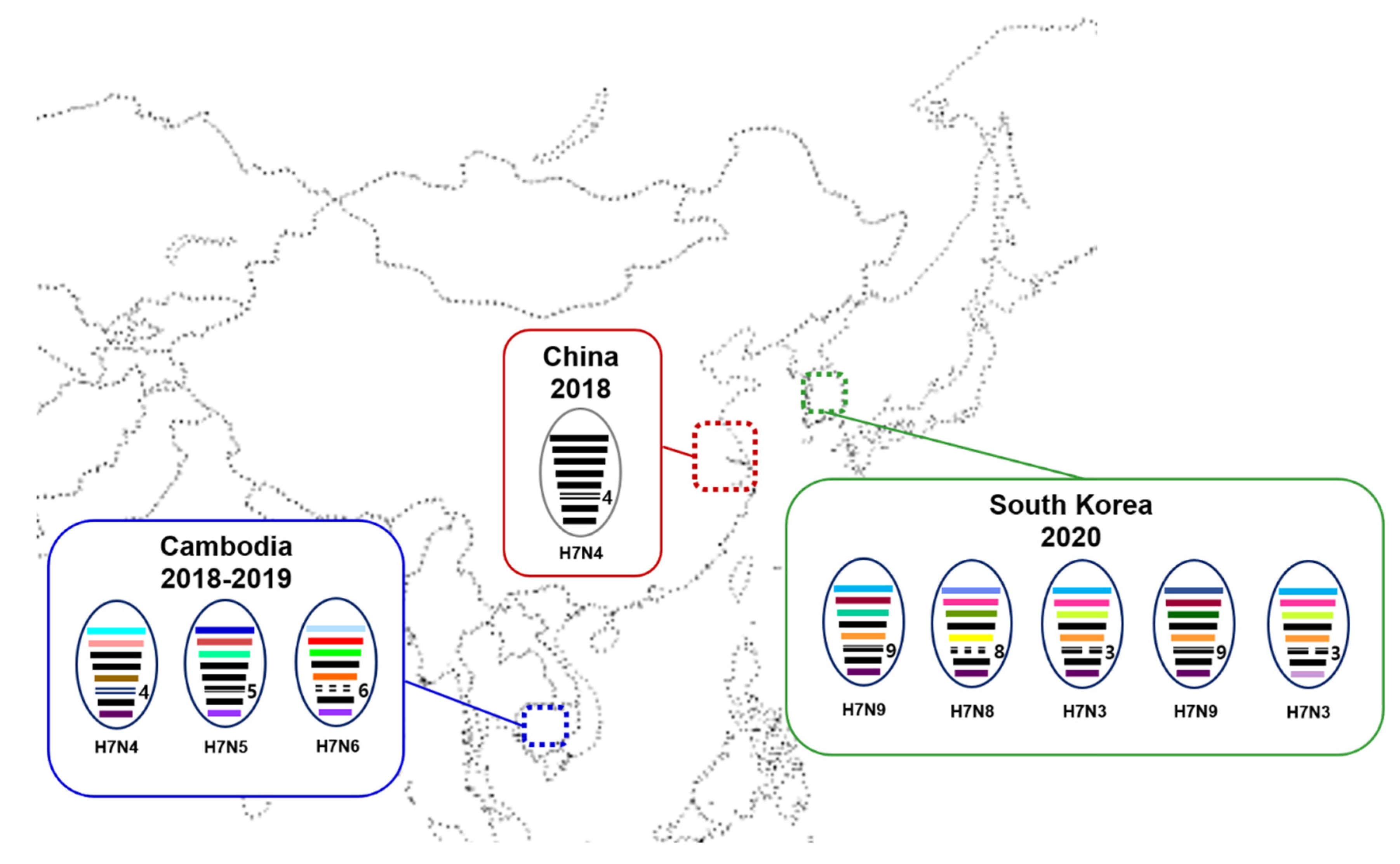
3. Results and Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abdelwhab, E.M.; Veits, J.; Mettenleiter, T.C. Prevalence and control of H7 avian influenza viruses in birds and humans. Epidemiol. Infect. 2014, 142, 896–920. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Cao, B.; Hu, Y.; Feng, Z.; Wang, D.; Hu, W.; Chen, J.; Jie, Z.; Qiu, H.; Xu, K.; et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 2013, 368, 1888–1897. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Zhu, W.; Li, X.; Chen, M.; Wu, J.; Yu, P.; Qi, S.; Huang, Y.; Shi, W.; Dong, J.; et al. Genesis and Spread of Newly Emerged Highly Pathogenic H7N9 Avian Viruses in Mainland China. J. Virol. 2017, 91, e01277-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Jiang, H.; Wu, P.; Uyeki, T.M.; Feng, L.; Lai, S.; Wang, L.; Huo, X.; Xu, K.; Chen, E.; et al. Epidemiology of avian influenza A H7N9 virus in human beings across five epidemics in mainland China, 2013–2017: An epidemiological study of laboratory-confirmed case series. Lancet Infect. Dis. 2017, 17, 822–832. [Google Scholar] [CrossRef]
- Shi, J.; Deng, G.; Ma, S.; Zeng, X.; Yin, X.; Li, M.; Zhang, B.; Cui, P.; Chen, Y.; Yang, H.; et al. Rapid Evolution of H7N9 Highly Pathogenic Viruses that Emerged in China in 2017. Cell Host Microbe 2018, 24, 558–568 e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bi, Y.; Li, J.; Li, S.; Fu, G.; Jin, T.; Zhang, C.; Yang, Y.; Ma, Z.; Tian, W.; Li, J.; et al. Dominant subtype switch in avian influenza viruses during 2016–2019 in China. Nat. Commun. 2020, 11, 5909. [Google Scholar] [CrossRef] [PubMed]
- Huo, X.; Cui, L.B.; Chen, C.; Wang, D.; Qi, X.; Zhou, M.H.; Guo, X.; Wang, F.; Liu, W.J.; Kong, W.; et al. Severe human infection with a novel avian-origin influenza A(H7N4) virus. Sci. Bull. (Beijing) 2018, 63, 1043–1050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, B.; Li, X.; Cardona, C.J.; Xing, Z. Reassortment and adaptive mutations of an emerging avian influenza virus H7N4 subtype in China. PLoS ONE 2020, 15, e0227597. [Google Scholar] [CrossRef] [Green Version]
- Vijaykrishna, D.; Deng, Y.M.; Grau, M.L.; Kay, M.; Suttie, A.; Horwood, P.F.; Kalpravidh, W.; Claes, F.; Osbjer, K.; Dussart, P.; et al. Emergence of Influenza A(H7N4) Virus, Cambodia. Emerg. Infect. Dis. 2019, 25, 1988–1991. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.N.; Lee, D.H.; Shin, J.I.; Si, Y.J.; Lee, J.H.; Baek, Y.G.; Hong, S.Y.; Bunnary, S.; Tum, S.; Park, M.; et al. Pathogenesis and genetic characteristics of a novel reassortant low pathogenic avian influenza A(H7N6) virus isolated in Cambodia in 2019. Transbound. Emerg. Dis. 2021, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.N.; Cheon, S.H.; Lee, E.K.; Heo, G.B.; Bae, Y.C.; Joh, S.J.; Lee, M.H.; Lee, Y.J. Pathogenesis and genetic characteristics of novel reassortant low-pathogenic avian influenza H7 viruses isolated from migratory birds in the Republic of Korea in the winter of 2016–2017. Emerg. Microbes Infect. 2018, 7, 182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, D.H.; Lee, H.J.; Lee, Y.J.; Kang, H.M.; Jeong, O.M.; Kim, M.C.; Kwon, J.S.; Kwon, J.H.; Kim, C.B.; Lee, J.B.; et al. DNA barcoding techniques for avian influenza virus surveillance in migratory bird habitats. J. Wildl. Dis. 2010, 46, 649–654. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H. Complete Genome Sequencing of Influenza A Viruses Using Next-Generation Sequencing. Methods Mol. Biol. 2020, 2123, 69–79. [Google Scholar]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [Green Version]
- Hasegawa, M.; Kishino, H.; Yano, T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef]
- Minin, V.N.; Bloomquist, E.W.; Suchard, M.A. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol. Biol. Evol. 2008, 25, 1459–1471. [Google Scholar] [CrossRef] [Green Version]
- Hill, S.C.; Lee, Y.J.; Song, B.M.; Kang, H.M.; Lee, E.K.; Hanna, A.; Gilbert, M.; Brown, I.H.; Pybus, O.G. Wild waterfowl migration and domestic duck density shape the epidemiology of highly pathogenic H5N8 influenza in the Republic of Korea. Infect. Genet. Evol. 2015, 34, 267–277. [Google Scholar] [CrossRef]
- Hebert, P.D.; Stoeckle, M.Y.; Zemlak, T.S.; Francis, C.M. Identification of Birds through DNA Barcodes. PLoS Biol. 2004, 2, e312. [Google Scholar] [CrossRef] [Green Version]
- Yoo, H.S.; Eah, J.Y.; Kim, J.S.; Kim, Y.J.; Min, M.S.; Paek, W.K.; Lee, H.; Kim, C.B. DNA barcoding Korean birds. Mol. Cells 2006, 22, 323–327. [Google Scholar]
- Dugan, V.G.; Chen, R.; Spiro, D.J.; Sengamalay, N.; Zaborsky, J.; Ghedin, E.; Nolting, J.; Swayne, D.E.; Runstadler, J.A.; Happ, G.M.; et al. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog. 2008, 4, e1000076. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.H.; Criado, M.F.; Swayne, D.E. Pathobiological Origins and Evolutionary History of Highly Pathogenic Avian Influenza Viruses. Cold Spring Harb. Perspect. Med. 2021, 11, a038679. [Google Scholar] [CrossRef] [Green Version]
- Neumann, G.; Kawaoka, Y. Transmission of influenza A viruses. Virology 2015, 479–480, 234–246. [Google Scholar] [CrossRef] [Green Version]
- Hatta, M.; Gao, P.; Halfmann, P.; Kawaoka, Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 2001, 293, 1840–1842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Virus Name | Subtype | Samples | Collection Date | Region | Latitude/Longitude | GISAID Isolate ID |
|---|---|---|---|---|---|---|
| A/wDK/Kr/H296/2020 | H7N9 | Feces | 2020-11-03 | JB | 35°46′4.9692″ N/126°38′22.6278″ E | EPI_ISL_3663323 |
| A/wDK/Kr/H333/2020 | H7N8 | Feces | 2020-11-17 | JB | 35°47′56″ N/126°46′16″ E | EPI_ISL_3663324 |
| A/wDK/Kr/H42/2021 | H7N3 | Feces | 2021-01-11 | JJ | 33°30′19.22″ N/126°53′31.18″ E | EPI_ISL_3663325 |
| A/mallard/Kr/H118/2021 | H7N9 | Feces | 2021-02-01 | CB | 36°39′9.5004″ N/127°22′40.2096″ E | EPI_ISL_3663326 |
| A/wDK/Kr/H214/2021 | H7N3 | Feces | 2021-03-02 | JJ | 33°30′22.27″ N/126°53′33.53″ E | EPI_ISL_3663327 |
| Gene | Virus with the Highest Nucleotide Sequence Identity [% Nucleotide Sequence Identity] | ||||
|---|---|---|---|---|---|
| H296/20 (H7N9) | H333/20 (H7N8) | H42/21 (H7N3) | H118/21 (H7N9) | H214/21 (H7N3) | |
| PB2 | A/Anas platyrhynchos/South Korea/JB29-91-95/2019 (H11N2) [99.61%] | A/barnacle goose/Netherlands/2/ 2014 (H3N6) [98.07%] | A/duck/Bangladesh/ 41797/2019 (H3N8) [98.90%] | A/duck/Kumamoto/ 431107/2014 (H1N4) [99.04%] | A/duck/Bangladesh/ 41797/2019 (H3N8) [98.90%] |
| PB1 | A/mallard/Anhui/3-683/ 2019 (H6N2) [99.47%] | A/Anas platyrhynchos/South Korea/JB31-96/2019 (H11N9) [98.94%] | A/mallard/Anhui/3-617/ 2019 (H6N1) [98.72%] | A/mallard/Anhui/3-683/ 2019 (H6N2) [98.94%] | A/mallard/Anhui/3-617/2019 (H6N1) [98.72%] |
| PA | A/mallard/South Korea/KNU2019-54/2019 (H5N3) [99.49%] | A/duck/Mongolia/543/ 2015 (H4N6) [99.63%] | A/duck/Guangdong/ H31/2020 (H3N8) [99.07%] | A/duck/Mongolia/398/ 2018 (H3N8) [98.79%] | A/duck/Guangdong/ H31/2020 (H3N8) [99.11%] |
| HA | A/Jiangsu/1/2018(H7N4) [97.74%] | A/Jiangsu/1/2018 (H7N4) [97.68%] | A/Jiangsu/1/2018(H7N4) [97.62%] | A/Jiangsu/1/2018(H7N4) [97.03%] | A/Jiangsu/1/2018 (H7N4) [97.50%] |
| NP | A/mallard/Anhui/2-549/ 2019 (H6N1) [99.33%] | A/mallard/South Korea/KNU2019-51/2019 (H5N3) [99.60%] | A/teal/Egypt/ MB-D-487OP/2016 (H7N3) [99.06%] | A/teal/Egypt/ MB-D-487OP/2016 (H7N3) [99.0%] | A/teal/Egypt/ MB-D-487OP/2016 (H7N3) [99.13%] |
| NA | A/mallard/Kagoshima/ KU-KGS6/2018 (H11N9) [98.73%] | A/mallard/China/T222(3)/2018 (H3N8) [98.73%] | A/mallard/South Korea/JB21-58/2019 (H5N3) [99.29%] | A/mallard/Kagoshima/ KU-KGS6/2018 (H11N9) [98.58%] | A/mallard/South Korea/JB21-58/2019 (H5N3) [99.36%] |
| M | A/duck/Mongolia/ MN18-1/2018 (H3N6) [100%] | A/duck/Mongolia/ MN18-1/2018 (H3N6) [99.59%] | A/duck/Mongolia/ MN18-1/2018 (H3N6) [99.69%] | A/duck/Mongolia/ MN18-1/2018 (H3N6) [99.69%] | A/duck/Mongolia/ MN18-1/2018 (H3N6) [99.39%] |
| NS | A/green-winged teal/South Korea/KNU2019-72/2019 (H3N8) [99.40%] | A/green-winged teal/South Korea/KNU2019-72/2019 (H3N8) [99.40%] | A/green-winged teal/South Korea/KNU2019-72/2019 (H3N8) [99. 28%] | A/green-winged teal/South Korea/KNU2019-72/2019 (H3N8) [98. 80%] | A/Environment/Jiangxi/12590/2019 (H10N3) [99.40%] |
| Virus † | Subtype | HA ‡ | PB2 | PB1 | PA | NA | M2 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cleavage Site | 226 | 228 | 627 | 701 | 368 | 409 | Stalk del. | 274 | 292 | 31 | ||
| A/wDK/Kr/H296/2020 | H7N9 | PELPKGR/GLF | Q | G | E | D | I | S | No | H | R | S |
| A/wDK/Kr/H333/2020 | H7N8 | PELPKGR/GLF | Q | G | E | D | I | S | No | H | R | S |
| A/wDK/Kr/H42/2021 | H7N3 | PELPKGR/GLF | Q | G | E | D | I | S | No | H | R | S |
| A/mallard/Kr/H118/2021 | H7N9 | PELPKGR/GLF | Q | G | E | D | I | S | No | H | R | S |
| A/wDK/Kr/H214/2021 | H7N3 | PELPKRR/GLF | Q | G | E | D | I | S | No | H | R | S |
| A/DK/Cambodia/CAM08a/2019 | H7N6 | PELPKGR/GLF | Q | G | E | D | I | S | No | H | R | S |
| A/DK/Cambodia/CAM09a/2019 | H7N6 | PELPKGR/GLF | Q | G | E | D | I | S | No | H | R | S |
| A/Jiangsu/1/2018 | H7N4 | PELPKGR/GLF | Q | G | K | D | I | S | No | H | R | S |
| A/CK/Jiangsu/1/2018 | H7N4 | PELPKGR/GLF | Q | G | E | D | I | S | No | H | R | S |
| A/DK/Jiangsu/4/2018 | H7N4 | PELPKGR/GLF | Q | G | E | D | I | S | No | H | R | S |
| A/DK/Cambodia/12T-24-1-D3/2018 | H7N4 | PELPKGR/GLF | Q | G | E | D | I | S | No | H | R | S |
| A/DK/Cambodia/C50W8M1/2018 | H7N4 | PELPKGR/GLF | Q | G | E | D | I | S | No | H | R | S |
| A/Anhui/1/2013 | H7N9 | PEIPKGR/GLF | L | G | E | D | V | N | Yes | H | R | N |
| A/Shanghai/1/2013 | H7N9 | PEIPKGR/GLF | Q | G | E | D | I | N | Yes | H | R/K | N |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, Y.-N.; Lee, D.-H.; Kwon, J.-H.; Shin, J.-I.; Hong, S.Y.; Cha, R.M.; Baek, Y.-G.; Lee, E.-K.; Sagong, M.; Heo, G.-B.; et al. Genetic Characterization of Novel H7Nx Low Pathogenic Avian Influenza Viruses from Wild Birds in South Korea during the Winter of 2020–2021. Viruses 2021, 13, 2274. https://doi.org/10.3390/v13112274
Lee Y-N, Lee D-H, Kwon J-H, Shin J-I, Hong SY, Cha RM, Baek Y-G, Lee E-K, Sagong M, Heo G-B, et al. Genetic Characterization of Novel H7Nx Low Pathogenic Avian Influenza Viruses from Wild Birds in South Korea during the Winter of 2020–2021. Viruses. 2021; 13(11):2274. https://doi.org/10.3390/v13112274
Chicago/Turabian StyleLee, Yu-Na, Dong-Hun Lee, Jung-Hoon Kwon, Jae-In Shin, Seo Yun Hong, Ra Mi Cha, Yoon-Gi Baek, Eun-Kyoung Lee, Mingeun Sagong, Gyeong-Beum Heo, and et al. 2021. "Genetic Characterization of Novel H7Nx Low Pathogenic Avian Influenza Viruses from Wild Birds in South Korea during the Winter of 2020–2021" Viruses 13, no. 11: 2274. https://doi.org/10.3390/v13112274
APA StyleLee, Y.-N., Lee, D.-H., Kwon, J.-H., Shin, J.-I., Hong, S. Y., Cha, R. M., Baek, Y.-G., Lee, E.-K., Sagong, M., Heo, G.-B., Lee, K.-N., & Lee, Y.-J. (2021). Genetic Characterization of Novel H7Nx Low Pathogenic Avian Influenza Viruses from Wild Birds in South Korea during the Winter of 2020–2021. Viruses, 13(11), 2274. https://doi.org/10.3390/v13112274






