Phage Cocktail Development against Aeromonas salmonicida subsp. salmonicida Strains Is Compromised by a Prophage
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Bacteriophages
2.3. Growth Curves of A. salmonicida subsp. salmonicida Strains in Presence of Phages
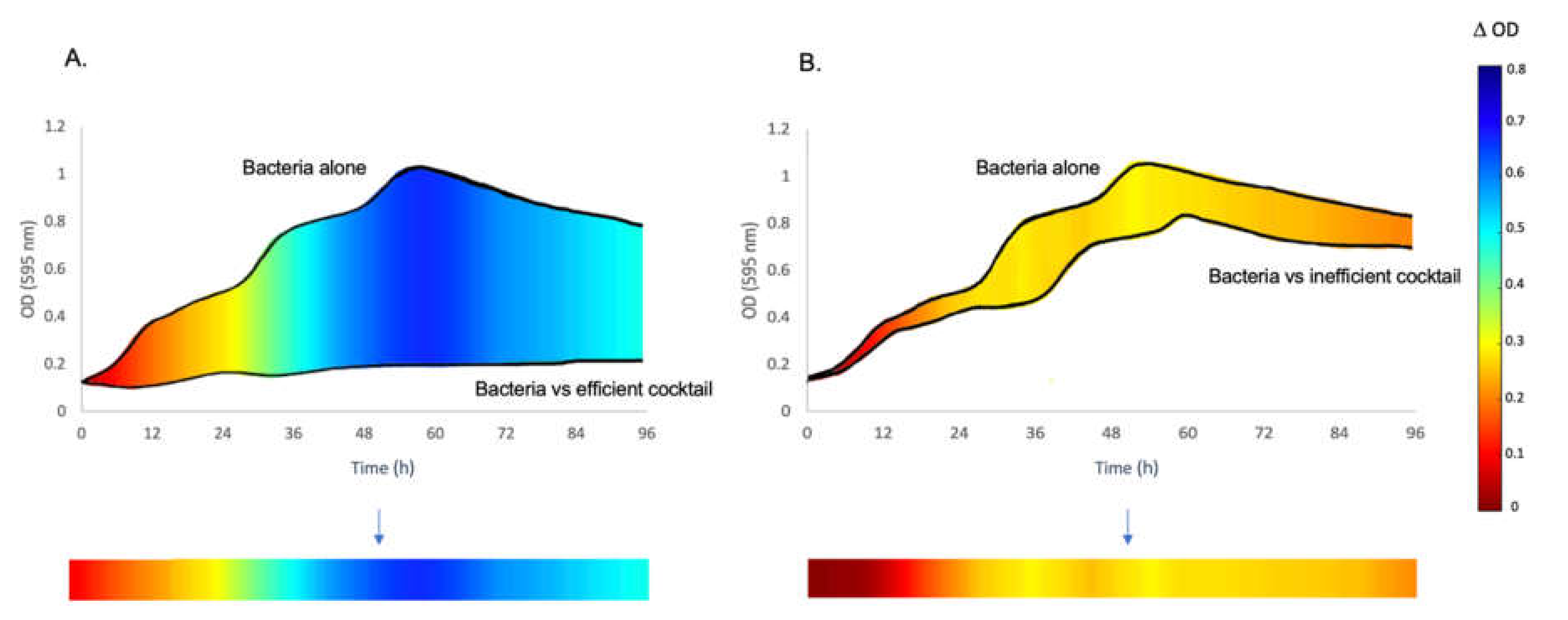
2.4. Data Analysis
3. Results and Discussion
3.1. The Inhibitory Effect of Phage Combinations on Bacterial Growth
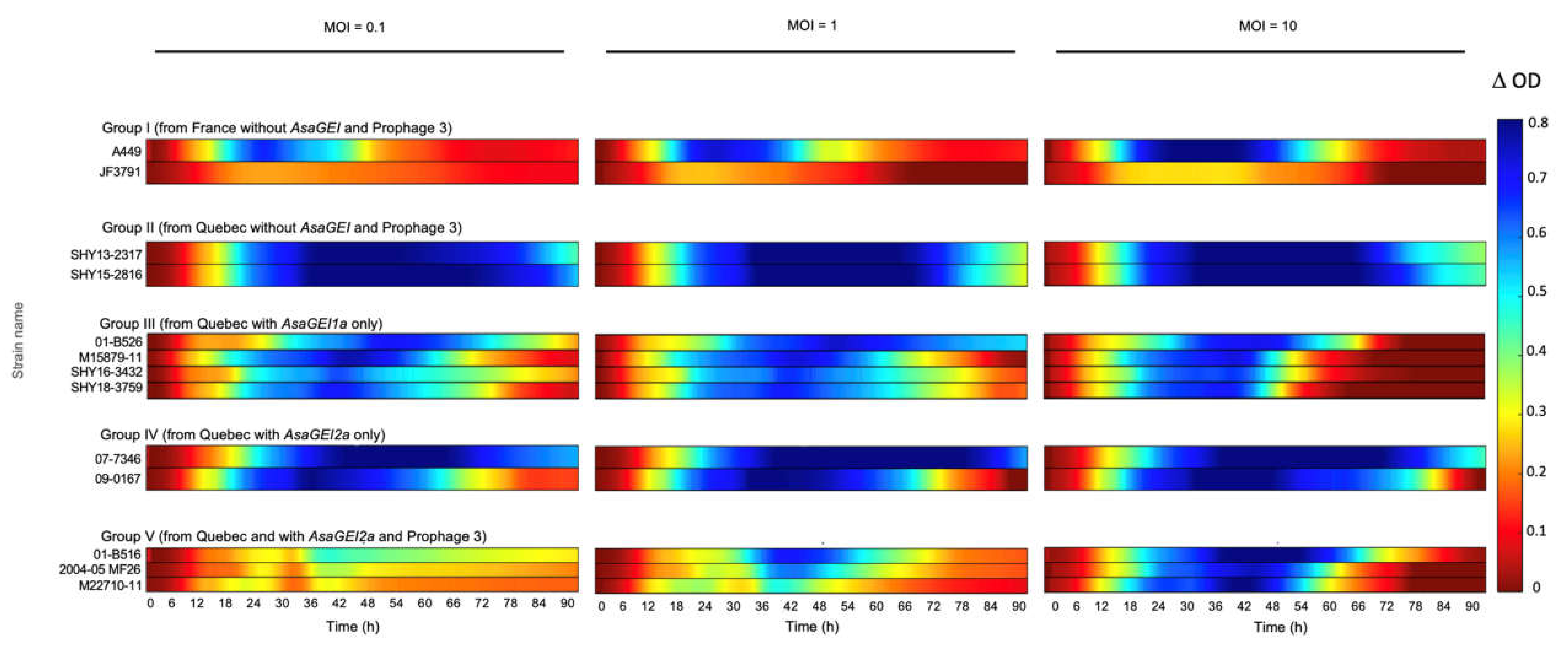
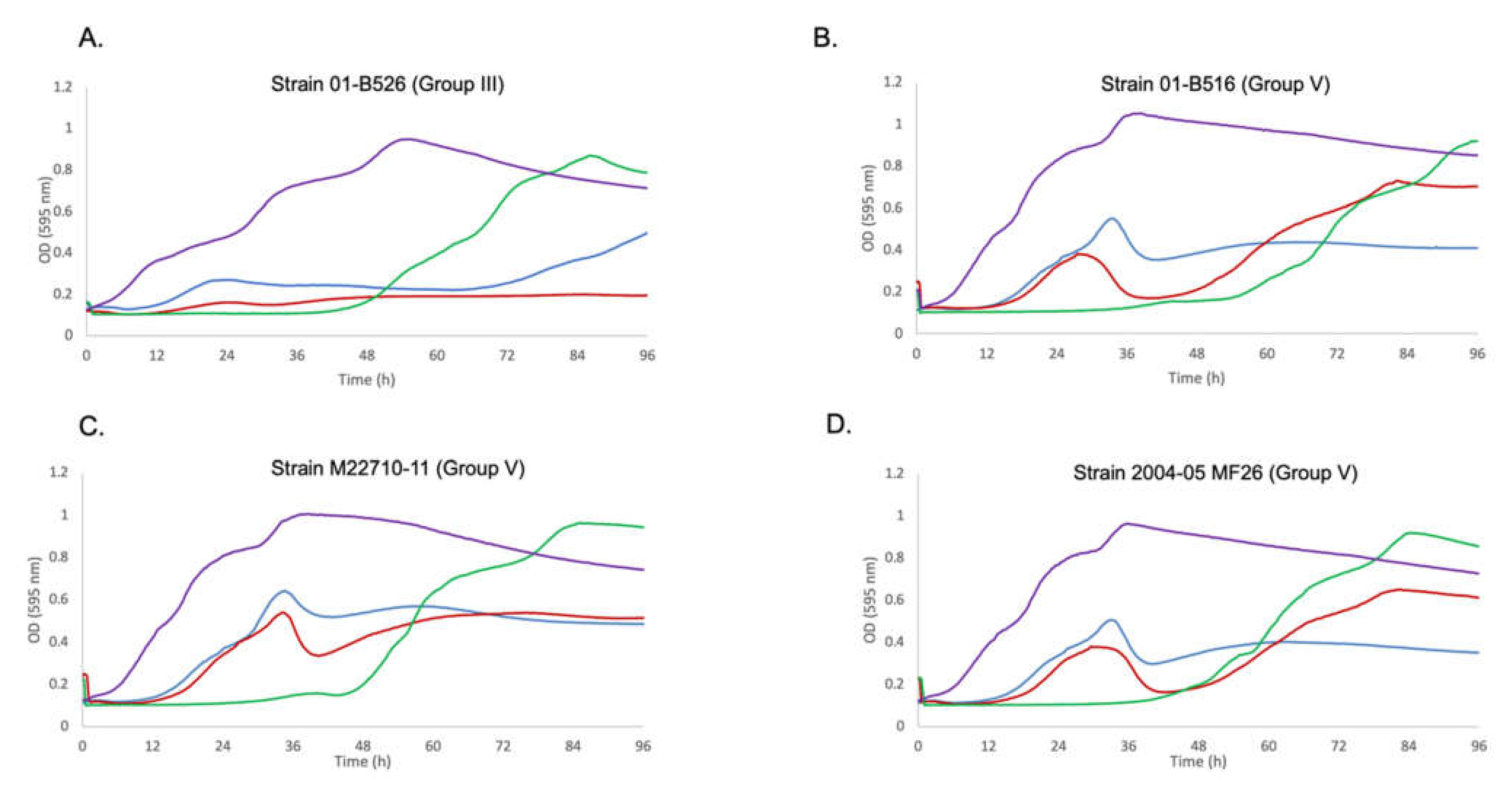
3.2. Effect of the Best Phage Cocktail on Various Strains with Different Genetic Backgrounds
3.3. Confirmation of the Role of Prophage 3 in Reducing the Effectiveness of Phage Growth Inhibition
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dallaire-Dufresne, S.; Tanaka, K.H.; Trudel, M.V.; Lafaille, A.; Charette, S.J. Virulence, genomic features, and plasticity of Aeromonas salmonicida subsp. salmonicida, the causative agent of fish furunculosis. Vet. Microbiol. 2014, 169, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Brenner, D.J.; Staley, J.T. Bergey’s Manual of Systematic Bacteriology. In The Proteobacteria. Part B, the Gammaproteobacteria, 2nd ed.; Springer: New York, NY, USA, 2005; Volume 2. [Google Scholar]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in agriculture and its consequential resistance in environmental sources: Potential public health implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef] [PubMed]
- Massicotte, M.-A.; Vincent, A.T.; Schneider, A.; Paquet, V.E.; Frenette, M.; Charette, S.J. One Aeromonas salmonicida subsp. salmonicida isolate with a pAsa5 variant bearing antibiotic resistance and a pRAS3 variant making a link with a swine pathogen. Sci. Total Environ. 2019, 690, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.T.; Trudel, M.V.; Paquet, V.E.; Boyle, B.; Tanaka, K.H.; Dallaire-Dufresne, S.; Daher, R.K.; Frenette, M.; Derome, N.; Charette, S.J. Detection of variants of the pRAS3, pAB5S9, and pSN254 plasmids in Aeromonas salmonicida subsp. salmonicida: Multidrug resistance, interspecies exchanges, and plasmid reshaping. Antimicrob. Agents Chemother. 2014, 58, 7367–7374. [Google Scholar] [CrossRef][Green Version]
- Vincent, A.T.; Hosseini, N.; Charette, S.J. The Aeromonas salmonicida plasmidome: A model of modular evolution and genetic diversity. Ann. N. Y. Acad. Sci. 2021, 1488, 16–32. [Google Scholar] [CrossRef]
- Nakai, T. Application of bacteriophages for control of infectious diseases in aquaculture. In Bacteriophages in the Control of Food- and Waterborne Pathogens; American Society for Microbiology: Washington, DC, USA, 2014; pp. 257–272. [Google Scholar]
- Feiner, R.; Argov, T.; Rabinovich, L.; Sigal, N.; Borovok, I.; Herskovits, A.A. A new perspective on lysogeny: Prophages as active regulatory switches of bacteria. Nat. Rev. Genet. 2015, 13, 641–650. [Google Scholar] [CrossRef]
- Emond-Rheault, J.-G.; Vincent, A.T.; Trudel, M.V.; Brochu, F.; Boyle, B.; Tanaka, K.H.; Attéré, S.A.; Jubinville, É.; Loch, T.P.; Winters, A.D.; et al. Variants of a genomic island in Aeromonas salmonicida subsp. salmonicida link isolates with their geographical origins. Vet. Microbiol. 2015, 175, 68–76. [Google Scholar] [CrossRef]
- Emond-Rheault, J.G.; Vincent, A.T.; Trudel, M.V.; Frey, J.; Frenette, M.; Charette, S.J. AsaGEI2b: A new variant of a genomic island identified in the Aeromonas salmonicida subsp. salmonicida JF3224 strain isolated from a wild fish in Switzerland. FEMS Microbiol. Lett. 2015, 362, fnv093. [Google Scholar] [CrossRef][Green Version]
- Long, M.; Nielsen, T.K.; Leisner, J.J.; Hansen, L.H.; Shen, Z.X.; Zhang, Q.Q.; Li, A. Aeromonas salmonicida subsp. salmonicida strains isolated from Chinese freshwater fish contain a novel genomic island and possible regional-specific mobile genetic elements profiles. FEMS Microbiol. Lett. 2016, 363, fnw190. [Google Scholar] [CrossRef]
- Reith, M.E.; Singh, R.K.; Curtis, B.A.; Boyd, J.M.; Bouevitch, A.; Kimball, J.; Munholland, J.; Murphy, C.; Sarty, D.; Williams, J.; et al. The genome of Aeromonas salmonicida subsp. salmonicida A449: Insights into the evolution of a fish pathogen. BMC Genom. 2008, 9, 427–15. [Google Scholar] [CrossRef]
- Vincent, A.T.; Intertaglia, L.; Loyer, V.; Paquet, E.V.; Adouane, É.; Martin, P.; Bérard, C.; Lami, R.; Charette, S.J. AsaGEI2d: A new variant of a genomic island identified in a group of Aeromonas salmonicida subsp. salmonicida isolated from France, which bears the pAsa7 plasmid. FEMS Microbiol. Lett. 2021, 368, fnab021. [Google Scholar] [CrossRef]
- Marcoux, P.É.; Vincent, A.T.; Massicotte, M.-A.; Paquet, V.E.; Doucet, É.J.; Hosseini, N.; Trudel, M.V.; Byatt, G.; Laurent, M.; Frenette, M.; et al. Systematic analysis of the stress-induced genomic instability of Type Three Secretion System in Aeromonas salmonicida subsp. salmonicida. Microorganisms 2020, 9, 85. [Google Scholar] [CrossRef]
- Chen, L.; Yuan, S.; Liu, Q.; Mai, G.; Yang, J.; Deng, D.; Zhang, B.; Liu, C.; Ma, Y. In vitro design and evaluation of phage cocktails against Aeromonas salmonicida. Front. Microbiol. 2018, 9, 1476. [Google Scholar] [CrossRef]
- Duarte, J.; Pereira, C.; Moreirinha, C.; Salvio, R.; Lopes, A.S.; Wang, D.; Almeida, A. New insights on phage efficacy to control Aeromonas salmonicida in aquaculture systems: An in vitro preliminary study. Aquaculture 2018, 495, 970–982. [Google Scholar] [CrossRef]
- Yang, Z.; Yuan, S.; Chen, L.; Liu, Q.; Zhang, H.; Ma, Y.; Wei, T.; Huang, S. Complete genome analysis of bacteriophage AsXd-1, a new member of the genus Hk97virus, family Siphoviridae. Arch. Virol. 2018, 163, 3195–3197. [Google Scholar] [CrossRef]
- Imbeault, S.; Parent, S.; Lagacé, M.; Uhland, C.F.; Blais, J.-F.O. Using bacteriophages to prevent furunculosis caused by Aeromonas salmonicida in farmed brook trout. J. Aquat. Anim. Health 2006, 18, 203–214. [Google Scholar] [CrossRef]
- Kim, J.H.; Choresca, C.H.; Shin, S.P.; Han, J.E.; Jun, J.W.; Park, S.C. Biological control of Aeromonas salmonicida subsp. salmonicida infection in rainbow trout (Oncorhynchus mykiss) using Aeromonas Phage PAS-1. Transbound. Emerg. Dis. 2015, 62, 81–86. [Google Scholar] [CrossRef]
- Nikapitiya, C.; Dananjaya, S.H.S.; Chandrarathna, H.P.S.U.; Senevirathne, A.; De Zoysa, M.; Lee, J. Isolation and characterization of multidrug resistance Aeromonas salmonicida subsp. salmonicida and its infecting novel phage ASP-1 from goldfish (Carassius auratus). Indian J. Microbiol. 2019, 59, 161–170. [Google Scholar] [CrossRef]
- Silva, Y.J.; Moreirinha, C.; Pereira, C.; Costa, L.; Rocha, R.; Cunha, A.; Gomes, N.; Calado, R.; Almeida, A. Biological control of Aeromonas salmonicida infection in juvenile Senegalese sole (Solea senegalensis) with Phage AS-A. Aquaculture 2016, 450, 225–233. [Google Scholar] [CrossRef]
- Vincent, A.T.; Paquet, V.E.; Bernatchez, A.; Tremblay, D.M.; Moineau, S.; Charette, S.J. Characterization and diversity of phages infecting Aeromonas salmonicida subsp. salmonicida. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Kendall, L.V.; Owiny, J.R.; Dohm, E.D.; Knapek, K.J.; Lee, E.S.; Kopanke, J.H.; Fink, M.; Hansen, S.A.; Ayers, J.D. Replacement, refinement, and Reduction in Animal studies with biohazardous agents. ILAR J. 2018, 59, 177–194. [Google Scholar] [CrossRef] [PubMed]
- Attéré, S.A.; Vincent, A.T.; Trudel, M.V.; Chanut, R.; Charette, S.J. Diversity and homogeneity among small plasmids of Aeromonas salmonicida subsp. salmonicida linked with geographical origin. Front. Microbiol. 2015, 6, 1274. [Google Scholar] [CrossRef] [PubMed]
- Trudel, M.V.; Vincent, A.T.; Attéré, S.A.; Labbé, M.; Derome, N.; Culley, A.I.; Charette, S.J. Diversity of antibiotic-resistance genes in Canadian isolates of Aeromonas salmonicida subsp. salmonicida: Dominance of pSN254b and discovery of pAsa8. Sci. Rep. 2016, 6, 35617. [Google Scholar] [CrossRef] [PubMed]
- Attéré, S.A.; Vincent, A.T.; Paccaud, M.; Frenette, M.; Charette, S.J. The role for the small cryptic plasmids as moldable vectors for genetic innovation in Aeromonas salmonicida subsp. salmonicida. Front. Genet. 2017, 8, 211. [Google Scholar] [CrossRef]
- Charette, S.J.; Brochu, F.; Boyle, B.; Filion, G.; Tanaka, K.H.; Derome, N. Draft genome sequence of the virulent strain 01-B526 of the fish pathogen Aeromonas salmonicida. J. Bacteriol. 2012, 194, 722–723. [Google Scholar] [CrossRef]
- Paquet, V.E.; Vincent, A.T.; Moineau, S.; Charette, S.J. Beyond the A-layer: Adsorption of lipopolysaccharides and characterization of bacteriophage-insensitive mutants of Aeromonas salmonicida subsp. salmonicida. Mol. Microbiol. 2019, 112, 667–677. [Google Scholar] [CrossRef]
- Storms, Z.J.; Teel, M.R.; Mercurio, K.; Sauvageau, D. The virulence index: A metric for quantitative analysis of phage virulence. PHAGE 2020, 1, 27–36. [Google Scholar] [CrossRef]
- Tanaka, K.H.; Vincent, A.T.; Emond-Rheault, J.-G.; Adamczuk, M.; Frenette, M.; Charette, S.J. Plasmid composition in Aeromonas salmonicida subsp. salmonicida 01-B526 unravels unsuspected type three secretion system loss patterns. BMC Genom. 2017, 18, 1–12. [Google Scholar] [CrossRef]
- Wolf, A.; Wiese, J.; Jost, G.; Witzel, K.-P. Wide geographic distribution of bacteriophages that lyse the same indigenous freshwater isolate (Sphingomonas sp. strain B18). Appl. Environ. Microbiol. 2003, 69, 2395–2398. [Google Scholar] [CrossRef]
- Ssekatawa, K.; Byarugaba, D.K.; Kato, C.D.; Wampande, E.M.; Ejobi, F.; Tweyongyere, R.; Nakavuma, J.L. A review of phage mediated antibacterial applications. Alex. J. Med. 2021, 57, 1–20. [Google Scholar] [CrossRef]
- Tanaka, K.H.; Vincent, A.T.; Trudel, M.V.; Paquet, V.E.; Frenette, M.; Charette, S.J. The mosaic architecture of Aeromonas salmonicida subsp. salmonicida pAsa4 plasmid and its consequences on antibiotic resistance. Peer J. 2016, 4, e2595. [Google Scholar] [CrossRef]
- Bondy-Denomy, J.; Qian, J.; Westra, E.R.; Buckling, A.; Guttman, D.S.; Davidson, A.R.; Maxwell, K.L. Prophages mediate defense against phage infection through diverse mechanisms. ISME J. 2016, 10, 2854–2866. [Google Scholar] [CrossRef]
- Castillo, D.; Andersen, N.; Kalatzis, P.G.; Middelboe, M. Large Phenotypic and genetic diversity of prophages induced from the fish pathogen Vibrio anguillarum. Viruses 2019, 11, 983. [Google Scholar] [CrossRef]
- Samson, J.E.; Magadan, A.H.; Sabri, M.; Moineau, S. Revenge of the phages: Defeating bacterial defences. Nat. Rev. Genet. 2013, 11, 675–687. [Google Scholar] [CrossRef]
- Ishiguro, E.; Kay, W.; Trust, T. Temperate bacteriophages for Aeromonas salmonicida. FEMS Microbiol. Lett. 1980, 8, 247–250. [Google Scholar] [CrossRef][Green Version]
- Ishiguro, E.E.; Ainsworth, T.; Harkness, R.E.; Kay, W.W.; Trust, T.J. A temperate bacteriophage specific for strains of Aeromonas salmonicida possessing A-layer, a cell surface virulence factor. Curr. Microbiol. 1984, 10, 199–202. [Google Scholar] [CrossRef]


| Strain | Characteristic | Origin | Reference a | Group b | AsaGEIc | Pro. 3 d | TTSS e | A-Layer | ARG f |
|---|---|---|---|---|---|---|---|---|---|
| A449 | Virulent | Fish infection, France | [12] | I | No | No | Yes | Yes | sul1, cat, tetA(E) |
| JF3791 | Virulent | Fish infection, Switzerland | [24] | I | No | No | No | Yes | No |
| SHY13-2317 | Virulent | Fish infection, Quebec, Canada | [25] | II | No | No | Yes | Yes | No |
| SHY15-2816 | Virulent | Fish infection, Quebec, Canada | [26] | II | No | No | Yes | Yes | No |
| 01-B526 | Virulent | Fish infection, Quebec, Canada | [27] | III | 1a | No | Yes | Yes | No |
| M15879-11 | Virulent | Fish infection, Quebec, Canada | [28] | III | 1a | No | Yes | Yes | sul1, sul2, floR, tetA |
| SHY16-3432 | Virulent | Fish infection, Quebec, Canada | [4] | III | 1a | No | Yes | Yes | sul1, sul2, floR, tetA |
| SHY18-3759 | Virulent | Fish infection, Quebec, Canada | [14] | III | 1a | No | Yes | Yes | tetA(C) |
| 07-7346 | Virulent | Fish infection, Quebec, Canada | [25] | IV | 2a | No | Yes | Yes | No |
| 09-0167 | Virulent | Fish infection, Quebec, Canada | [25] | IV | 2a | No | Yes | Yes | No |
| 01-B516 | Virulent | Fish infection, Quebec, Canada | [25] | V | 2a | Yes | Yes | Yes | No |
| 2004-05 MF26 | Virulent | Fish infection, New-Brunswick, Canada | [25] | V | 2a | Yes | Yes | Yes | sul1, sul2, floR, tetA |
| M22710-11 | Virulent | Fish infection, Quebec, Canada | [25] | V | 2a | Yes | Yes | Yes | No |
| HER1098 | Phage host | Brook trout, USA | FHRC | N/A | No | No | No | No | No |
| HER1110 | Phage host | Trout, Japan | FHRC | N/A | No | No | No | No | No |
| AS-R5 | Phage host | Rearranged strain from m15879-11 | [28] | N/A | 1a | No | No | No | sul1, sul2, floR, tetA |
| AS-19-R1 | Phage host | Rearranged strain from 08-2783 | This study and [14] | N/A | 1a | No | No | Yes | sul1, sul2, floR, tetA |
| Bacteriophage | Family (Genus) | Genome Size (bp) | GenBank | Origin | Host Strain | Reference a |
|---|---|---|---|---|---|---|
| HER98 (44RR2.8t.2) b | Myoviridae (Biquartavirus) | 173,590 | KY290948 | Water, Ontario, Canada | HER1098 | FHRC |
| HER110 (65.2) b | Myoviridae | 236,567 | KY290955 | River, Saône-et-Loire, France | HER1110 | FHRC |
| L9-6 | Myoviridae | 173,578 | KY290956 | Lake, Quebec, Canada | AS-19-R1 | [22] |
| Riv-10 | Myoviridae | 174,311 | KY290957 | River, Quebec, Canada | AS-R5 | [22] |
| SW69-9 | Myoviridae | 173,097 | KY290958 | Source, Quebec, Canada | AS-R5 | [22] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hosseini, N.; Paquet, V.E.; Chehreghani, M.; Moineau, S.; Charette, S.J. Phage Cocktail Development against Aeromonas salmonicida subsp. salmonicida Strains Is Compromised by a Prophage. Viruses 2021, 13, 2241. https://doi.org/10.3390/v13112241
Hosseini N, Paquet VE, Chehreghani M, Moineau S, Charette SJ. Phage Cocktail Development against Aeromonas salmonicida subsp. salmonicida Strains Is Compromised by a Prophage. Viruses. 2021; 13(11):2241. https://doi.org/10.3390/v13112241
Chicago/Turabian StyleHosseini, Nava, Valérie E. Paquet, Mahdi Chehreghani, Sylvain Moineau, and Steve J. Charette. 2021. "Phage Cocktail Development against Aeromonas salmonicida subsp. salmonicida Strains Is Compromised by a Prophage" Viruses 13, no. 11: 2241. https://doi.org/10.3390/v13112241
APA StyleHosseini, N., Paquet, V. E., Chehreghani, M., Moineau, S., & Charette, S. J. (2021). Phage Cocktail Development against Aeromonas salmonicida subsp. salmonicida Strains Is Compromised by a Prophage. Viruses, 13(11), 2241. https://doi.org/10.3390/v13112241







