Disassembling the Nature of Capsid: Biochemical, Genetic, and Imaging Approaches to Assess HIV-1 Capsid Functions
Abstract
1. Introduction
2. Properties of Capsid
2.1. Overview of HIV-1 Capsid Functions during Early Replication Events
2.2. Assessing Capsid Stability Using Biochemical, Genetic, and Imaging Approaches
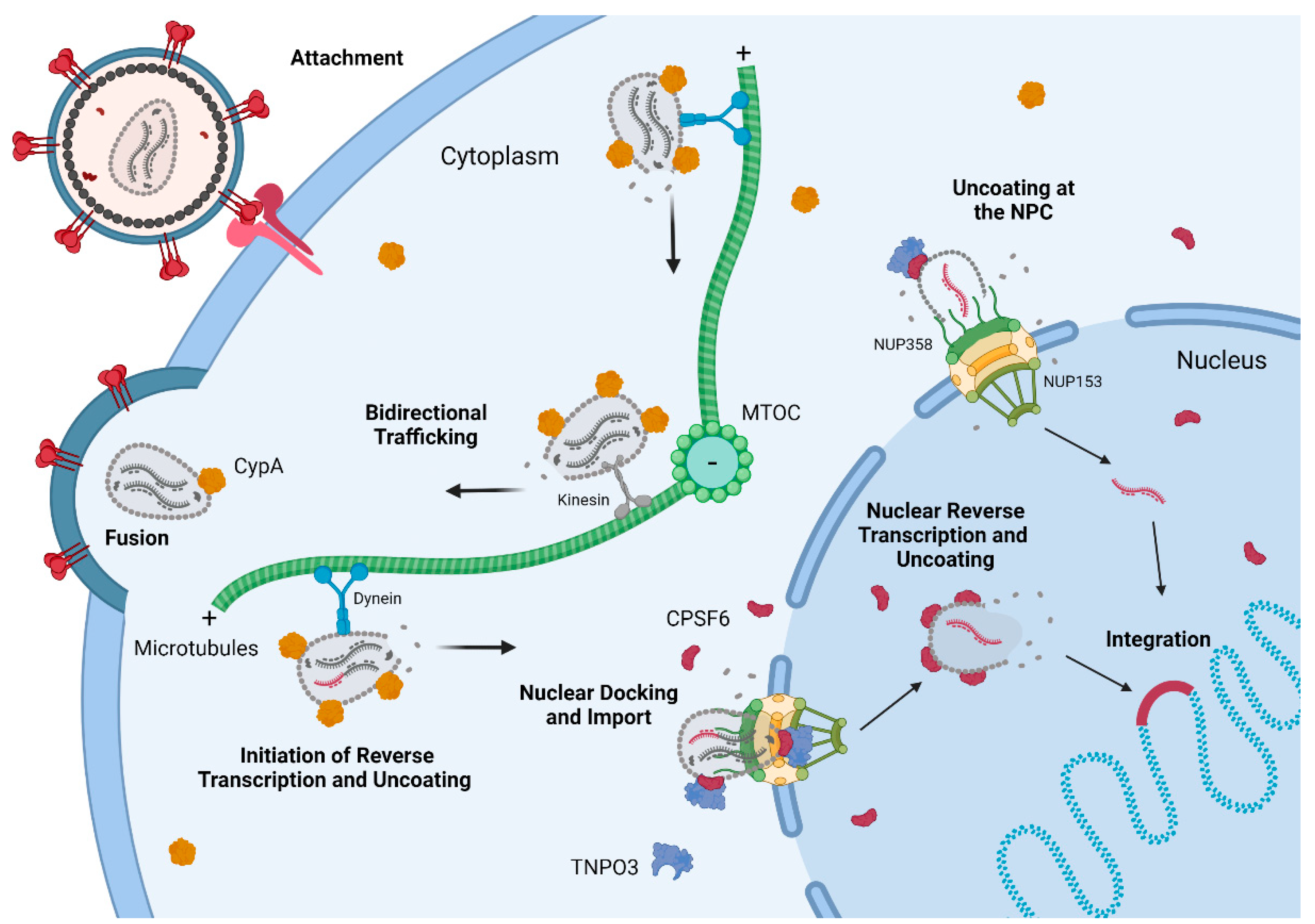
3. Capsid Trafficking to the Nucleus and Nuclear Import
3.1. Host Factors Involved in Capsid Trafficking
3.2. Host Proteins That Bind to Capsid in the Cytoplasm
3.3. Capsids at the Nuclear Pore
3.4. CPSF6, TNPO3, and Nuclear Import
4. HIV-1 Capsid Uncoating and Reverse Transcription Complete in the Nucleus
4.1. Limits of HIV-1 CA Detection
4.2. Capsids in the Nucleus
4.3. Reverse Transcription in the Nucleus
4.4. Biphasic Uncoating
5. Nuclear CPSF6, Capsid, and Integration
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- WHO. HIV/AIDS. Available online: https://www.who.int/news-room/fact-sheets/detail/hiv-aids (accessed on 1 July 2021).
- Pornillos, O.; Ganser-Pornillos, B.K. Maturation of retroviruses. Curr. Opin. Virol. 2019, 36, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Briggs, J.; Wilk, T.; Welker, R.; Kräusslich, H.; Fuller, S.D. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 2003, 22, 1707–1715. [Google Scholar] [CrossRef]
- Pornillos, O.; Ganser-Pornillos, B.K.; Yeager, M. Atomic-level modelling of the HIV capsid. Nat. Cell Biol. 2011, 469, 424–427. [Google Scholar] [CrossRef]
- Zhang, P.; Meng, X.; Zhao, G. Tubular Crystals and Helical Arrays: Structural Determination of HIV-1 Capsid Assemblies Using Iterative Helical Real-Space Reconstruction. Methods Mol. Biol. 2013, 955, 381–399. [Google Scholar] [CrossRef] [PubMed]
- Perilla, J.; Schulten, K. Physical properties of the HIV-1 capsid from all-atom molecular dynamics simulations. Nat. Commun. 2017, 8, 15959. [Google Scholar] [CrossRef]
- Yamashita, M.; Engelman, A.N. Capsid-Dependent Host Factors in HIV-1 Infection. Trends Microbiol. 2017, 25, 741–755. [Google Scholar] [CrossRef] [PubMed]
- Rossi, E.; Meuser, M.; Cunanan, C.; Cocklin, S. Structure, Function, and Interactions of the HIV-1 Capsid Protein. Life 2021, 11, 100. [Google Scholar] [CrossRef]
- Stremlau, M.; Owens, C.M.; Perron, M.J.; Kiessling, M.; Autissier, P.; Sodroski, J. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 2004, 427, 848–853. [Google Scholar] [CrossRef]
- Pertel, T.; Hausmann, S.; Morger, D.; Züger, S.; Guerra, J.; Lascano, J.; Reinhard, C.; Santoni, F.A.; Uchil, P.D.; Chatel, L.; et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 2011, 472, 361–365. [Google Scholar] [CrossRef]
- OhAinle, M.; Kim, K.; Keceli, S.K.; Felton, A.; Campbell, E.; Luban, J.; Emerman, M. TRIM34 restricts HIV-1 and SIV capsids in a TRIM5α-dependent manner. PLoS Pathog. 2020, 16, e1008507. [Google Scholar] [CrossRef]
- Nisole, S.; Lynch, C.; Stoye, J.P.; Yap, M.W. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc. Natl. Acad. Sci. 2004, 101, 13324–13328. [Google Scholar] [CrossRef]
- Goujon, C.; Moncorge, O.; Bauby, H.; Doyle, T.; Ward, C.C.; Schaller, T.; Hue, S.; Barclay, W.; Schulz, R.; Malim, M.H. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nat. Cell Biol. 2013, 502, 559–562. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.M.; Hope, T.J. HIV-1 capsid: The multifaceted key player in HIV-1 infection. Nat. Rev. Microbiol. 2015, 13, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Rasaiyaah, J.; Tan, C.P.; Fletcher, A.J.; Price, A.J.; Blondeau, C.; Hilditch, L.; Jacques, D.; Selwood, D.; James, L.C.; Noursadeghi, M.; et al. HIV-1 evades innate immune recognition through specific cofactor recruitment. Nat. Cell Biol. 2013, 503, 402–405. [Google Scholar] [CrossRef]
- Lahaye, X.; Satoh, T.; Gentili, M.; Cerboni, S.; Conrad, C.; Hurbain, I.; El Marjou, A.; Lacabaratz, C.; Lelièvre, J.-D.; Manel, N. The Capsids of HIV-1 and HIV-2 Determine Immune Detection of the Viral cDNA by the Innate Sensor cGAS in Dendritic Cells. Immunity 2013, 39, 1132–1142. [Google Scholar] [CrossRef]
- Eschbach, J.E.; Elliott, J.L.; Li, W.; Zadrozny, K.K.; Davis, K.; Mohammed, S.J.; Lawson, D.Q.; Pornillos, O.; Engelman, A.N.; Kutluay, S.B. Capsid Lattice Destabilization Leads to Premature Loss of the Viral Genome and Integrase Enzyme during HIV-1 Infection. J. Virol. 2020, 95. [Google Scholar] [CrossRef] [PubMed]
- Stremlau, M.; Perron, M.; Lee, M.; Li, Y.; Song, B.; Javanbakht, H.; Diaz-Griffero, F.; Anderson, D.J.; Sundquist, W.I.; Sodroski, J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5 restriction factor. Proc. Natl. Acad. Sci. USA 2006, 103, 5514–5519. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Langer, S.; Zhang, Z.; Herbert, K.M.; Yoh, S.; König, R.; Chanda, S.K. Sensor Sensibility—HIV-1 and the Innate Immune Response. Cells 2020, 9, 254. [Google Scholar] [CrossRef]
- Sumner, R.P.; Harrison, L.; Touizer, E.; Peacock, T.P.; Spencer, M.; Zuliani-Alvarez, L.; Towers, G.J. Disrupting HIV -1 capsid formation causes cGAS sensing of viral DNA. EMBO J. 2020, 39. [Google Scholar] [CrossRef]
- Gao, D.; Wu, J.; Wu, Y.-T.; Du, F.; Aroh, C.; Yan, N.; Sun, L.; Chen, Z.J. Cyclic GMP-AMP Synthase Is an Innate Immune Sensor of HIV and Other Retroviruses. Science 2013, 341, 903–906. [Google Scholar] [CrossRef]
- von Schwedler, U.K.; Stray, K.M.; Garrus, J.E.; Sundquist, W.I. Functional Surfaces of the Human Immunodeficiency Virus Type 1 Capsid Protein. J. Virol. 2003, 77, 5439–5450. [Google Scholar] [CrossRef] [PubMed]
- Rihn, S.J.; Wilson, S.J.; Loman, N.J.; Alim, M.; Bakker, S.E.; Bhella, D.; Gifford, R.J.; Rixon, F.J.; Bieniasz, P.D. Extreme Genetic Fragility of the HIV-1 Capsid. PLoS Pathog. 2013, 9, e1003461. [Google Scholar] [CrossRef]
- Lee, K.; Ambrose, Z.; Martin, T.; Oztop, I.; Mulky, A.; Julias, J.G.; Vandegraaff, N.; Baumann, J.G.; Wang, R.; Yuen, W.; et al. Flexible Use of Nuclear Import Pathways by HIV-1. Cell Host Microbe 2010, 7, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Fischer, D.K.; Saito, A.; Kline, C.; Cohen, R.; Watkins, S.; Yamashita, M.; Ambrose, Z. CA Mutation N57A Has Distinct Strain-Specific HIV-1 Capsid Uncoating and Infectivity Phenotypes. J. Virol. 2019, 93, e00214-19. [Google Scholar] [CrossRef] [PubMed]
- Forshey, B.M.; von Schwedler, U.; Sundquist, W.I.; Aiken, C. Formation of a Human Immunodeficiency Virus Type 1 Core of Optimal Stability Is Crucial for Viral Replication. J. Virol. 2002, 76, 5667–5677. [Google Scholar] [CrossRef]
- Ganser, B.K.; Li, S.; Klishko, V.Y.; Finch, J.T.; Sundquist, W.I. Assembly and Analysis of Conical Models for the HIV-1 Core. Science 1999, 283, 80–83. [Google Scholar] [CrossRef]
- Li, S.; Hill, C.P.; Sundquist, W.I.; Finch, J.T. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nat. Cell Biol. 2000, 407, 409–413. [Google Scholar] [CrossRef]
- Yamashita, M.; Perez, O.; Hope, T.J.; Emerman, M. Evidence for Direct Involvement of the Capsid Protein in HIV Infection of Nondividing Cells. PLoS Pathog. 2007, 3, e156-10. [Google Scholar] [CrossRef]
- Diaz-Griffero, F.; Vandegraaff, N.; Li, Y.; McGee-Estrada, K.; Stremlau, M.; Welikala, S.; Si, Z.; Engelman, A.; Sodroski, J. Requirements for capsid-binding and an effector function in TRIMCyp-mediated restriction of HIV-1. Virology 2006, 351, 404–419. [Google Scholar] [CrossRef]
- Rankovic, S.; Varadarajan, J.; Ramalho, R.; Aiken, C.; Rousso, I. Reverse Transcription Mechanically Initiates HIV-1 Capsid Disassembly. J. Virol. 2017, 91, e00289-17. [Google Scholar] [CrossRef]
- Hulme, A.E.; Perez, O.; Hope, T.J. Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc. Natl. Acad. Sci. 2011, 108, 9975–9980. [Google Scholar] [CrossRef]
- Márquez, C.L.; Lau, D.; Walsh, J.; Shah, V.; McGuinness, C.; Wong, A.; Aggarwal, A.; Parker, M.W.; Jacques, D.A.; Turville, S.; et al. Kinetics of HIV-1 capsid uncoating revealed by single-molecule analysis. eLife 2018, 7, e34772. [Google Scholar] [CrossRef]
- Xu, C.; Fischer, D.K.; Rankovic, S.; Li, W.; Dick, R.A.; Runge, B.; Zadorozhnyi, R.; Ahn, J.; Aiken, C.; Polenova, T.; et al. Permeability of the HIV-1 capsid to metabolites modulates viral DNA synthesis. PLoS Biol. 2020, 18, e3001015. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Franks, T.; Gibson, G.; Huber, K.; Rahm, N.; De Castillia, C.S.; Luban, J.; Aiken, C.; Watkins, S.; Sluis-Cremer, N.; et al. Evidence for biphasic uncoating during HIV-1 infection from a novel imaging assay. Retrovirology 2013, 10, 70. [Google Scholar] [CrossRef]
- Francis, A.C.; Marin, M.; Shi, J.; Aiken, C.; Melikyan, G.B. Time-Resolved Imaging of Single HIV-1 Uncoating In Vitro and in Living Cells. PLoS Pathog. 2016, 12, e1005709. [Google Scholar] [CrossRef]
- Mamede, J.I.; Cianci, G.C.; Anderson, M.R.; Hope, T.J. Early cytoplasmic uncoating is associated with infectivity of HIV-1. Proc. Natl. Acad. Sci. 2017, 114, E7169–E7178. [Google Scholar] [CrossRef]
- Burdick, R.C.; Li, C.; Munshi, M.; Rawson, J.M.O.; Nagashima, K.; Hu, W.-S.; Pathak, V.K. HIV-1 uncoats in the nucleus near sites of integration. Proc. Natl. Acad. Sci. 2020, 117, 5486–5493. [Google Scholar] [CrossRef]
- Hulme, A.E.; Kelley, Z.; Okocha, E.A.; Hope, T.J. Identification of Capsid Mutations That Alter the Rate of HIV-1 Uncoating in Infected Cells. J. Virol. 2014, 89, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Francis, A.C.; Melikyan, G.B. Single HIV-1 Imaging Reveals Progression of Infection through CA-Dependent Steps of Docking at the Nuclear Pore, Uncoating, and Nuclear Transport. Cell Host Microbe 2018, 23, 536–548.e6. [Google Scholar] [CrossRef] [PubMed]
- Pornillos, O.; Ganser-Pornillos, B.K.; Banumathi, S.; Hua, Y.; Yeager, M. Disulfide Bond Stabilization of the Hexameric Capsomer of Human Immunodeficiency Virus. J. Mol. Biol. 2010, 401, 985–995. [Google Scholar] [CrossRef]
- Huang, P.-T.; Summers, B.J.; Xu, C.; Perilla, J.R.; Malikov, V.; Naghavi, M.H.; Xiong, Y. FEZ1 Is Recruited to a Conserved Cofactor Site on Capsid to Promote HIV-1 Trafficking. Cell Rep. 2019, 28, 2373–2385.e7. [Google Scholar] [CrossRef]
- Dharan, A.; Opp, S.; Abdel-Rahim, O.; Keceli, S.K.; Imam, S.; Diaz-Griffero, F.; Campbell, E.M. Bicaudal D2 facilitates the cytoplasmic trafficking and nuclear import of HIV-1 genomes during infection. Proc. Natl. Acad. Sci. 2017, 114, E10707–E10716. [Google Scholar] [CrossRef]
- Delaney, M.K.; Malikov, V.; Chai, Q.; Zhao, G.; Naghavi, M.H. Distinct functions of diaphanous-related formins regulate HIV-1 uncoating and transport. Proc. Natl. Acad. Sci. 2017, 114, E6932–E6941. [Google Scholar] [CrossRef]
- Carnes, S.K.; Zhou, J.; Aiken, C. HIV-1 Engages a Dynein-Dynactin-BICD2 Complex for Infection and Transport to the Nucleus. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Lukic, Z.; Dharan, A.; Fricke, T.; Diaz-Griffero, F.; Campbell, E.M. HIV-1 Uncoating Is Facilitated by Dynein and Kinesin 1. J. Virol. 2014, 88, 13613–13625. [Google Scholar] [CrossRef]
- McDonald, D.; Vodicka, M.A.; Lucero, G.; Svitkina, T.M.; Borisy, G.G.; Emerman, M.; Hope, T.J. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 2002, 159, 441–452. [Google Scholar] [CrossRef]
- Pawlica, P.; Berthoux, L. Cytoplasmic Dynein Promotes HIV-1 Uncoating. Viruses 2014, 6, 4195–4211. [Google Scholar] [CrossRef]
- Malikov, V.; Naghavi, M.H. Localized Phosphorylation of a Kinesin-1 Adaptor by a Capsid-Associated Kinase Regulates HIV-1 Motility and Uncoating. Cell Rep. 2017, 20, 2792–2799. [Google Scholar] [CrossRef][Green Version]
- Malikov, V.; Da, D.S.A.V.; Jovasevic, V.; Bennett, G.; Vieira, D.A.D.S.A.; Schulte, B.; Diaz-Griffero, F.; Walsh, D.; Naghavi, M.H. HIV-1 capsids bind and exploit the kinesin-1 adaptor FEZ1 for inward movement to the nucleus. Nat. Commun. 2015, 6, 1–13. [Google Scholar] [CrossRef]
- Zakeri, B.; Fierer, J.O.; Celik, E.; Chittock, E.C.; Schwarz-Linek, U.; Moy, V.T.; Howarth, M. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. 2012, 109, E690–E697. [Google Scholar] [CrossRef]
- Summers, B.J.; Digianantonio, K.M.; Smaga, S.S.; Huang, P.-T.; Zhou, K.; Gerber, E.E.; Wang, W.; Xiong, Y. Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms. Cell Host Microbe 2019, 26, 203–216.e6. [Google Scholar] [CrossRef] [PubMed]
- Ferro, L.S.; Can, S.; Turner, M.; Elshenawy, M.M.; Yildiz, A. Kinesin and dynein use distinct mechanisms to bypass obstacles. eLife 2019, 8, 8. [Google Scholar] [CrossRef]
- McCaffrey, P.; Perrino, B.; Soderling, T.; Rao, A. NF-ATp, a T lymphocyte DNA-binding protein that is a target for calcineurin and immunosuppressive drugs. J. Biol. Chem. 1993, 268, 3747–3752. [Google Scholar] [CrossRef]
- Luban, J.; Bossolt, K.L.; Franke, E.K.; Kalpana, G.V.; Goff, S.P. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 1993, 73, 1067–1078. [Google Scholar] [CrossRef]
- Vajdos, F.F.; Yoo, S.; Houseweart, M.; Sundquist, W.I.; Hill, C.P. Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein. Protein Sci. 2008, 6, 2297–2307. [Google Scholar] [CrossRef]
- Yoo, S.; Myszka, D.G.; Yeh, C.-Y.; McMurray, M.; Hill, C.P.; I Sundquist, W. Molecular recognition in the HIV-1 capsid/cyclophilin A complex. J. Mol. Biol. 1997, 269, 780–795. [Google Scholar] [CrossRef]
- Braaten, D.; Ansari, H.; Luban, J. The hydrophobic pocket of cyclophilin is the binding site for the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 1997, 71, 2107–2113. [Google Scholar] [CrossRef]
- Gamble, T.R.; Vajdos, F.; Yoo, S.; Worthylake, D.K.; Houseweart, M.; I Sundquist, W.; Hill, C.P. Crystal Structure of Human Cyclophilin A Bound to the Amino-Terminal Domain of HIV-1 Capsid. Cell 1996, 87, 1285–1294. [Google Scholar] [CrossRef]
- Braaten, D.; Franke, E.K.; Luban, J. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J. Virol. 1996, 70, 3551–3560. [Google Scholar] [CrossRef]
- Sokolskaja, E.; Sayah, D.M.; Luban, J. Target Cell Cyclophilin A Modulates Human Immunodeficiency Virus Type 1 Infectivity. J. Virol. 2004, 78, 12800–12808. [Google Scholar] [CrossRef] [PubMed]
- Hatziioannou, T.; Perez-Caballero, D.; Cowan, S.; Bieniasz, P.D. Cyclophilin Interactions with Incoming Human Immunodeficiency Virus Type 1 Capsids with Opposing Effects on Infectivity in Human Cells. J. Virol. 2005, 79, 176–183. [Google Scholar] [CrossRef]
- Kim, K.; Dauphin, A.; Komurlu, S.; McCauley, S.M.; Yurkovetskiy, L.; Carbone, C.; Diehl, W.E.; De Castillia, C.S.; Campbell, E.M.; Luban, J. Cyclophilin A protects HIV-1 from restriction by human TRIM5α. Nat. Microbiol. 2019, 4, 2044–2051. [Google Scholar] [CrossRef]
- Selyutina, A.; Persaud, M.; Simons, L.M.; Bulnes-Ramos, A.; Buffone, C.; Martinez-Lopez, A.; Scoca, V.; Di Nunzio, F.; Hiatt, J.; Marson, A.; et al. Cyclophilin A Prevents HIV-1 Restriction in Lymphocytes by Blocking Human TRIM5α Binding to the Viral Core. Cell Rep. 2020, 30, 3766–3777. [Google Scholar] [CrossRef] [PubMed]
- Ganser-Pornillos, B.K.; Chandrasekaran, V.; Pornillos, O.; Sodroski, J.G.; Sundquist, W.I.; Yeager, M. Hexagonal assembly of a restricting TRIM5 protein. Proc. Natl. Acad. Sci. USA 2011, 108, 534–539. [Google Scholar] [CrossRef]
- Diaz-Griffero, F.; Qin, X.-R.; Hayashi, F.; Kigawa, T.; Finzi, A.; Sarnak, Z.; Lienlaf, M.; Yokoyama, S.; Sodroski, J. A B-Box 2 Surface Patch Important for TRIM5α Self-Association, Capsid Binding Avidity, and Retrovirus Restriction. J. Virol. 2009, 83, 10737–10751. [Google Scholar] [CrossRef]
- Zhong, Z.; Ning, J.; Boggs, E.A.; Jang, S.; Wallace, C.; Telmer, C.; Bruchez, M.; Ahn, J.; Engelman, A.N.; Zhang, P.; et al. Cytoplasmic CPSF6 Regulates HIV-1 Capsid Trafficking and Infection in a Cyclophilin A-Dependent Manner. mBio 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Neve, J.; Patel, R.; Wang, Z.; Louey, A.; Furger, A.M. Cleavage and polyadenylation: Ending the message expands gene regulation. RNA Biol. 2017, 14, 865–890. [Google Scholar] [CrossRef]
- Price, A.J.; Fletcher, A.J.; Schaller, T.; Elliott, T.; Lee, K.; KewalRamani, V.N.; Chin, J.W.; Towers, G.; James, L.C. CPSF6 Defines a Conserved Capsid Interface that Modulates HIV-1 Replication. PLoS Pathog. 2012, 8, e1002896. [Google Scholar] [CrossRef]
- Price, A.J.; Jacques, D.; McEwan, W.; Fletcher, A.J.; Essig, S.; Chin, J.W.; Halambage, U.; Aiken, C.; James, L.C. Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly. PLoS Pathog. 2014, 10, e1004459. [Google Scholar] [CrossRef]
- Ning, J.; Zhong, Z.; Fischer, D.K.; Harris, G.; Watkins, S.C.; Ambrose, Z.; Zhang, P. Truncated CPSF6 Forms Higher-Order Complexes That Bind and Disrupt HIV-1 Capsid. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Toccafondi, E.; Lener, D.; Negroni, M. HIV-1 Capsid Core: A Bullet to the Heart of the Target Cell. Front. Microbiol. 2021, 12, 652486. [Google Scholar] [CrossRef]
- Kalderon, D.; Richardson, W.D.; Markham, A.F.; Smith, A.E. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nat. Cell Biol. 1984, 311, 33–38. [Google Scholar] [CrossRef]
- Lanford, R.E.; Butel, J. Construction and characterization of an SV40 mutant defective in nuclear transport of T antigen. Cell 1984, 37, 801–813. [Google Scholar] [CrossRef]
- Yamashita, M.; Emerman, M. Capsid Is a Dominant Determinant of Retrovirus Infectivity in Nondividing Cells. J. Virol. 2004, 78, 5670–5678. [Google Scholar] [CrossRef]
- Di Nunzio, F.; Danckaert, A.; Fricke, T.; Perez, P.; Fernandez, J.; Perret, E.; Roux, P.; Shorte, S.; Charneau, P.; Diaz-Griffero, F.; et al. Human Nucleoporins Promote HIV-1 Docking at the Nuclear Pore, Nuclear Import and Integration. PLoS ONE 2012, 7, e46037. [Google Scholar] [CrossRef] [PubMed]
- Burdick, R.C.; Delviks-Frankenberry, K.A.; Chen, J.; Janaka, S.K.; Sastri, J.; Hu, W.-S.; Pathak, V.K. Dynamics and regulation of nuclear import and nuclear movements of HIV-1 complexes. PLoS Pathog. 2017, 13, e1006570. [Google Scholar] [CrossRef]
- Francis, A.C.; Melikyan, G.B. Live-Cell Imaging of Early Steps of Single HIV-1 Infection. Viruses 2018, 10, 275. [Google Scholar] [CrossRef] [PubMed]
- Bichel, K.; Price, A.J.; Schaller, T.; Towers, G.J.; Freund, S.M.; James, L.C. HIV-1 capsid undergoes coupled binding and isomerization by the nuclear pore protein NUP358. Retrovirology 2013, 10, 81. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Mehla, R.; Chauhan, A. Perturbation of Host Nuclear Membrane Component RanBP2 Impairs the Nuclear Import of Human Immunodeficiency Virus -1 Preintegration Complex (DNA). PLoS ONE 2010, 5, e15620. [Google Scholar] [CrossRef]
- Di Nunzio, F.; Fricke, T.; Miccio, A.; Valle-Casuso, J.C.; Perez, P.; Souque, P.; Rizzi, E.; Severgnini, M.; Mavilio, F.; Charneau, P.; et al. Nup153 and Nup98 bind the HIV-1 core and contribute to the early steps of HIV-1 replication. Virology 2013, 440, 8–18. [Google Scholar] [CrossRef]
- Matreyek, K.; Engelman, A. The Requirement for Nucleoporin NUP153 during Human Immunodeficiency Virus Type 1 Infection Is Determined by the Viral Capsid. J. Virol. 2011, 85, 7818–7827. [Google Scholar] [CrossRef]
- Matreyek, K.; Yücel, S.S.; Li, X.; Engelman, A. Nucleoporin NUP153 Phenylalanine-Glycine Motifs Engage a Common Binding Pocket within the HIV-1 Capsid Protein to Mediate Lentiviral Infectivity. PLoS Pathog. 2013, 9, e1003693. [Google Scholar] [CrossRef]
- Dharan, A.; Talley, S.; Tripathi, A.; Mamede, J.I.; Majetschak, M.; Hope, T.J.; Campbell, E.M. KIF5B and Nup358 Cooperatively Mediate the Nuclear Import of HIV-1 during Infection. PLoS Pathog. 2016, 12, e1005700. [Google Scholar] [CrossRef]
- Lin, D.H.; Zimmermann, S.; Stuwe, T.; Stuwe, E.; Hoelz, A. Structural and Functional Analysis of the C-Terminal Domain of Nup358/RanBP2. J. Mol. Biol. 2013, 425, 1318–1329. [Google Scholar] [CrossRef]
- Schaller, T.; Ocwieja, K.; Rasaiyaah, J.; Price, A.J.; Brady, T.L.; Roth, S.L.; Hué, S.; Fletcher, A.J.; Lee, K.; KewalRamani, V.N.; et al. HIV-1 Capsid-Cyclophilin Interactions Determine Nuclear Import Pathway, Integration Targeting and Replication Efficiency. PLoS Pathog. 2011, 7, e1002439. [Google Scholar] [CrossRef] [PubMed]
- Meehan, A.M.; Saenz, D.T.; Guevara, R.; Morrison, J.H.; Peretz, M.; Fadel, H.J.; Hamada, M.; Van Deursen, J.; Poeschla, E.M. A Cyclophilin Homology Domain-Independent Role for Nup358 in HIV-1 Infection. PLoS Pathog. 2014, 10, e1003969. [Google Scholar] [CrossRef] [PubMed]
- Christ, F.; Thys, W.; De Rijck, J.; Gijsbers, R.; Albanese, A.; Arosio, D.; Emiliani, S.; Rain, J.-C.; Benarous, R.; Cereseto, A.; et al. Transportin-SR2 Imports HIV into the Nucleus. Curr. Biol. 2008, 18, 1192–1202. [Google Scholar] [CrossRef]
- Brass, A.L.; Dykxhoorn, D.M.; Benita, Y.; Yan, N.; Engelman, A.; Xavier, R.J.; Lieberman, J.; Elledge, S.J. Identification of Host Proteins Required for HIV Infection Through a Functional Genomic Screen. Science 2008, 319, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Maertens, G.; Cook, N.J.; Wang, W.; Hare, S.; Gupta, S.S.; Oztop, I.; Lee, K.; E Pye, V.; Cosnefroy, O.; Snijders, B.; et al. Structural basis for nuclear import of splicing factors by human Transportin 3. Proc. Natl. Acad. Sci. 2014, 111, 2728–2733. [Google Scholar] [CrossRef] [PubMed]
- De Iaco, A.; Santoni, F.; Vannier, A.; Guipponi, M.; Antonarakis, S.; Luban, J. TNPO3 protects HIV-1 replication from CPSF6-mediated capsid stabilization in the host cell cytoplasm. Retrovirology 2013, 10, 20. [Google Scholar] [CrossRef]
- Casuso, J.C.V.; Di Nunzio, F.; Yang, Y.; Reszka, N.; Lienlaf, M.; Arhel, N.; Perez, P.; Brass, A.L.; Diaz-Griffero, F. TNPO3 Is Required for HIV-1 Replication after Nuclear Import but prior to Integration and Binds the HIV-1 Core. J. Virol. 2012, 86, 5931–5936. [Google Scholar] [CrossRef]
- Fricke, T.; Valle-Casuso, J.C.; E White, T.; Brandariz-Nuñez, A.; Bosche, W.J.; Reszka, N.; Gorelick, R.; Diaz-Griffero, F. The ability of TNPO3-depleted cells to inhibit HIV-1 infection requires CPSF6. Retrovirology 2013, 10, 46. [Google Scholar] [CrossRef] [PubMed]
- Saito, A.; Henning, M.S.; Serrao, E.; Dubose, B.N.; Teng, S.; Huang, J.; Li, X.; Saito, N.; Roy, S.P.; Siddiqui, M.A.; et al. Capsid-CPSF6 Interaction Is Dispensable for HIV-1 Replication in Primary Cells but Is Selected during Virus Passage In Vivo. J. Virol. 2016, 90, 6918–6935. [Google Scholar] [CrossRef] [PubMed]
- Bejarano, D.A.; Peng, K.; Laketa, V.; Börner, K.; Jost, K.L.; Lucic, B.; Glass, B.; Lusic, M.; Müller, B.; Kräusslich, H.-G. HIV-1 nuclear import in macrophages is regulated by CPSF6-capsid interactions at the nuclear pore complex. eLife 2019, 8, e41800. [Google Scholar] [CrossRef]
- Kane, M.; Rebensburg, S.V.; A Takata, M.; Zang, T.M.; Yamashita, M.; Kvaratskhelia, M.; Bieniasz, P.D. Nuclear pore heterogeneity influences HIV-1 infection and the antiviral activity of MX2. eLife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.D.; Farnet, C.M.; Bushman, F.D. Human immunodeficiency virus type 1 preintegration complexes: Studies of organization and composition. J. Virol. 1997, 71, 5382–5390. [Google Scholar] [CrossRef]
- Fassati, A.; Goff, S.P. Characterization of Intracellular Reverse Transcription Complexes of Human Immunodeficiency Virus Type 1. J. Virol. 2001, 75, 3626–3635. [Google Scholar] [CrossRef]
- Sayah, D.M.; Sokolskaja, E.; Berthoux, L.; Luban, J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 2004, 430, 569–573. [Google Scholar] [CrossRef]
- Panté, N.; Kann, M. Nuclear Pore Complex Is Able to Transport Macromolecules with Diameters of ∼39 nm. Mol. Biol. Cell 2002, 13, 425–434. [Google Scholar] [CrossRef]
- Lowe, A.R.; Siegel, J.J.; Kalab, P.; Siu, M.; Weis, K.; Liphardt, J.T. Selectivity mechanism of the nuclear pore complex characterized by single cargo tracking. Nat. Cell Biol. 2010, 467, 600–603. [Google Scholar] [CrossRef]
- Zila, V.; Margiotta, E.; Turoňová, B.; Müller, T.G.; Zimmerli, C.E.; Mattei, S.; Allegretti, M.; Börner, K.; Rada, J.; Müller, B.; et al. Cone-shaped HIV-1 capsids are transported through intact nuclear pores. Cell 2021, 184, 1032–1046.e18. [Google Scholar] [CrossRef]
- Li, C.; Burdick, R.C.; Nagashima, K.; Hu, W.-S.; Pathak, V.K. HIV-1 cores retain their integrity until minutes before uncoating in the nucleus. Proc. Natl. Acad. Sci. 2021, 118. [Google Scholar] [CrossRef]
- Dharan, A.; Bachmann, N.; Talley, S.; Zwikelmaier, V.; Campbell, E.M. Nuclear pore blockade reveals that HIV-1 completes reverse transcription and uncoating in the nucleus. Nat. Microbiol. 2020, 5, 1–8. [Google Scholar] [CrossRef]
- Trono, D. Partial reverse transcripts in virions from human immunodeficiency and murine leukemia viruses. J. Virol. 1992, 66, 4893–4900. [Google Scholar] [CrossRef]
- Katz, R.A.; Skalka, A.M. THE RETROVIRAL ENZYMES. Annu. Rev. Biochem. 1994, 63, 133–173. [Google Scholar] [CrossRef]
- Ambrose, Z.; Lee, K.; Ndjomou, J.; Xu, H.; Oztop, I.; Matous, J.; Takemura, T.; Unutmaz, D.; Engelman, A.; Hughes, S.H.; et al. Human Immunodeficiency Virus Type 1 Capsid Mutation N74D Alters Cyclophilin A Dependence and Impairs Macrophage Infection. J. Virol. 2012, 86, 4708–4714. [Google Scholar] [CrossRef] [PubMed]
- Jacques, D.; McEwan, W.A.; Hilditch, L.; Price, A.J.; Towers, G.J.; James, L.C. HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis. Nat. Cell Biol. 2016, 536, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Mallery, D.L.; Márquez, C.L.; A McEwan, W.; Dickson, C.F.; A Jacques, D.; Anandapadamanaban, M.; Bichel, K.; Towers, G.; Saiardi, A.; Böcking, T.; et al. IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis. eLife 2018, 7, e35335. [Google Scholar] [CrossRef]
- Lavigne, M.; Roux, P.; Buc, H.; Schaeffer, F. DNA curvature controls termination of plus strand DNA synthesis at the centre of HIV-1 genome. J. Mol. Biol. 1997, 266, 507–524. [Google Scholar] [CrossRef]
- Cosnefroy, O.; Murray, P.J.; Bishop, K.N. HIV-1 capsid uncoating initiates after the first strand transfer of reverse transcription. Retrovirology 2016, 13, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Stultz, R.D.; Cenker, J.J.; McDonald, D. Imaging HIV-1 Genomic DNA from Entry through Productive Infection. J. Virol. 2017, 91, 91. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Muranyi, W.; Glass, B.; Laketa, V.; Yant, S.R.; Tsai, L.; Cihlar, T.; Müller, B.; Kräusslich, H.-G. Quantitative microscopy of functional HIV post-entry complexes reveals association of replication with the viral capsid. eLife 2014, 3, e04114. [Google Scholar] [CrossRef]
- Francis, A.; Marin, M.; Prellberg, M.; Palermino-Rowland, K.; Melikyan, G. HIV-1 Uncoating and Nuclear Import Precede the Completion of Reverse Transcription in Cell Lines and in Primary Macrophages. Viruses 2020, 12, 1234. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Rodriguez, G.; Gazi, A.; Monel, B.; Frabetti, S.; Scoca, V.; Mueller, F.; Schwartz, O.; Krijnse-Locker, J.; Charneau, P.; Di Nunzio, F. Remodeling of the Core Leads HIV-1 Preintegration Complex into the Nucleus of Human Lymphocytes. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Rankovic, S.; Deshpande, A.; Harel, S.; Aiken, C.; Rousso, I. HIV-1 Uncoating Occurs via a Series of Rapid Biomechanical Changes in the Core Related to Individual Stages of Reverse Transcription. J. Virol. 2021, 95. [Google Scholar] [CrossRef]
- Selyutina, A.; Diaz-Griffero, F. Biochemical detection of capsid in the nucleus during HIV-1 infection. STAR Protoc. 2021, 2, 100323. [Google Scholar] [CrossRef]
- Müller, T.G.; Zila, V.; Peters, K.; Schifferdecker, S.; Stanic, M.; Lucic, B.; Laketa, V.; Lusic, M.; Müller, B.; Kräusslich, H.-G. HIV-1 uncoating by release of viral cDNA from capsid-like structures in the nucleus of infected cells. eLife 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Engelman, A.; Mizuuchi, K.; Craigie, R. HIV-1 DNA integration: Mechanism of viral DNA cleavage and DNA strand transfer. Cell 1991, 67, 1211–1221. [Google Scholar] [CrossRef]
- Bushman, F.D.; Fujiwara, T.; Craigie, R. Retroviral DNA Integration Directed by HIV Integration Protein in Vitro. Science 1990, 249, 1555–1558. [Google Scholar] [CrossRef]
- Farnet, C.; A Haseltine, W. Circularization of human immunodeficiency virus type 1 DNA in vitro. J. Virol. 1991, 65, 6942–6952. [Google Scholar] [CrossRef]
- Li, L.; Olvera, J.M.; Yoder, K.E.; Mitchell, R.S.; Butler, S.L.; Lieber, M.; Martin, S.L.; Bushman, F.D. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 2001, 20, 3272–3281. [Google Scholar] [CrossRef] [PubMed]
- Schröder, A.R.; Shinn, P.; Chen, H.; Berry, C.; Ecker, J.; Bushman, F. HIV-1 Integration in the Human Genome Favors Active Genes and Local Hotspots. Cell 2002, 110, 521–529. [Google Scholar] [CrossRef]
- Wang, G.P.; Ciuffi, A.; Leipzig, J.; Berry, C.C.; Bushman, F.D. HIV integration site selection: Analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007, 17, 1186–1194. [Google Scholar] [CrossRef]
- Chin, C.; Perreira, J.M.; Savidis, G.; Portmann, J.M.; Aker, A.M.; Feeley, E.; Smith, M.; Brass, A.L. Direct Visualization of HIV-1 Replication Intermediates Shows that Capsid and CPSF6 Modulate HIV-1 Intra-nuclear Invasion and Integration. Cell Rep. 2015, 13, 1717–1731. [Google Scholar] [CrossRef]
- Achuthan, V.; Perreira, J.M.; Sowd, G.A.; Puray-Chavez, M.; McDougall, W.M.; Paulucci-Holthauzen, A.; Wu, X.; Fadel, H.J.; Poeschla, E.M.; Multani, A.S.; et al. Capsid-CPSF6 Interaction Licenses Nuclear HIV-1 Trafficking to Sites of Viral DNA Integration. Cell Host Microbe 2018, 24, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Sowd, G.A.; Serrao, E.; Wang, H.; Wang, W.; Fadel, H.J.; Poeschla, E.M.; Engelman, A.N. A critical role for alternative polyadenylation factor CPSF6 in targeting HIV-1 integration to transcriptionally active chromatin. Proc. Natl. Acad. Sci. 2016, 113, E1054–E1063. [Google Scholar] [CrossRef]
- Francis, A.C.; Marin, M.; Singh, P.K.; Achuthan, V.; Prellberg, M.J.; Palermino-Rowland, K.; Lan, S.; Tedbury, P.R.; Sarafianos, S.G.; Engelman, A.N.; et al. HIV-1 replication complexes accumulate in nuclear speckles and integrate into speckle-associated genomic domains. Nat. Commun. 2020, 11, 3505. [Google Scholar] [CrossRef]
| Category | Assay (Lab) | Principle | Pros | Cons |
|---|---|---|---|---|
| Biochemical | In vitro capsid stability assay (Aiken) [26] | Isolated virions are ultracentrifuged over a sucrose gradient to separate intact capsids from disassembled capsids; western blot of fractions provides a bulk readout of capsid populations | Direct measurement of intact capsids Not technically challenging | Does not determine infectivity Some intact capsids are lost during the sample prep and ultracentrifugation Population measurement that does not allow visualization of individual particles or their kinetics |
| Fate of the capsid (Sodroski/Diaz-Griffero) [30] | Western blot of pelletable vs. soluble CA isolated from infected cells | Theoretically not technically challenging Cellular assay that can be performed in any cells | Does not differentiate between intact capsids or aggregated CA Does not determine infectivity Population measurement that does not allow visualization of individual particles or their kinetics | |
| Atomic force microscopy of capsids (Rousso) [31] | Pressures of individual capsids are measured by atomic force microscopy | Analysis of individual capsids Quantifies structural changes over time | Specific equipment required Does not determine infectivity | |
| Infectivity | CsA washout assay (Hope) [32] | Cells that express TRIMCyp are infected in the presence of cyclosporine A (CsA), which is washed out at different time points prior to measurement of infectivity | Not technically challenging and no special equipment needed Measures infectivity | Requires cells expressing TRIMCyp Population measurement that does not allow visualization of individual particles May not accurately reflect uncoating of CsA dependent CA mutants Limited to cytoplasmic capsids |
| In vitro imaging | Single-molecular fluorescence imaging of Gag-iGFP (Böcking) [33] | HIV-1 containing Gag-iGFP (with or without CypA-dsRed) is imaged for retention of iGFP by total internal reflection fluorescence (TIRF) microscopy | Provides kinetics of many individual capsids Can evaluate contribution of cell factors and drugs on capsid integrity | Requires confocal microscopy Does not determine infectivity Quantifies loss of fluorescence |
| CA retention assay (Ambrose) [34] | Fixation and permeabilization of HIV-1 containing a fluorescent capsid marker, followed by CA staining and TIRF imaging for CA retention | Provides kinetics of many individual capsids Can evaluate contribution of cell factors and drugs on capsid integrity | Requires confocal microscopy Does not determine infectivity Quantifies loss of fluorescence |
| Category | Assay (Lab) | Principle | Pros | Cons |
|---|---|---|---|---|
| Fixed Cell Imaging | In Situ uncoating assay (Hope) [29] | Cells are infected with HIV-1, followed by fixation and staining of CA protein | Cellular assay that can be performed in any cells Visualization of actual CA/capsid | Staining may be variable depending upon antibody used Does not allow visualization of individual particle kinetics Quantifies loss of fluorescence |
| EU staining assay (Ambrose) [35] | Cells are infected with HIV-1 produced in the presence of 5-ethynyl uridine (EU) and a second marker, followed by fixation and staining of EU | Cellular assay that can be performed in any cells Measures gain of fluorescence signal | Does not allow visualization of individual particle kinetics | |
| Live Cell Imaging | CypA-DsRed live cell imaging (Melikian) [36] | Cells are infected with HIV-1 made in the presence of CypA-DsRed and a second marker, followed by imaging of loss of CypA-DsRed signal | Cellular assay that can be performed in any cells Provides kinetics of many individual capsids | Quantifies loss of fluorescence Does not reflect uncoating of CypA independent CA mutants Limits CA tracking to the cytoplasm |
| Gag-iGFP live cell imaging assay (Hope) [37] | Cells are infected with HIV-1 containing Gag-internal GFP (Gag-iGFP) and second marker, followed by imaging of loss of GFP signal | Cellular assay that can be performed in any cells Provides kinetics of many individual capsids | Quantifies loss of fluorescence | |
| GFP-CA live cell imaging assay (Pathak) [38] | HIV-1 is produced by phenotypic mixing of WT CA and GFP-CA and second marker, followed by imaging of loss of GFP-CA signal | Cellular assay that can be performed in any cells Provides kinetics of many individual capsids | Virus has decreased infectivity Not all CA is labelled with GFP Quantifies loss of fluorescence |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ingram, Z.; Fischer, D.K.; Ambrose, Z. Disassembling the Nature of Capsid: Biochemical, Genetic, and Imaging Approaches to Assess HIV-1 Capsid Functions. Viruses 2021, 13, 2237. https://doi.org/10.3390/v13112237
Ingram Z, Fischer DK, Ambrose Z. Disassembling the Nature of Capsid: Biochemical, Genetic, and Imaging Approaches to Assess HIV-1 Capsid Functions. Viruses. 2021; 13(11):2237. https://doi.org/10.3390/v13112237
Chicago/Turabian StyleIngram, Zachary, Douglas K. Fischer, and Zandrea Ambrose. 2021. "Disassembling the Nature of Capsid: Biochemical, Genetic, and Imaging Approaches to Assess HIV-1 Capsid Functions" Viruses 13, no. 11: 2237. https://doi.org/10.3390/v13112237
APA StyleIngram, Z., Fischer, D. K., & Ambrose, Z. (2021). Disassembling the Nature of Capsid: Biochemical, Genetic, and Imaging Approaches to Assess HIV-1 Capsid Functions. Viruses, 13(11), 2237. https://doi.org/10.3390/v13112237






