Stephan Oroszlan and the Proteolytic Processing of Retroviral Proteins: Following A Pro
Abstract
:1. Creating a Framework
2. Defining Protease Cleavage Sites
3. The Oroszlan Pro1 Amino Acid Regulates the Structure and Function of the Viral Capsid
4. Turning the P1’ Pro Cleavage Site Question Around
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Oroszlan, S.I.; Rizvi, S.; O’Connor, T.E.; Mora, P.T. Use of Synthetic Polyglucose for Density-Gradient Centrifugation of Viruses. Nat. Cell Biol. 1964, 202, 780–781. [Google Scholar] [CrossRef] [PubMed]
- Oroszlan, S.; Johns, L.W.; Rich, M.A. Ultracentrifugation of a murine leukemia virus in polymer density gradients. Virology 1965, 26, 638–645. [Google Scholar] [CrossRef]
- Gilden, R.V.; Lee, Y.K.; Oroszlan, S.; Walker, J.L.; Huebner, R.J. Reptilian C-type virus: Biophysical, biological, and immunological properties. Virology 1970, 41, 187–190. [Google Scholar] [CrossRef]
- Oroszlan, S.; Fisher, C.L.; Stanley, T.B.; Gilden, R.V. Proteins of the Murine C-Type RNA Tumour Viruses: Isolation of a Group-specific Antigen by Isoelectric Focusing. J. Gen. Virol. 1970, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Oroszlan, S.; Foreman, C.; Kelloff, G.; Gilden, R.V. The group-specific antigen and other structural proteins of hamster and mouse C-type viruses. Virology 1971, 43, 665–674. [Google Scholar] [CrossRef]
- Oroszlan, S.; Huebner, R.J.; Gilden, R.V. Species-Specific and Interspecific Antigenic Determinants Associated with the Structural Protein of Feline C-Type Virus. Proc. Natl. Acad. Sci. USA 1971, 68, 901–904. [Google Scholar] [CrossRef] [Green Version]
- Oroszlan, S.; Copeland, T.; Summers, M.R.; Gilden, R.V. Feline Leukemia and RD-114 Virus Group-Specific Proteins: Comparison of Amino Terminal Sequence. Science 1973, 181, 454–456. [Google Scholar] [CrossRef]
- Oroszlan, S.; Summers, M.; Gilden, R.V. Amino-terminal sequence of baboon type C virus p30. Virology 1975, 64, 581–583. [Google Scholar] [CrossRef]
- Oroszlan, S.; Copeland, T.; Smythers, G.; Summers, M.R.; Gilden, R.V. Comparative primary structure analysis of the p30 protein of woolly monkey and gibbon type C viruses. Virology 1977, 77, 413–417. [Google Scholar] [CrossRef]
- Oroszlan, S.; Copeland, T.D.; Henderson, L.E.; Stephenson, J.R.; Gilden, R.V. Amino-terminal sequence of bovine leukemia virus major internal protein: Homology with mammalian type C virus p30 structural proteins. Proc. Natl. Acad. Sci. USA 1979, 76, 2996–3000. [Google Scholar] [CrossRef] [Green Version]
- Copeland, T.D.; Grandgenett, D.P.; Oroszlan, S. Amino Acid Sequence Analysis of Reverse Transcriptase Subunits from Avian Myeloblastosis Virus. J. Virol. 1980, 36, 115–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Copeland, T.D.; Henderson, L.E.; Vanlaningham-Miller, E.S.; Stephenson, J.R.; Smythers, G.W.; Oroszlan, S. Amino- and carboxyl-terminal sequences of proteins coded by gag gene of endogenous baboon and cat type c viruses. Virology 1981, 109, 13–24. [Google Scholar] [CrossRef]
- Oroszlan, S.; Barbacid, M.; Copeland, T.D.; Aaronson, S.A.; Gilden, R.V. Chemical and Immunological characterization of the major structural protein (p28) of MMC-1, a rhesus monkey endogenous type C virus: Homology with the major structural protein of avian reticuloendotheliosis virus. J. Virol. 1981, 39, 845–854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oroszlan, S.; Copeland, T.D.; Gilden, R.V.; Todaro, G.J. Structural homology of the major internal proteins of endogenous type C viruses of two distantly related species of old world monkeys: Macaca arctoides and Colobus polykomos. Virology 1981, 115, 262–271. [Google Scholar] [CrossRef]
- Oroszlan, S.; Sarngadharan, M.G.; Copeland, T.D.; Kalyanaraman, V.S.; Gilden, R.V.; Gallo, R.C. Primary structure analysis of the major internal protein p24 of human type C T-cell leukemia virus. Proc. Natl. Acad. Sci. USA 1982, 79, 1291–1294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henderson, L.E.; Sowder, R.; Smythers, G.; Benveniste, R.E.; Oroszlan, S. Purification and N-terminal amino acid sequence comparisons of structural proteins from retrovirus-D/Washington and Mason-Pfizer monkey virus. J. Virol. 1985, 55, 778–787. [Google Scholar] [CrossRef] [Green Version]
- Henderson, L.E.; Sowder, R.C.; Smythers, G.W.; Oroszlan, S. Chemical and immunological characterizations of equine infectious anemia virus gag-encoded proteins. J. Virol. 1987, 61, 1116–1124. [Google Scholar] [CrossRef] [Green Version]
- Hizi, A.; Henderson, L.E.; Copeland, T.D.; Sowder, R.C.; Hixson, C.V.; Oroszlan, S. Characterization of mouse mammary tumor virus gag-pro gene products and the ribosomal frameshift site by protein sequencing. Proc. Natl. Acad. Sci. USA 1987, 84, 7041–7045. [Google Scholar] [CrossRef] [Green Version]
- Henderson, L.E.; Benveniste, R.E.; Sowder, R.; Copeland, T.D.; Schultz, A.M.; Oroszlan, S. Molecular characterization of gag proteins from simian immunodeficiency virus (SIVMne). J. Virol. 1988, 62, 2587–2595. [Google Scholar] [CrossRef] [Green Version]
- Henderson, L.E.; Copeland, T.D.; Sowder, R.C.; Schultz, A.M.; Oroszlan, S. Analysis of proteins and peptides purified from sucrose gradient-banded HTLV-III. UCLA Symp. Mol. Cell. Biol. 1988, 71, 135–147. [Google Scholar]
- Gilden, R.V.; Oroszlan, S.; Huebner, R.J. Coexistence of Intraspecies and Interspecies Specific Antigenic Determinants on the Major Structural Polypeptide of Mammalian C-type Viruses. Nat. New Biol. 1971, 231, 107–108. [Google Scholar] [CrossRef] [PubMed]
- Gilden, R.V.; Oroszlan, S. Group-Specific Antigens of RNA Tumor Viruses as Markers for Subinfectious Expression of the RNA Virus Genome. Proc. Natl. Acad. Sci. USA 1972, 69, 1021–1025. [Google Scholar] [CrossRef] [Green Version]
- Leis, J.; Baltimore, D.; Bishop, J.M.; Coffin, J.; Fleissner, E.; Goff, S.P.; Oroszlan, S.; Robinson, H.; Skalka, A.M.; Temin, H.M. Standardized and simplified nomenclature for proteins common to all retroviruses. J. Virol. 1988, 62, 1808–1809. [Google Scholar] [CrossRef] [Green Version]
- Coffin, J.; Haase, A.; Levy, J.A.; Montagnier, L.; Oroszlan, S.; Teich, N.; Temin, H.; Toyoshima, K.; Varmus, H.; Vogt, P.; et al. Human Immunodeficiency Viruses. Science 1986, 232, 697. [Google Scholar] [CrossRef]
- Coffin, J.; Haase, A.; Levy, J.A.; Montagnier, L.; Oroszlan, S.; Teich, N.; Temin, H.; Toyoshima, K.; Varmus, H.; Vogt, P.; et al. What to call the AIDS virus? Nat. Cell Biol. 1986, 321, 10. [Google Scholar] [CrossRef] [Green Version]
- Henderson, L.E.; Krutzsch, H.C.; Oroszlan, S. Myristyl amino-terminal acylation of murine retrovirus proteins: An unusual post-translational protein modification. Proc. Natl. Acad. Sci. USA 1983, 80, 339–343. [Google Scholar] [CrossRef] [Green Version]
- Schultz, A.M.; Oroszlan, S. In vivo modification of retroviral gag gene-encoded polyproteins by myristic acid. J. Virol. 1983, 46, 355–361. [Google Scholar] [CrossRef] [Green Version]
- Oroszlan, S.; Copeland, T.; Summers, M.; Gilden, R. Amino terminal sequences of mammalian Type C RNA tumor virus group-specific antigens. Biochem. Biophys. Res. Commun. 1972, 48, 1549–1555. [Google Scholar] [CrossRef]
- Oroszlan, S.; Copeland, T.; Summers; Smythers, G.; Gilden, R. Amino acid sequence homology of mammalian type C RNA virus major internal proteins. J. Biol. Chem. 1975, 250, 6232–6239. [Google Scholar] [CrossRef]
- Oroszlan, S.; Henderson, L.E.; Stephenson, J.R.; Copeland, T.D.; Long, C.W.; Ihle, J.N.; Gilden, R.V. Amino- and carboxyl-terminal amino acid sequences of proteins coded by gag gene of murine leukemia virus. Proc. Natl. Acad. Sci. USA 1978, 75, 1404–1408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henderson, L.; Copeland, T.; Sowder, R.; Smythers, G.; Oroszlan, S. Primary structure of the low molecular weight nucleic acid-binding proteins of murine leukemia viruses. J. Biol. Chem. 1981, 256, 8400–8406. [Google Scholar] [CrossRef]
- Versteegen, R.J.; Copeland, T.D.; Oroszlan, S. Complete amino acid sequence of the group-specific antigen gene-encoded phosphorylated proteins of mouse leukemia viruses. J. Biol. Chem. 1982, 257, 3007–3013. [Google Scholar] [CrossRef]
- Copeland, T.D.; Oroszlan, S.; Kalyanaraman, V.; Sarngadharan, M.; Gallo, R.C. Complete amino acid sequence of human T-cell leukemia virus structural protein p15. FEBS Lett. 1983, 162, 390–395. [Google Scholar] [CrossRef] [Green Version]
- Tsai, W.-P.; Copeland, T.D.; Oroszlan, S. Purification and chemical and immunological characterization of avian reticuloendotheliosis virus gag-gene-encoded structural proteins. Virology 1985, 140, 289–312. [Google Scholar] [CrossRef]
- Hizi, A.; Henderson, L.E.; Copeland, T.D.; Sowder, R.C.; Krutzsch, H.C.; Oroszlan, S. Analysis of gag proteins from mouse mammary tumor virus. J. Virol. 1989, 63, 2543–2549. [Google Scholar] [CrossRef] [Green Version]
- Vogt, V.M.; Eisenman, R.; Diggelmann, H. Generation of avian myeloblastosis virus structural proteins by proteolytic cleavage of a precursor polypeptide. J. Mol. Biol. 1975, 96, 471–493. [Google Scholar] [CrossRef]
- Vogt, V.M.; Eisenman, R. Identification of a Large Polypeptide Precursor of Avian Oncornavirus Proteins. Proc. Natl. Acad. Sci. USA 1973, 70, 1734–1738. [Google Scholar] [CrossRef] [Green Version]
- Yoshinaka, Y.; Luftig, R. Properties of a P70 proteolytic factor of murine leukemia viruses. Cell 1977, 12, 709–720. [Google Scholar] [CrossRef]
- Vogt, V.M.; Wight, A.; Eisenman, R. In vitro cleavage of avian retrovirus gag proteins by viral protease p15. Virology 1979, 98, 154–167. [Google Scholar] [CrossRef]
- Tozser, J.; Bláha, I.; Copeland, T.D.; Wondrak, E.M.; Oroszlan, S. Comparison of the HIV-1 and HIV-2 proteinases using oligopeptide substrates representing cleavage sites in Gag and Gag-Pol polyproteins. FEBS Lett. 1991, 281, 77–80. [Google Scholar] [CrossRef] [Green Version]
- Tőzsér, J.; Gustchina, A.; Weber, I.T.; Bláha, I.; Wondrak, E.M.; Oroszlan, S. Studies on the role of the S4 substrate binding site of HIV proteinases. FEBS Lett. 1991, 279, 356–360. [Google Scholar] [CrossRef] [Green Version]
- Poorman, R.A.; Tomasselli, A.G.; Heinrikson, R.L.; Kezdy, F.J. A cumulative specificity model for proteases from human im-munodeficiency virus types 1 and 2, inferred from statistical analysis of an extended substrate data base. J. Biol. Chem. 1991, 266, 14554–14561. [Google Scholar] [CrossRef]
- Pettit, S.C.; Simsic, J.; Loeb, D.D.; Everitt, L.; Hutchison, C.A., 3rd; Swanstrom, R. Analysis of retroviral protease cleavage sites reveals two types of cleavage sites and the structural requirements of the P1 amino acid. J. Biol. Chem. 1991, 266, 14539–14547. [Google Scholar] [CrossRef]
- Bayro, M.; Ganser-Pornillos, B.K.; Zadrozny, K.K.; Yeager, M.; Tycko, R. Helical Conformation in the CA-SP1 Junction of the Immature HIV-1 Lattice Determined from Solid-State NMR of Virus-like Particles. J. Am. Chem. Soc. 2016, 138, 12029–12032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, S.; Louis, J.M.; Tycko, R. Effects of an HIV-1 maturation inhibitor on the structure and dynamics of CA-SP1 junction helices in virus-like particles. Proc. Natl. Acad. Sci. USA 2020, 117, 10286–10293. [Google Scholar] [CrossRef] [PubMed]
- Keller, P.W.; Adamson, C.S.; Heymann, J.B.; Freed, E.O.; Steven, A.C. HIV-1 Maturation Inhibitor Bevirimat Stabilizes the Immature Gag Lattice. J. Virol. 2011, 85, 1420–1428. [Google Scholar] [CrossRef] [Green Version]
- Purdy, M.D.; Shi, D.; Chrustowicz, J.; Hattne, J.; Gonen, T.; Yeager, M. MicroED structures of HIV-1 Gag CTD-SP1 reveal binding interactions with the maturation inhibitor bevirimat. Proc. Natl. Acad. Sci. USA 2018, 115, 13258–13263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schur, F.K.M.; Obr, M.; Hagen, W.J.H.; Wan, W.; Jakobi, A.J.; Kirkpatrick, J.M.; Sachse, C.; Kräusslich, H.-G.; Briggs, J.A.G. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science 2016, 353, 506–508. [Google Scholar] [CrossRef]
- Wagner, J.M.; Zadrozny, K.K.; Chrustowicz, J.; Purdy, M.D.; Yeager, M.; Ganser-Pornillos, B.K.; Pornillos, O. Crystal structure of an HIV assembly and maturation switch. eLife 2016, 5, e17063. [Google Scholar] [CrossRef]
- Kelly, B.N.; Howard, B.R.; Wang, H.; Robinson, H.; Sundquist, W.I.; Hill, C.P. Implications for Viral Capsid Assembly from Crystal Structures of HIV-1 Gag1-278 and CAN(133-278). Biochemistry 2006, 45, 11257–11266. [Google Scholar] [CrossRef]
- Schur, F.K.M.; Hagen, W.; Rumlová, M.; Ruml, T.; Müller, B.; Kräusslich, H.-G.; Briggs, J. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 Å resolution. Nat. Cell Biol. 2015, 517, 505–508. [Google Scholar] [CrossRef]
- Tang, C.; Ndassa, Y.; Summers, M.F. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat. Genet. 2002, 9, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Gitti, R.K.; Lee, B.M.; Walker, J.; Summers, M.F.; Yoo, S.; Sundquist, W.I. Structure of the Amino-Terminal Core Domain of the HIV-1 Capsid Protein. Science 1996, 273, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Vajdos, F.F.; Yoo, S.; Houseweart, M.; Sundquist, W.I.; Hill, C.P. Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein. Protein Sci. 2008, 6, 2297–2307. [Google Scholar] [CrossRef] [Green Version]
- Hedstrom, L.; Lin, T.-Y.; Fast, W. Hydrophobic Interactions Control Zymogen Activation in the Trypsin Family of Serine Proteases. Biochemistry 1996, 35, 4515–4523. [Google Scholar] [CrossRef] [PubMed]
- Fitzon, T.; Leschonsky, B.; Bieler, K.; Paulus, C.; Schröder, J.; Wolf, H.; Wagner, R. Proline Residues in the HIV-1 NH2-Terminal Capsid Domain: Structure Determinants for Proper Core Assembly and Subsequent Steps of Early Replication. Virology 2000, 268, 294–307. [Google Scholar] [CrossRef] [Green Version]
- Von Schwedler, U.K.; Stemmler, T.; Klishko, V.Y.; Li, S.; Albertine, K.H.; Davis, D.R.; Sundquist, W.I. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 1998, 17, 1555–1568. [Google Scholar] [CrossRef] [Green Version]
- Tang, S.; Murakami, T.; Agresta, B.E.; Campbell, S.; Freed, E.O.; Levin, J.G. Human Immunodeficiency Virus Type 1 N-Terminal Capsid Mutants That Exhibit Aberrant Core Morphology and Are Blocked in Initiation of Reverse Transcription in Infected Cells. J. Virol. 2001, 75, 9357–9366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, S.; Murakami, T.; Cheng, N.; Steven, A.C.; Freed, E.O.; Levin, J.G. Human Immunodeficiency Virus Type 1 N-Terminal Capsid Mutants Containing Cores with Abnormally High Levels of Capsid Protein and Virtually No Reverse Transcriptase. J. Virol. 2003, 77, 12592–12602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganser, B.K.; Li, S.; Klishko, V.Y.; Finch, J.T.; Sundquist, W.I. Assembly and Analysis of Conical Models for the HIV-1 Core. Science 1999, 283, 80–83. [Google Scholar] [CrossRef]
- Li, S.; Hill, C.P.; Sundquist, W.I.; Finch, J.T. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nat. Cell Biol. 2000, 407, 409–413. [Google Scholar] [CrossRef]
- Pornillos, O.; Ganser-Pornillos, B.K.; Kelly, B.N.; Hua, Y.; Whitby, F.G.; Stout, C.D.; Sundquist, W.I.; Hill, C.P.; Yeager, M. X-Ray Structures of the Hexameric Building Block of the HIV Capsid. Cell 2009, 137, 1282–1292. [Google Scholar] [CrossRef] [Green Version]
- Pornillos, O.; Ganser-Pornillos, B.K.; Yeager, M. Atomic-level modelling of the HIV capsid. Nat. Cell Biol. 2011, 469, 424–427. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Perilla, J.; Yufenyuy, E.L.; Meng, X.; Chen, B.; Ning, J.; Ahn, J.; Gronenborn, A.M.; Schulten, K.; Aiken, C.; et al. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nat. Cell Biol. 2013, 497, 643–646. [Google Scholar] [CrossRef]
- Pornillos, O.; Ganser-Pornillos, B.K. Maturation of retroviruses. Curr. Opin. Virol. 2019, 36, 47–55. [Google Scholar] [CrossRef]
- Ganser-Pornillos, B.K.; Cheng, A.; Yeager, M. Structure of Full-Length HIV-1 CA: A Model for the Mature Capsid Lattice. Cell 2007, 131, 70–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattei, S.; Glass, B.; Hagen, W.J.H.; Kräusslich, H.-G.; Briggs, J.A.G. The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science 2016, 354, 1434–1437. [Google Scholar] [CrossRef]
- Mattei, S.; Tan, A.; Glass, B.; Müller, B.; Kräusslich, H.-G.; Briggs, J.A.G. High-resolution structures of HIV-1 Gag cleavage mutants determine structural switch for virus maturation. Proc. Natl. Acad. Sci. USA 2018, 115, E9401–E9410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zila, V.; Margiotta, E.; Turoňová, B.; Müller, T.G.; Zimmerli, C.E.; Mattei, S.; Allegretti, M.; Börner, K.; Rada, J.; Müller, B.; et al. Cone-shaped HIV-1 capsids are transported through intact nuclear pores. Cell 2021, 184, 1032–1046.e18. [Google Scholar] [CrossRef]
- Burdick, R.C.; Li, C.; Munshi, M.; Rawson, J.M.O.; Nagashima, K.; Hu, W.-S.; Pathak, V.K. HIV-1 uncoats in the nucleus near sites of integration. Proc. Natl. Acad. Sci. USA 2020, 117, 5486–5493. [Google Scholar] [CrossRef]
- Christensen, D.E.; Ganser-Pornillos, B.K.; Johnson, J.S.; Pornillos, O.; Sundquist, W.I. Reconstitution and visualization of HIV-1 capsid-dependent replication and integration in vitro. Science 2020, 370, eabc8420. [Google Scholar] [CrossRef]
- Jacques, D.; McEwan, W.A.; Hilditch, L.; Price, A.J.; Towers, G.J.; James, L.C. HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis. Nat. Cell Biol. 2016, 536, 349–353. [Google Scholar] [CrossRef]
- Dick, R.A.; Mallery, D.L.; Vogt, V.M.; James, L.C. IP6 Regulation of HIV Capsid Assembly, Stability, and Uncoating. Viruses 2018, 10, 640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dick, R.A.; Zadrozny, K.K.; Xu, C.; Schur, F.K.M.; Lyddon, T.D.; Ricana, C.; Wagner, J.M.; Perilla, J.R.; Ganser-Pornillos, B.K.; Johnson, M.C.; et al. Inositol phosphates are assembly co-factors for HIV-1. Nat. Cell Biol. 2018, 560, 509–512. [Google Scholar] [CrossRef] [PubMed]
- Mallery, D.L.; Faysal, K.R.; Kleinpeter, A.; Wilson, M.S.; Vaysburd, M.; Fletcher, A.; Novikova, M.; Böcking, T.; Freed, E.O.; Saiardi, A.; et al. Cellular IP6 Levels Limit HIV Production while Viruses that Cannot Efficiently Package IP6 Are Attenuated for Infection and Replication. Cell Rep. 2019, 29, 3983–3996.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mallery, D.L.; Márquez, C.L.; A McEwan, W.; Dickson, C.F.; A Jacques, D.; Anandapadamanaban, M.; Bichel, K.; Towers, G.; Saiardi, A.; Böcking, T.; et al. IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis. eLife 2018, 7, e35335. [Google Scholar] [CrossRef]
- Renner, N.; Mallery, D.L.; Faysal, K.M.R.; Peng, W.; Jacques, D.A.; Böcking, T.; James, L.C. A lysine ring in HIV capsid pores coordinates IP6 to drive mature capsid assembly. PLoS Pathog. 2021, 17, e1009164. [Google Scholar] [CrossRef] [PubMed]
- Pettit, S.; Moody, M.D.; Wehbie, R.S.; Kaplan, A.H.; Nantermet, P.V.; Klein, C.A.; Swanstrom, R. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J. Virol. 1994, 68, 8017–8027. [Google Scholar] [CrossRef] [Green Version]
- Potempa, M.; Lee, S.-K.; Yilmaz, N.K.; Nalivaika, E.A.; Rogers, A.; Spielvogel, E.; Carter, C.W.; Schiffer, C.A.; Swanstrom, R. HIV-1 Protease Uses Bi-Specific S2/S2′ Subsites to Optimize Cleavage of Two Classes of Target Sites. J. Mol. Biol. 2018, 430, 5182–5195. [Google Scholar] [CrossRef]
- Prabu-Jeyabalan, M.; Nalivaika, E.; Schiffer, C.A. Substrate Shape Determines Specificity of Recognition for HIV-1 Protease: Analysis of Crystal Structures of Six Substrate Complexes. Structure 2002, 10, 369–381. [Google Scholar] [CrossRef]
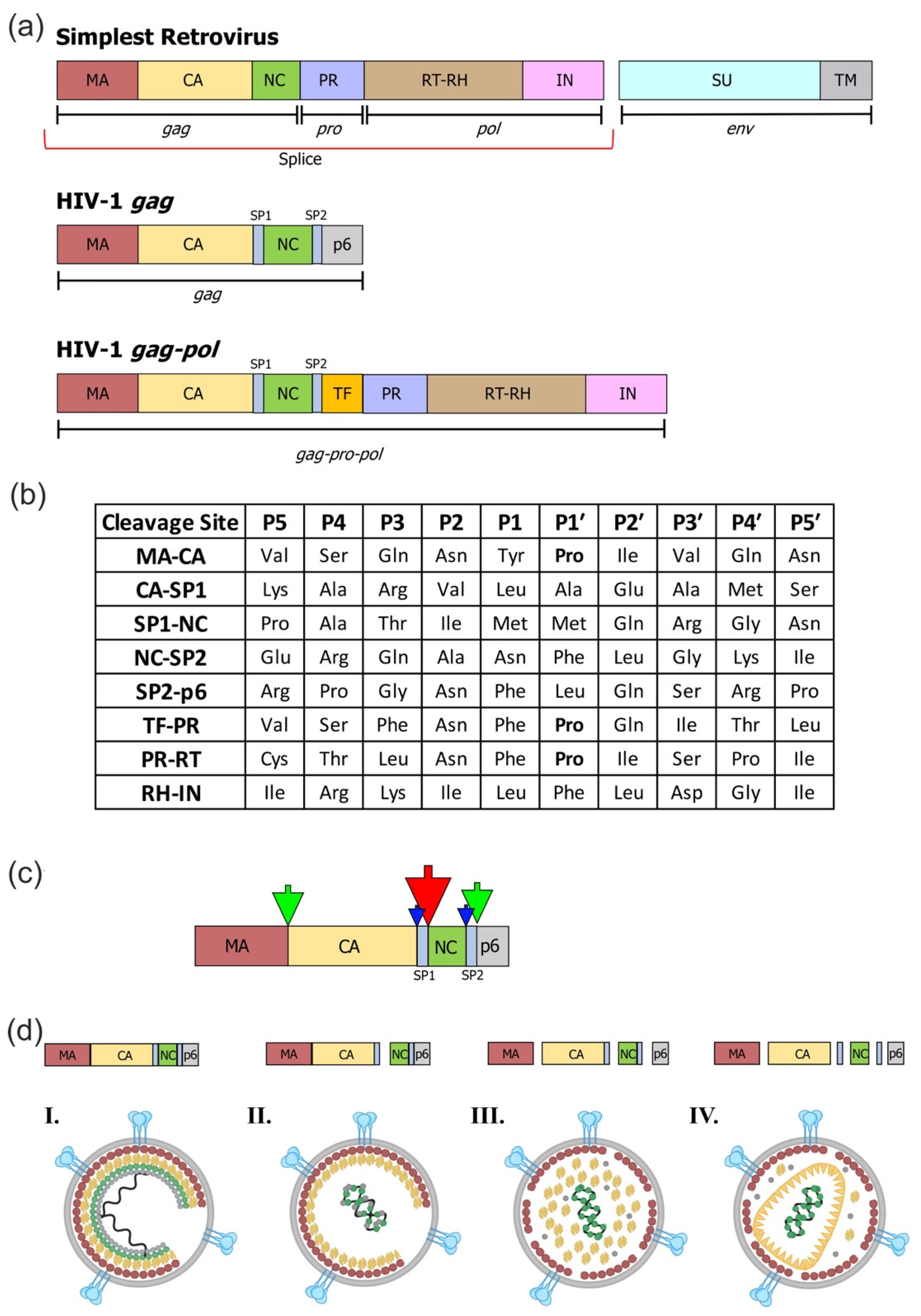
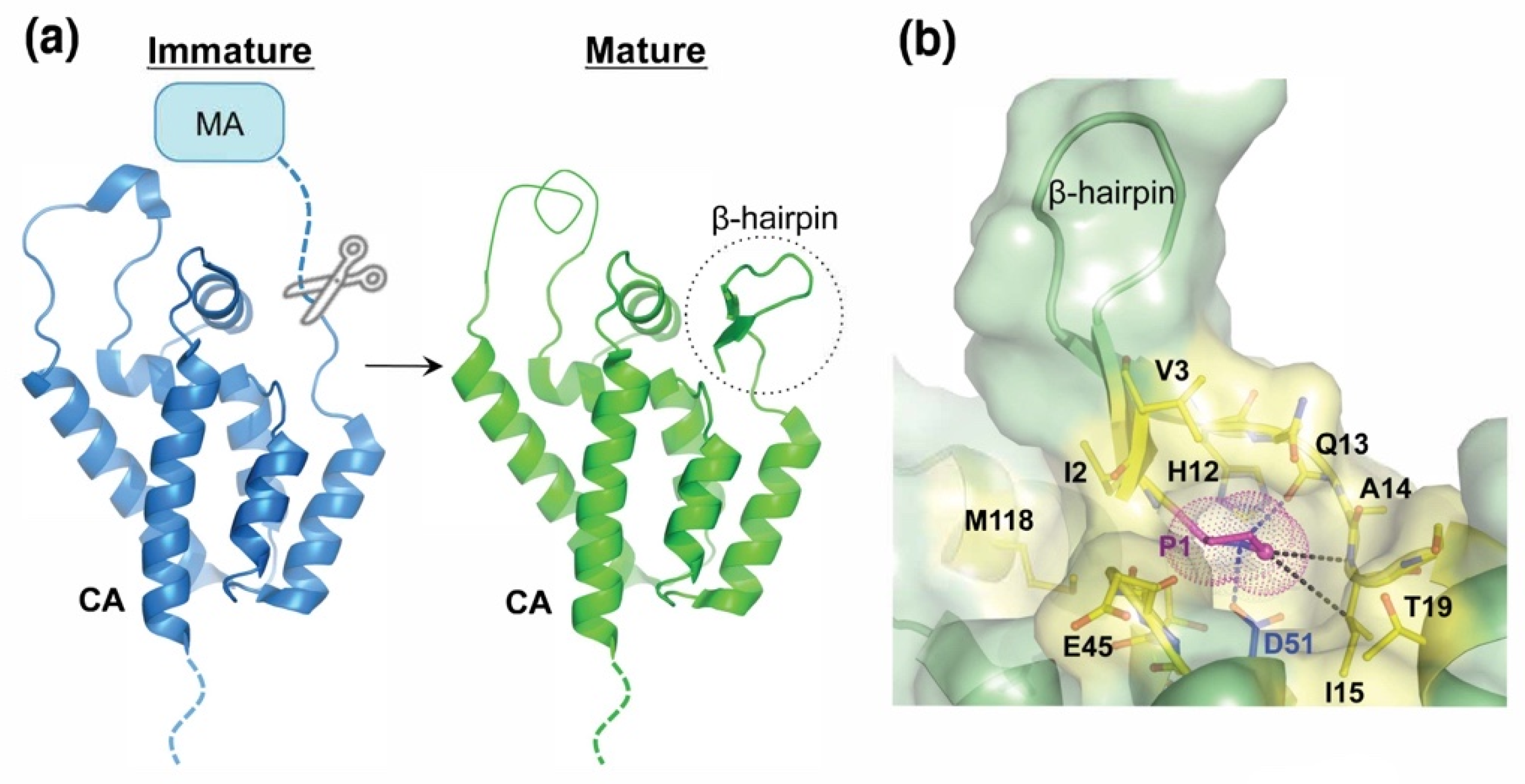
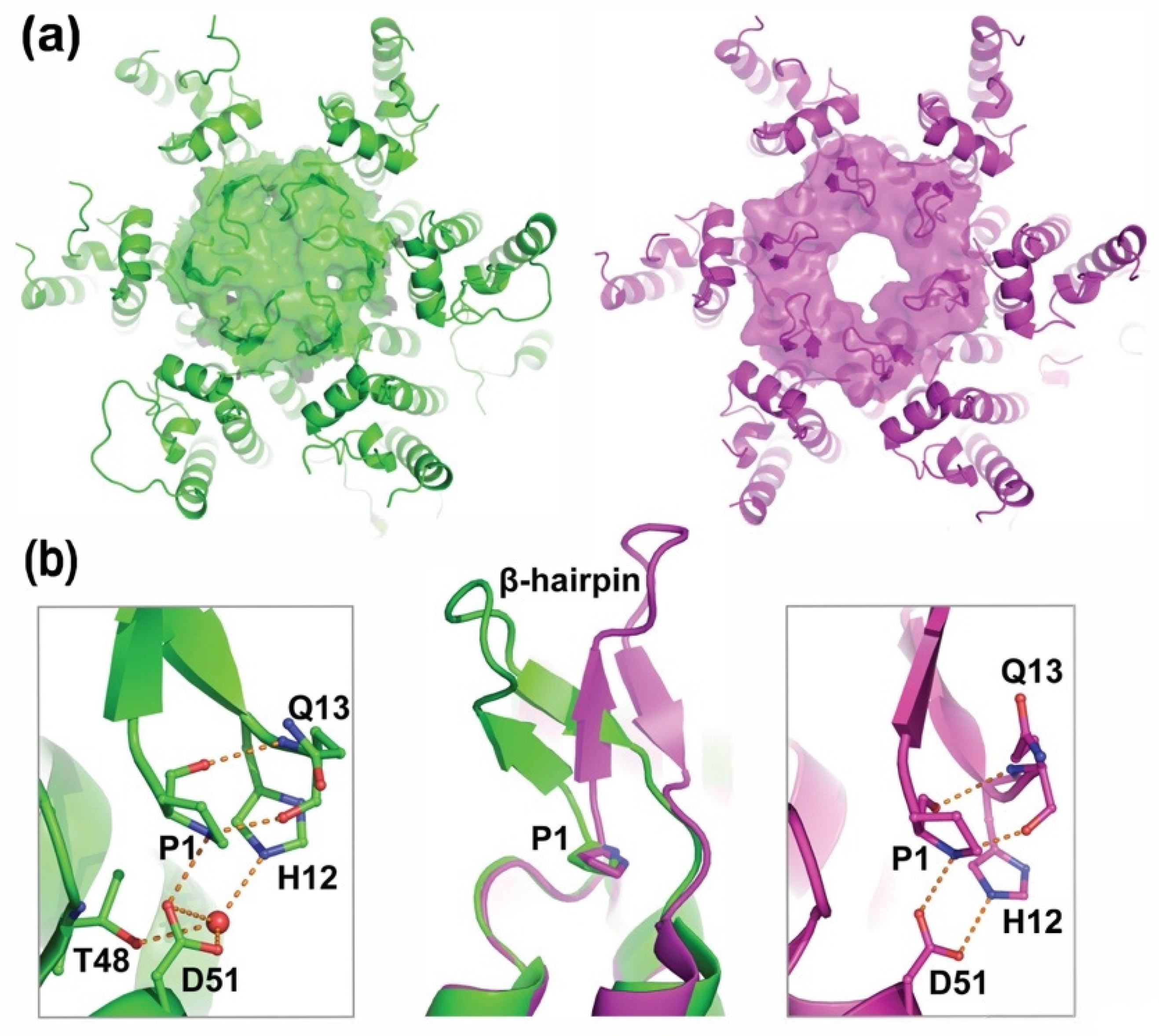
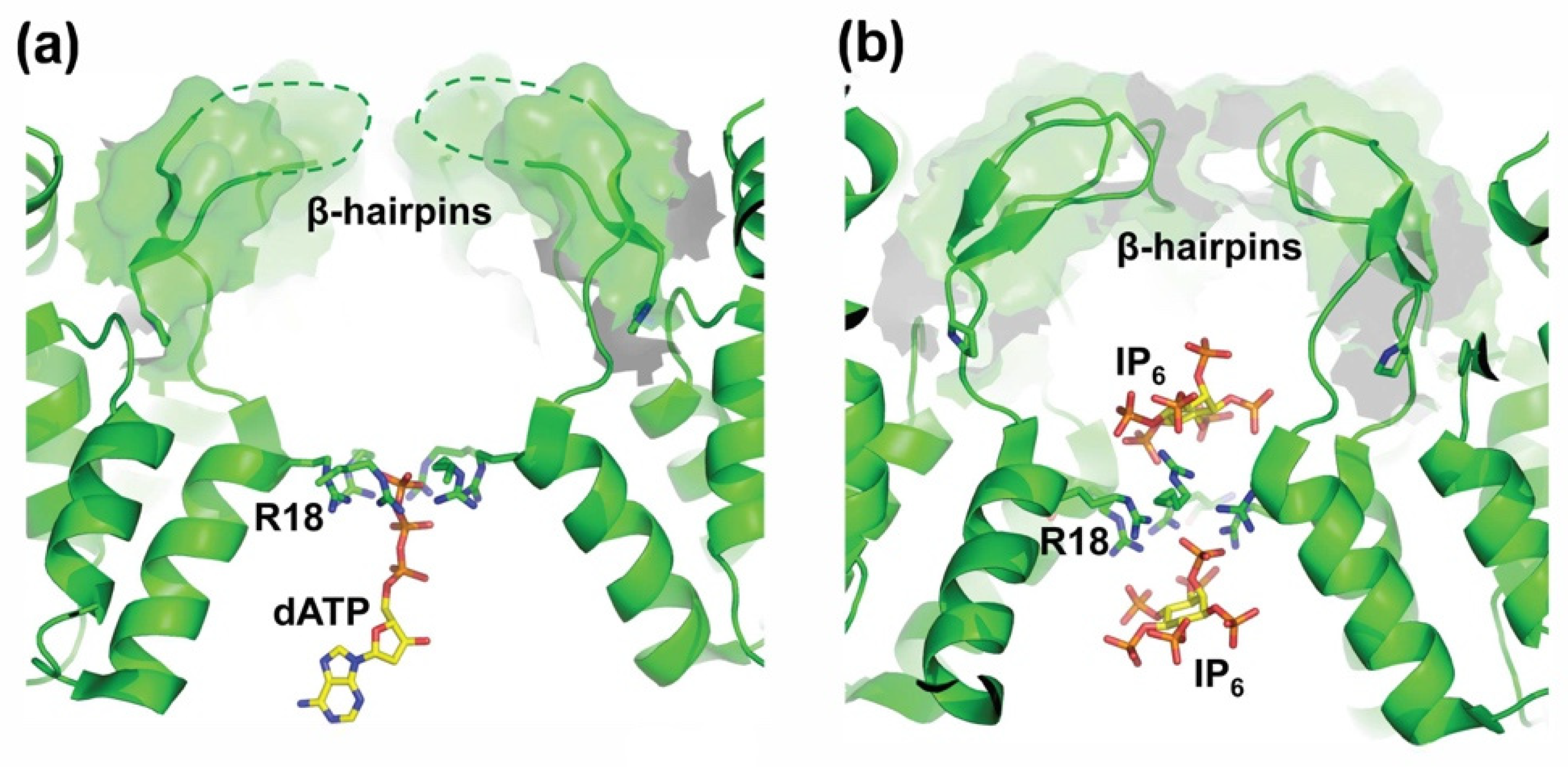
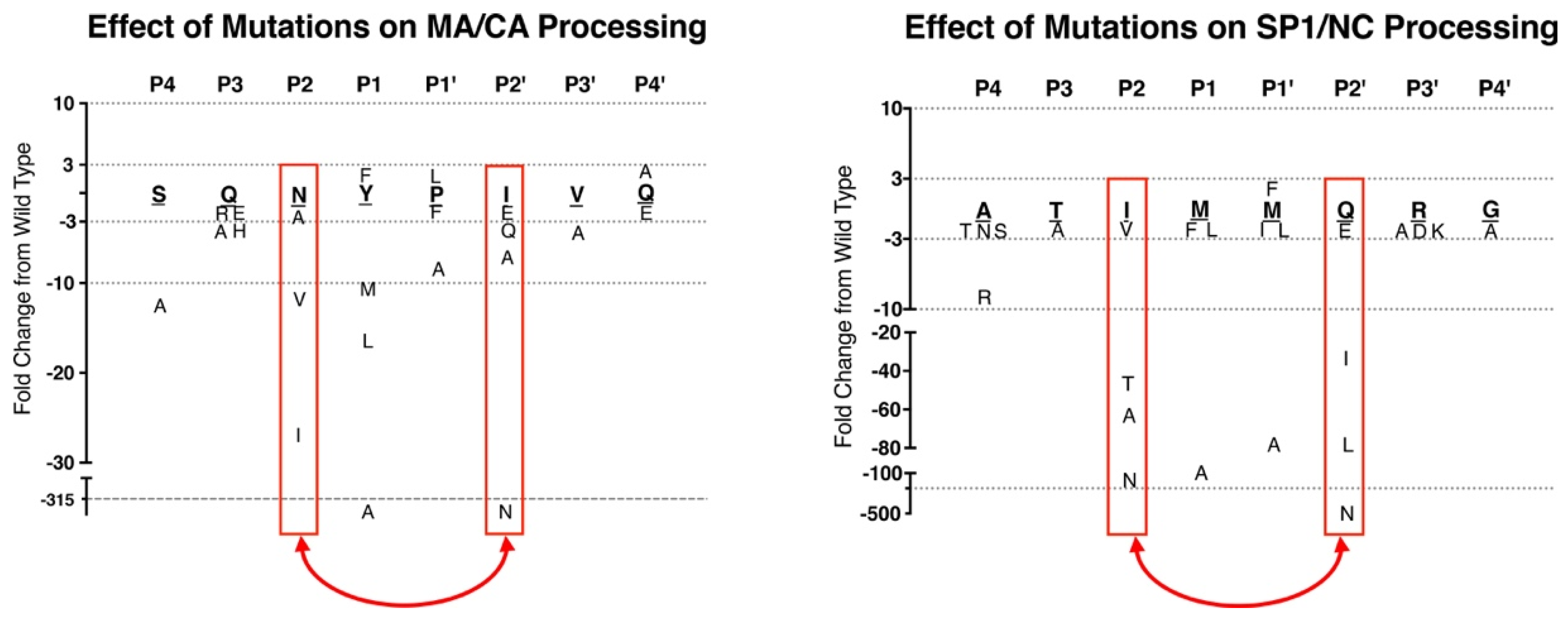
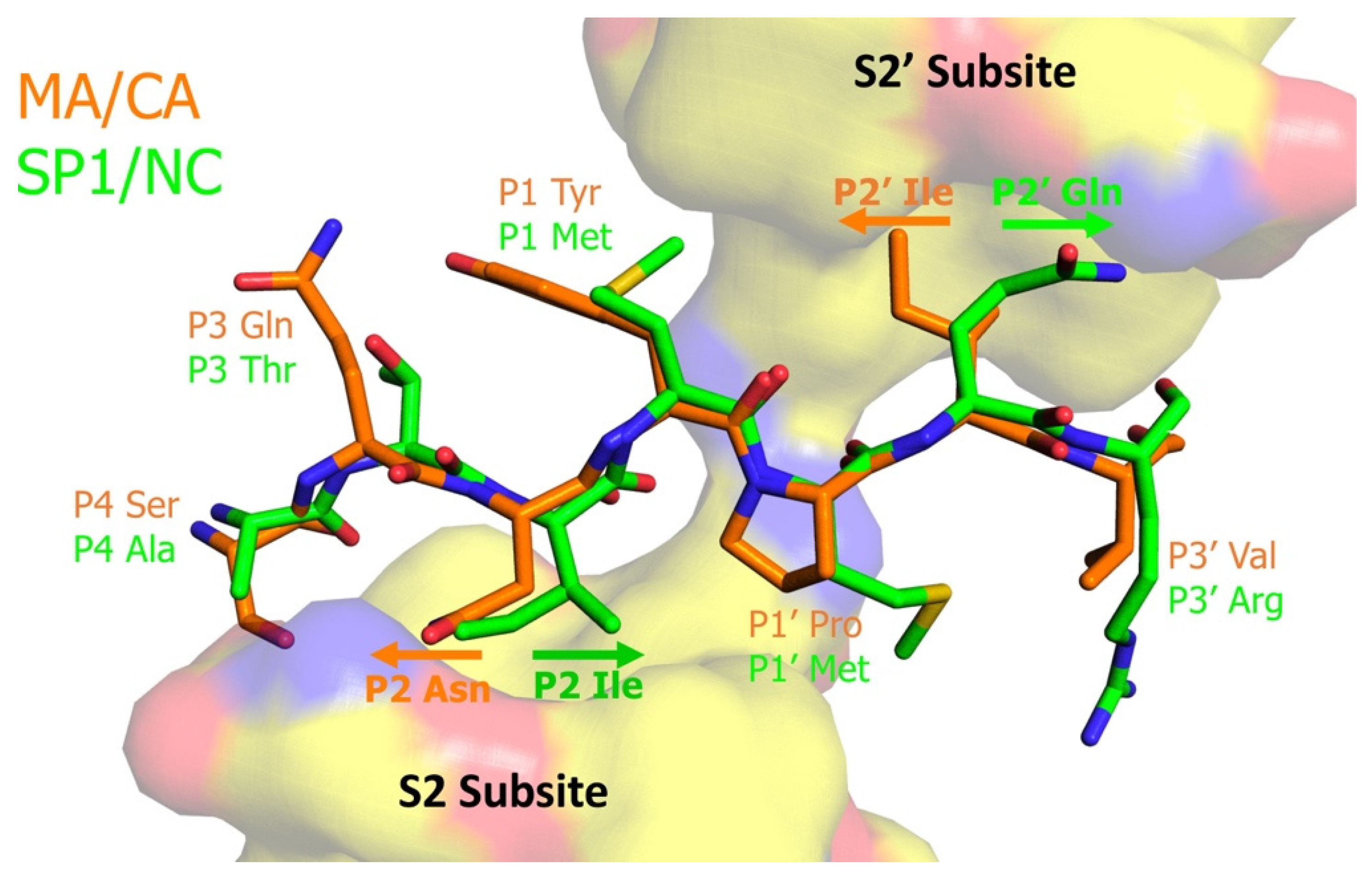
| BLV | MA/CA | PAIL/PIIS |
| EIAV | MA/CA | SEEY/PIMI |
| HTLV-I | MA/CA | PQVL/PVMH |
| HTLV-II | MA/CA | TQCF/PILH |
| HIV-1 | MA/CA | SQNY/PIVQ |
| MMTV | n/CA | TFTF/PVVF |
| MPMV | p12/CA | KDIF/PVTE |
| MuLV | p12/CA | SQAF/PLRA |
| RSV | p10/CA | VVAM/PVVI |
| SIVmac | MA/CA | GGNY/PVQQ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Swanstrom, R.; Sundquist, W.I. Stephan Oroszlan and the Proteolytic Processing of Retroviral Proteins: Following A Pro. Viruses 2021, 13, 2218. https://doi.org/10.3390/v13112218
Swanstrom R, Sundquist WI. Stephan Oroszlan and the Proteolytic Processing of Retroviral Proteins: Following A Pro. Viruses. 2021; 13(11):2218. https://doi.org/10.3390/v13112218
Chicago/Turabian StyleSwanstrom, Ronald, and Wesley I. Sundquist. 2021. "Stephan Oroszlan and the Proteolytic Processing of Retroviral Proteins: Following A Pro" Viruses 13, no. 11: 2218. https://doi.org/10.3390/v13112218
APA StyleSwanstrom, R., & Sundquist, W. I. (2021). Stephan Oroszlan and the Proteolytic Processing of Retroviral Proteins: Following A Pro. Viruses, 13(11), 2218. https://doi.org/10.3390/v13112218





