First Report on Detection and Molecular Characterization of Adenoviruses in the Small Indian Mongoose (Urva auropunctata)
Abstract
:1. Introduction
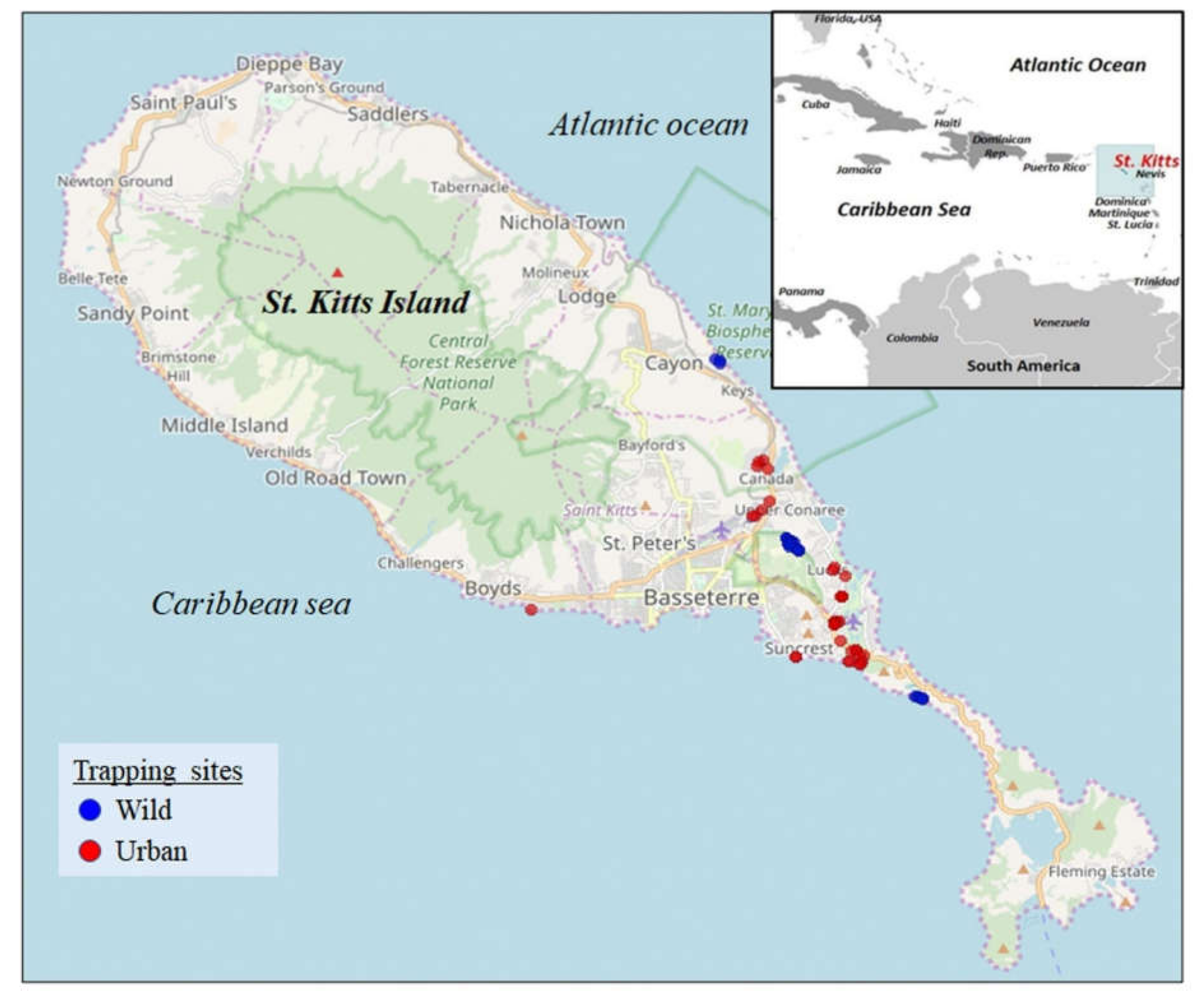
2. Materials and Methods
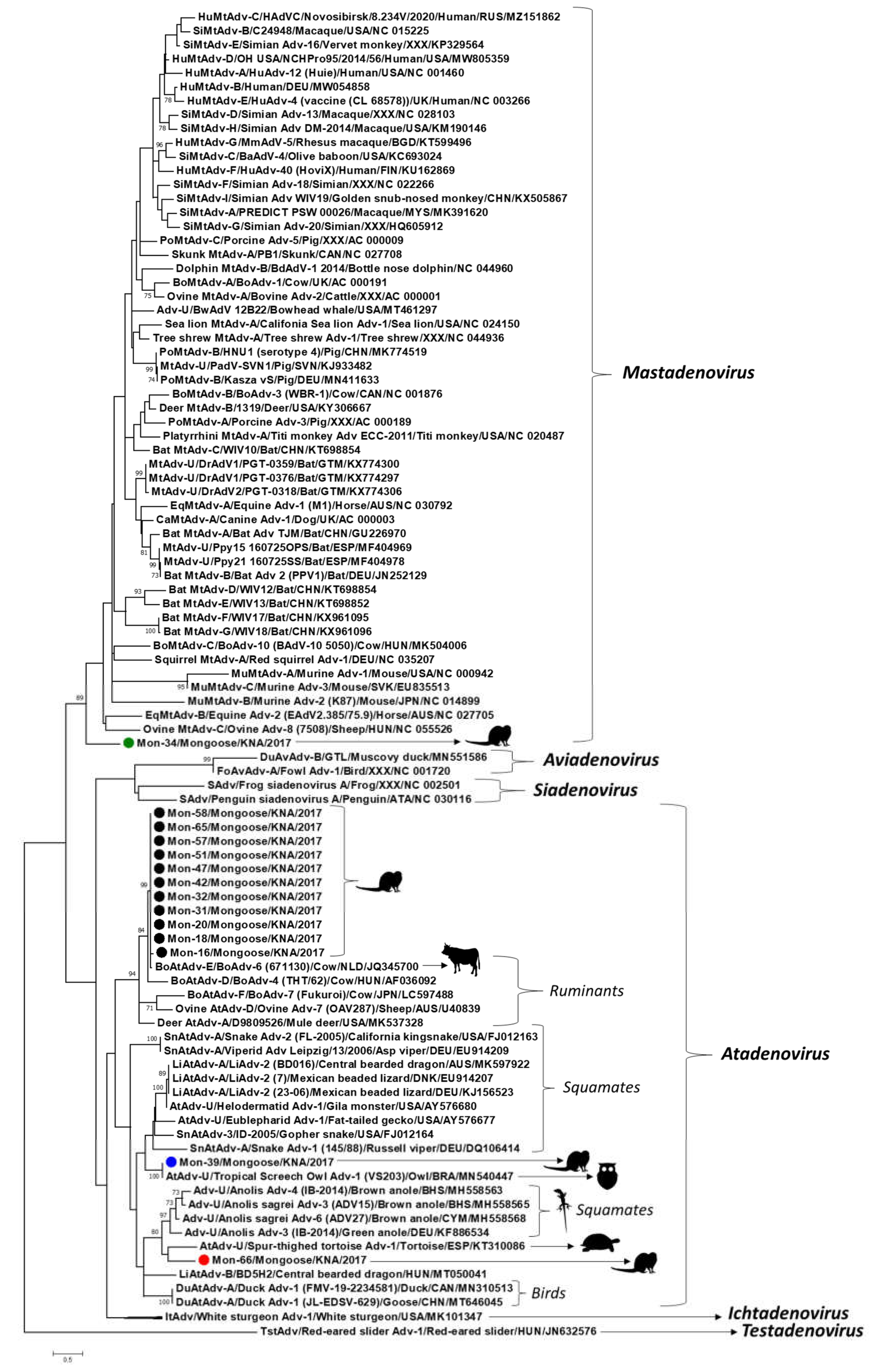
3. Results and Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harrach, B.; Benkő, M. Adenoviruses (Adenoviridae). In Encyclopedia of Virology; Bamford, D.H., Zuckerman, M., Eds.; Academic Press: Amsterdam, The Netherlands, 2021; pp. 3–16. [Google Scholar] [CrossRef]
- Harrach, B.; Tarján, Z.L.; Benkő, M. Adenoviruses across the animal kingdom: A walk in the zoo. FEBS Lett. 2019, 593, 3660–3673. [Google Scholar] [CrossRef]
- Machlachlan, J.; Dubovi, E. Chapter 10—Adenoviridae. In Fenner’s Veterinary Virology, 5th ed.; Academic Press: Amsterdam, The Netherlands, 2016; pp. 217–227. [Google Scholar] [CrossRef]
- Kaján, G.L.; Doszpoly, A.; Tarján, Z.L.; Vidovszky, M.Z.; Papp, T. Virus–Host Coevolution with a Focus on Animal and Human DNA Viruses. J. Mol. Evol. 2020, 88, 41–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benkö, M.; Harrach, B. Molecular evolution of adenoviruses. Curr. Top. Microbiol. Immunol. 2003, 272, 3–35. [Google Scholar] [CrossRef] [PubMed]
- Vaz, F.F.; Raso, T.F.; Agius, J.E.; Hunt, T.; Leishman, A.; Eden, J.S.; Phalen, D.N. Opportunistic sampling of wild native and invasive birds reveals a rich diversity of adenoviruses in Australia. Virus Evol. 2020, 6, veaa024. [Google Scholar] [CrossRef] [PubMed]
- Louppe, V.; Leroy, B.; Herrel, A.; Veron, G. The globally invasive small Indian mongoose Urva auropunctata is likely to spread with climate change. Sci. Rep. 2020, 10, 7461. [Google Scholar] [CrossRef] [PubMed]
- Veron, G.; Jennings, A.P. Javan mongoose or small Indian mongoose—who is where? Mamm. Biol. 2017, 87, 62–70. [Google Scholar] [CrossRef]
- Nidaira, M.; Takahashi, K.; Ogura, G.; Taira, K.; Okano, S.; Kudaka, J.; Itokazu, K.; Mishiro, S.; Nakamura, M. Detection and phylogenetic analysis of Hepatitis E Viruses from mongooses in Okinawa, Japan. J. Vet. Med. Sci. 2012, 74, 1665–1668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sauvé, C.C.; Rees, E.E.; Gilbert, A.T.; Berentsen, A.R.; Allibert, A.; Leighton, P.A. Modeling mongoose rabies in the caribbean: A model-guided fieldwork approach to identify research priorities. Viruses 2021, 13, 323. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.M.; Chen, C.C. Molecular characteristics of carnivore protoparvovirus 1 with high sequence similarity between wild and domestic carnivores in taiwan. Pathogens 2021, 10, 671. [Google Scholar] [CrossRef] [PubMed]
- Conceição-Neto, N.; Zeller, M.; Heylen, E.; Lefrère, H.; Mesquita, J.R.; Matthijnssens, J. Fecal virome analysis of three carnivores reveals a novel nodavirus and multiple gemycircularviruses. Virol. J. 2015, 12, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duarte, M.D.; Henriques, A.M.; Barros, S.C.; Fagulha, T.; Mendonça, P.; Carvalho, P.; Monteiro, M.; Fevereiro, M.; Basto, M.P.; Rosalino, L.M.; et al. Snapshot of Viral Infections in Wild Carnivores Reveals Ubiquity of Parvovirus and Susceptibility of Egyptian Mongoose to Feline Panleukopenia Virus. PLoS ONE 2013, 8, e59399. [Google Scholar] [CrossRef] [PubMed]
- Gainor, K.; Becker, A.A.M.J.; Malik, Y.S.; Ghosh, S. Detection and complete genome analysis of circoviruses and cycloviruses in the small indian mongoose (Urva auropunctata): Identification of novel species. Viruses 2021, 13, 1700. [Google Scholar] [CrossRef]
- Kleymann, A.; Becker, A.A.M.J.; Malik, Y.S.; Kobayashi, N.; Ghosh, S. Detection and molecular characterization of picobirnaviruses (PBVs) in the mongoose: Identification of a novel PBV using an alternative genetic code. Viruses 2020, 12, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogen-Odoi, A.; Miller, B.R.; Happ, C.M.; Maupin, G.O.; Burkot, T.R. Isolation of thogoto virus (Orthomyxoviridae) from the banded mongoose, Mongos mungo (herpestidae), in Uganda. Am. J. Trop. Med. Hyg. 1999, 60, 439–440. [Google Scholar] [CrossRef] [Green Version]
- Schmiedeknecht, G.; Eickmann, M.; Köhler, K.; Herden, C.E.; Kolesnikova, L.; Förster, C.; Burkhardt, E.H.; König, M.; Thiel, M.; Reinacher, M. Fatal cowpox virus infection in captive banded mongooses (Mungos mungo). Vet. Pathol. 2010, 47, 547–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becker, A.A.M.J.; Hill, K.C.; Butaye, P. Unraveling the gut microbiome of the invasive small indian mongoose (Urva auropunctata) in the caribbean. Microorganisms 2021, 9, 465. [Google Scholar] [CrossRef] [PubMed]
- Wellehan, J.F.X.; Johnson, A.J.; Harrach, B.; Benkö, M.; Pessier, A.P.; Johnson, C.M.; Garner, M.M.; Childress, A.; Jacobson, E.R. Detection and Analysis of Six Lizard Adenoviruses by Consensus Primer PCR Provides Further Evidence of a Reptilian Origin for the Atadenoviruses. J. Virol. 2004, 78, 13366–13369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0. molecular biology and evolution. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, T.; Halper, B.; Siebert, J.; Cruz-Martinez, L.; Chapwanya, A.; Kelly, P.; Ketzis, J.K.; Vessell, J.; Köster, L.; Yao, C. Parasites of small Indian mongoose, Herpestes auropunctatus, on St. Kitts, West Indies. Parasitol. Res. 2018, 117, 989–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ballmann, M.Z.; Harrach, B. Detection and partial genetic characterisation of novel avi- and siadenoviruses in racing and fancy pigeons (Columba livia domestica). Acta Vet. Hung. 2016, 64, 514–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salzmann, E.; Müller, E.; Marschang, R.E. Detection of testadenoviruses and atadenoviruses in tortoises and turtles in Europe. J. Zoo Wildl. Med. 2021, 52, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Rondhuis, P.R. A new bovine adenovirus. Archiv Gesamte Virusforschung 1968, 25, 235–236. [Google Scholar] [CrossRef]
- Sibley, S.D.; Goldberg, T.L.; Pedersen, J.A. Detection of known and novel adenoviruses in cattle wastes via broad-spectrum primers. Appl. Environ. Microbiol. 2011, 77, 5001–5008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jejesky de Oliveira, A.P.; Valdetaro Rangel, M.C.; Z Vidovszky, M.; Rossi, J.L.; Vicentini, F.; Harrach, B.; L Kaján, G. Identification of two novel adenoviruses in smooth-billed ani and tropical screech owl. PLoS ONE 2020, 15, e0229415. [Google Scholar] [CrossRef]
- Ball, I.; Behncke, H.; Schmidt, V.; Papp, T.; Stöhr, A.C.; Marschang, R.E. Partial characterization of new adenoviruses found in lizards. J. Zoo Wildl. Med. 2014, 45, 287–297. [Google Scholar] [CrossRef]
- Garcia-Morante, B.; Pénzes, J.J.; Costa, T.; Martorell, J.; Martínez, J. Hyperplastic stomatitis and esophagitis in a tortoise (Testudo graeca) associated with an adenovirus infection. J. Vet. Diagn. Investig. 2016, 28, 579–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Luca, E.; Stimmelmayr, R.; Rotstein, D.S.; Sanchez, S. A novel adenovirus detected in bering-chukchi-beaufort seas bowhead whale (Balaena mysticetus): Epidemiologic data and phylogenetic characterization. J. Wildl. Dis. 2021, 57, 652–656. [Google Scholar] [CrossRef] [PubMed]
- Wray, A.K.; Olival, K.J.; Morán, D.; Lopez, M.R.; Alvarez, D.; Navarrete-Macias, I.; Liang, E.; Simmons, N.B.; Lipkin, W.I.; Daszak, P.; et al. Viral Diversity, Prey Preference, and Bartonella Prevalence in Desmodus rotundus in Guatemala. Ecohealth 2016, 13, 761–774. [Google Scholar] [CrossRef]
- Hierholzer, J.C.; Halonen, P.E.; Dahlen, P.O.; Bingham, P.G.; McDonough, M.M. Detection of adenovirus in clinical specimens by polymerase chain reaction and liquid-phase hybridization quantitated by time-resolved fluorometry. J. Clin. Microbiol. 1993, 31, 1886–1891. [Google Scholar] [CrossRef] [Green Version]

| Mongoose-Associated Adenovirus | GenBank Accession Number/s | Genus 1 | Maximum/Significant Identity with Cognate Adenovirus Sequences (Species 2/Organism (Isolate/Strain)/Host/Country/GenBank Accession Number) from Other Host Species | |
|---|---|---|---|---|
| Nucleotide Sequence Identity (%) 3 | Deduced aa Sequence Identity (%) 4 | |||
| Mon-16, Mon-18, Mon-20, Mon-31, Mon-32, Mon-42, Mon-47, Mon-51, Mon-57, Mon-58, Mon-65 | OK381854–OK381858, OK381861–OK381863, OK381865–OK381867 | Atadenovirus | 98.3–99.6% with Bovine atadenovirus E/ Bovine adenovirus-6 (671130)/Cow/Netherlands/JQ345700 | 97.0–98.9% with Bovine atadenovirus E/ Bovine adenovirus-6 (671130)/Cow/Netherlands/JQ345700 |
| Mon-34 | OK381859 | Mastadenovirus | 72.0% with Unclassified Mastadenovirus/Desmodus rotundus Adenovirus-1 (DrAdV1/PGT-0359)/Bat/Guatemala/>KX774300 70.5% with Unclassified Mastadenovirus/Desmodus rotundus Adenovirus-2 (DrAdV2/PGT-0318)/Bat/Guatemala/KX774306 | 72.6% with Unclassified Adenoviridae/Bowhead whale adenovirus (BwAdV 12B22)/Bowhead whale/USA/MT461297 72.3% with Unclassified Mastadenovirus/Desmodus rotundus Adenovirus-1 (DrAdV1/PGT-0359)/Bat/Guatemala/KX774300 and Unclassified Mastadenovirus/Desmodus rotundus Adenovirus-2 (DrAdV2/PGT-0318)/Bat/Guatemala/KX774306 |
| Mon-39 | OK381860 | Atadenovirus | 100% with Unclassified Atadenovirus/Tropical screech owl adenovirus-1 (VS203)/Owl/ Brazil/MN540447 72.7% with Lizard Atadenovirus-A/Lizard adenovirus-2 (23-06)/Mexican beaded lizard/ Germany/KJ156523 | 100% with Unclassified Atadenovirus/Tropical screech owl adenovirus-1 (VS203)/Owl/ Brazil/MN540447 80.0% with Unclassified Atadenovirus/ Helodermatid adenovirus-1/Gila monster/USA/AY576680 78.9% with Lizard Atadenovirus-A/Lizard adenovirus-2 (23-06)/Mexican beaded lizard/ Germany/KJ156523 |
| Mon-66 | OK381868 | Atadenovirus | 69.0% with Unclassified Atadenovirus/ Spur-thighed tortoise adenovirus-1 (6211) /Tortoise/Spain/KT310086 5 68.7% with Unclassified Adenoviridae/ Anolis sagrei adenovirus-3 (ADV15)/Brown anole/Bahamas/MH558565 68.2% with Unclassified Adenoviridae/ Anolis sagrei adenovirus-6 (ADV27)/Brown anole/Cayman Islands/MH558568 | 71.4% with Unclassified Adenoviridae/Anolis sagrei adenovirus-6 (ADV27)/Brown anole/Cayman Islands/MH558568 70.0% with Snake Atadenovirus-A/Snake adenovirus-2 (FL-2005)/California kingsnake/USA/FJ012163 70.0% with Unclassified Atadenovirus/ Spur-thighed tortoise adenovirus-1 (6211)/ Tortoise/Spain/KT310086 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gainor, K.; Becker, A.A.M.J.; Malik, Y.S.; Ghosh, S. First Report on Detection and Molecular Characterization of Adenoviruses in the Small Indian Mongoose (Urva auropunctata). Viruses 2021, 13, 2194. https://doi.org/10.3390/v13112194
Gainor K, Becker AAMJ, Malik YS, Ghosh S. First Report on Detection and Molecular Characterization of Adenoviruses in the Small Indian Mongoose (Urva auropunctata). Viruses. 2021; 13(11):2194. https://doi.org/10.3390/v13112194
Chicago/Turabian StyleGainor, Kerry, Anne A. M. J. Becker, Yashpal S. Malik, and Souvik Ghosh. 2021. "First Report on Detection and Molecular Characterization of Adenoviruses in the Small Indian Mongoose (Urva auropunctata)" Viruses 13, no. 11: 2194. https://doi.org/10.3390/v13112194
APA StyleGainor, K., Becker, A. A. M. J., Malik, Y. S., & Ghosh, S. (2021). First Report on Detection and Molecular Characterization of Adenoviruses in the Small Indian Mongoose (Urva auropunctata). Viruses, 13(11), 2194. https://doi.org/10.3390/v13112194








