Norovirus Protease Structure and Antivirals Development
Abstract
1. Introduction
2. Structural Features of HuNoV Protease
2.1. Overall Structure of HuNoV Protease—Similarities and Differences
2.2. Active Site of HuNoV Protease
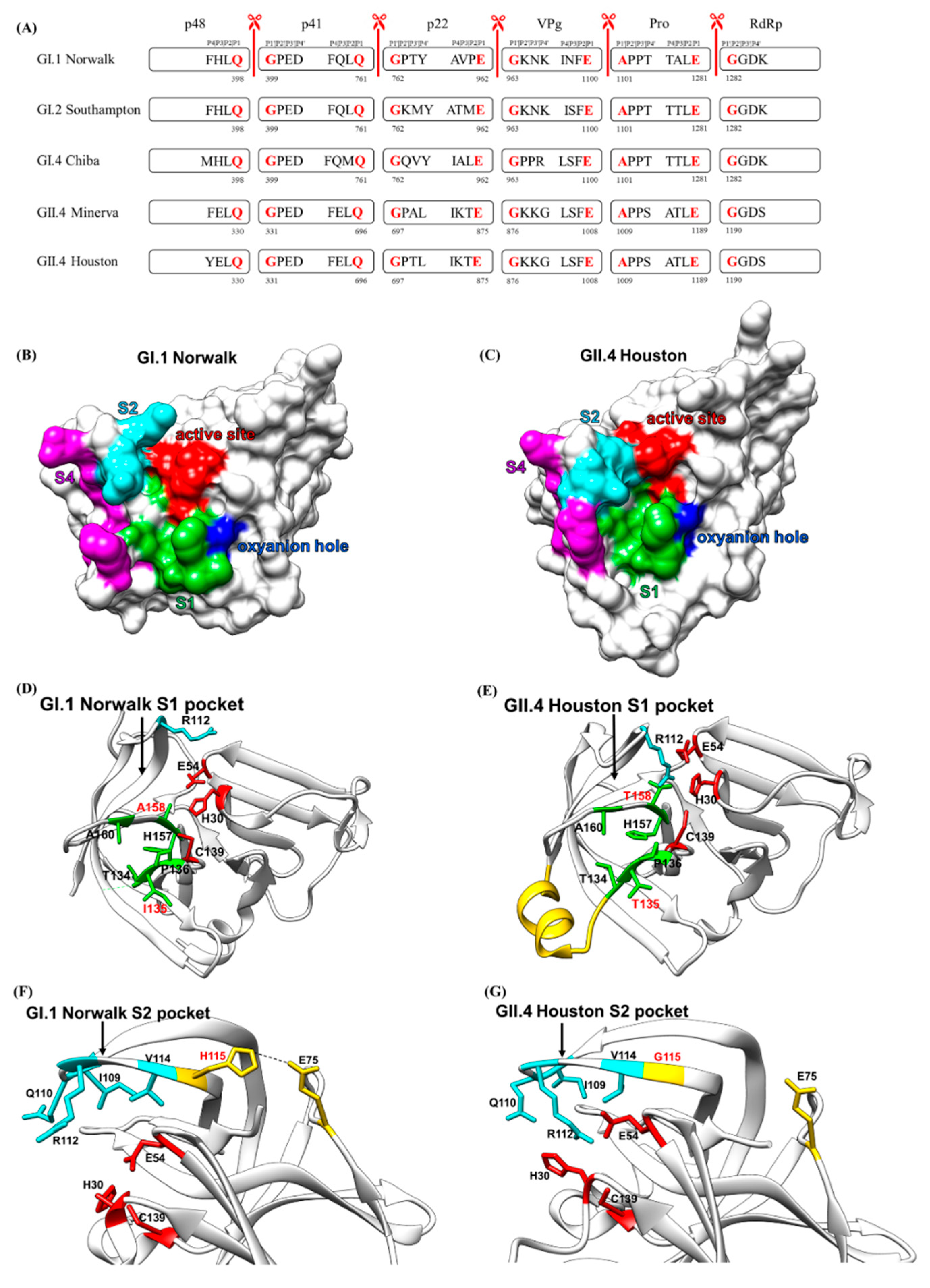
2.3. Cleavage Sites and Substrate-Binding Pockets of HuNoV Protease
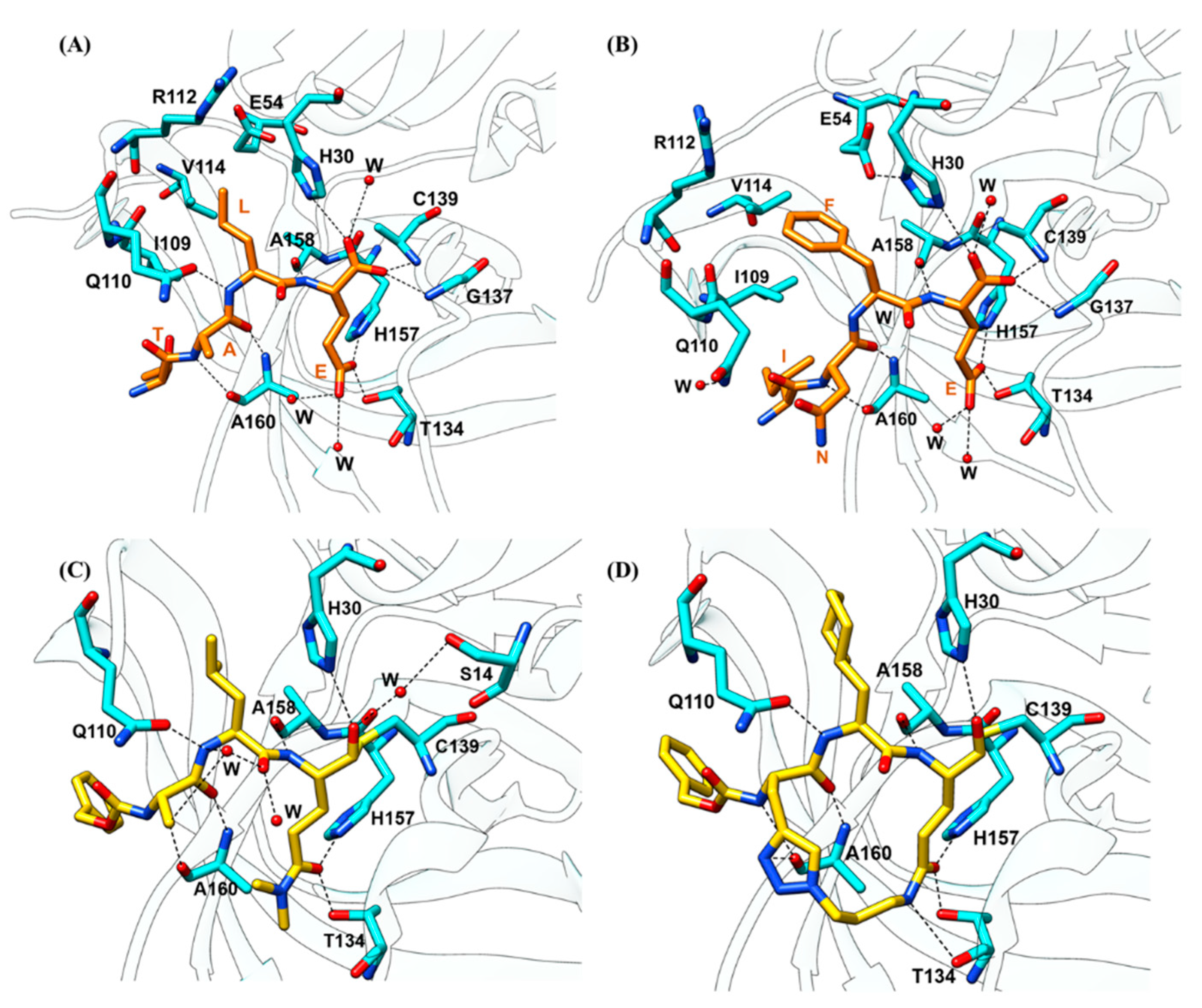
2.3.1. Substrate Recognition by the HuNoV Protease
2.3.2. Substrate-Induced Conformational Changes
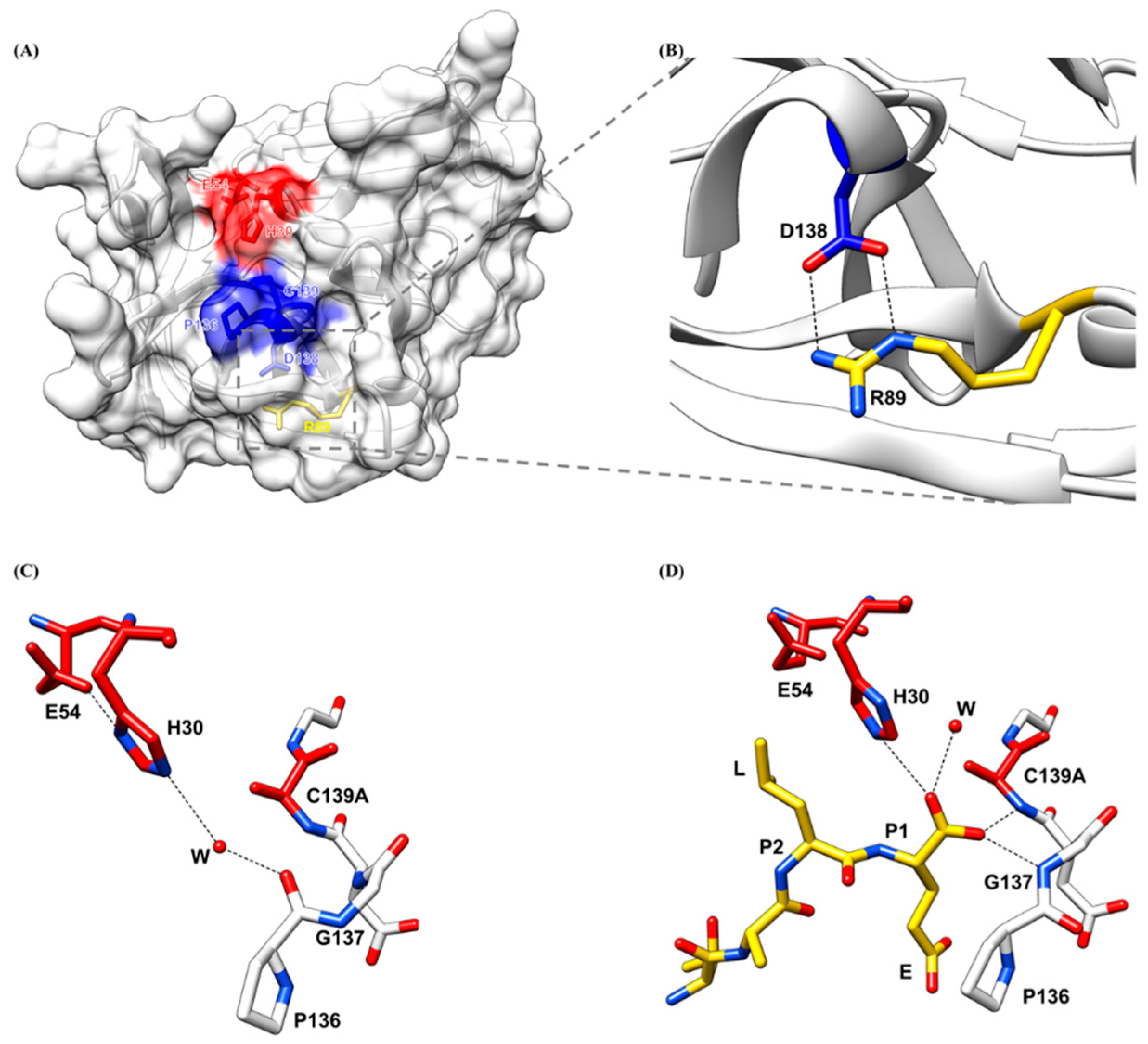
2.4. Oxyanion Hole of HuNoV Protease
3. Development of Current HuNoV Protease Inhibitors
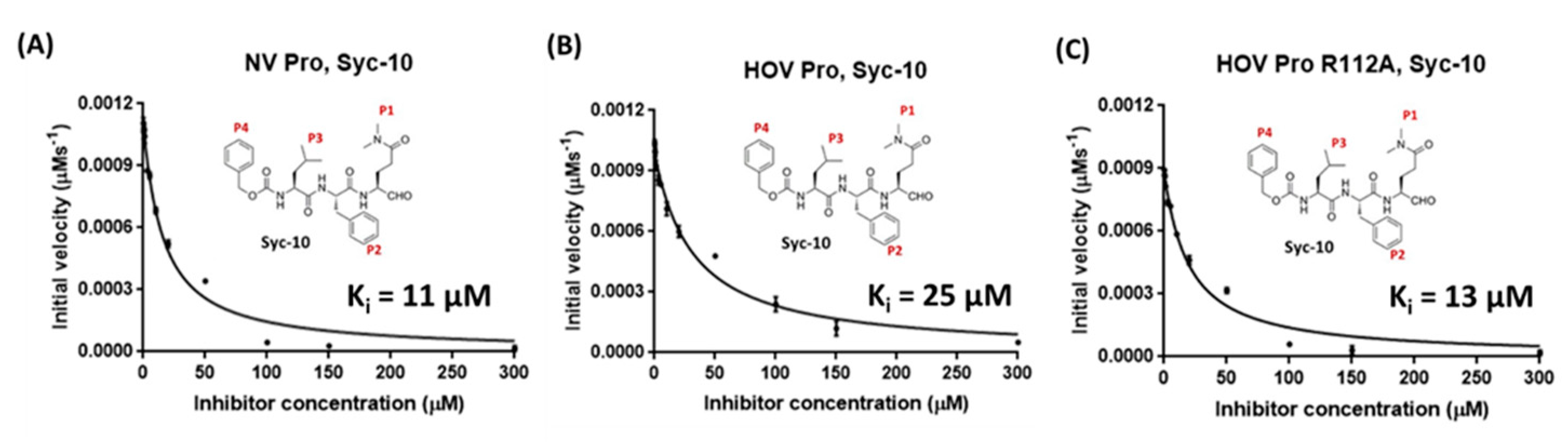
3.1. Linear Peptidomimetic Inhibitors
3.2. Prodrug
3.3. Macrocyclic Inhibitors
3.4. Commercially Available Inhibitors
4. Perspectives and Future Directions
4.1. Screening Approach—Lead Identification
4.2. Function Group Modification—Lead Optimization
4.3. In Vivo Testing—Lead Validation
4.4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ahmed, S.M.; Hall, A.J.; Robinson, A.E.; Verhoef, L.; Premkumar, P.; Parashar, U.D.; Koopmans, M.; Lopman, B.A. Global prevalence of norovirus in cases of gastroenteritis: A systematic review and meta-analysis. Lancet Infect. Dis. 2014, 14, 725–730. [Google Scholar] [CrossRef]
- Bartsch, S.M.; Lopman, B.A.; Ozawa, S.; Hall, A.J.; Lee, B.Y. Global Economic Burden of Norovirus Gastroenteritis. PLoS ONE 2016, 11, e0151219. [Google Scholar] [CrossRef] [PubMed]
- Belliot, G.; Lopman, B.; Ambert-Balay, K.; Pothier, P. The burden of norovirus gastroenteritis: An important foodborne and healthcare-related infection. Clin. Microbiol. Infect. 2014, 20, 724–730. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.M.; Widdowson, M.-A.; Glass, R.I.; Akazawa, K.; Vinje, J.; Parashar, U.D. Systematic Literature Review of Role of Noroviruses in Sporadic Gastroenteritis. Emerg. Infect. Dis. 2008, 14, 1224–1231. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.J.; Lopman, B.A.; Payne, D.C.; Patel, M.M.; Gastañaduy, P.A.; Vinje, J.; Parashar, U.D. Norovirus Disease in the United States. Emerg. Infect. Dis. 2013, 19, 1198–1205. [Google Scholar] [CrossRef] [PubMed]
- Krones, E.; Högenauer, C. Diarrhea in the Immunocompromised Patient. Gastroenterol. Clin. N. Am. 2012, 41, 677–701. [Google Scholar] [CrossRef]
- Schwartz, S.; Vergoulidou, M.; Schreier, E.; Loddenkemper, C.; Reinwald, M.; Schmidt-Hieber, M.; Flegel, W.A.; Thiel, E.; Schneider, T. Norovirus gastroenteritis causes severe and lethal complications after chemotherapy and hematopoietic stem cell transplantation. Blood 2011, 117, 5850–5856. [Google Scholar] [CrossRef]
- Xerry, J.; Gallimore, C.I.; Cubitt, D.; Gray, J.J. Tracking Environmental Norovirus Contamination in a Pediatric Primary Immunodeficiency Unit. J. Clin. Microbiol. 2010, 48, 2552–2556. [Google Scholar] [CrossRef]
- Casto, A.; Adler, A.L.; Makhsous, N.; Crawford, K.; Qin, X.; Kuypers, J.M.; Huang, M.-L.; Zerr, D.M.; Greninger, A.L. Prospective, Real-time Metagenomic Sequencing During Norovirus Outbreak Reveals Discrete Transmission Clusters. Clin. Infect. Dis. 2018, 69, 941–948. [Google Scholar] [CrossRef]
- Alejo-Cancho, I.; Avilés, F.F.; Capon, A.; Rodríguez, C.; Barrachina, J.; Salvador, P.; Valls, M.E.; Álvarez-Martinez, M.J.; Zboromyrska, Y.; Vila, J.; et al. Evaluation of a multiplex panel for the diagnosis of acute infectious diarrhea in immunocompromised hematologic patients. PLoS ONE 2017, 12, e0187458. [Google Scholar] [CrossRef]
- Atmar, R.L.; Bernstein, D.I.; Harro, C.D.; Al-Ibrahim, M.S.; Chen, W.H.; Ferreira, J.; Estes, M.K.; Graham, D.Y.; Opekun, A.R.; Richardson, C.; et al. Norovirus Vaccine against Experimental Human Norwalk Virus Illness. N. Engl. J. Med. 2011, 365, 2178–2187. [Google Scholar] [CrossRef]
- Atmar, R.L.; Bernstein, D.I.; Lyon, G.M.; Treanor, J.J.; Al-Ibrahim, M.S.; Graham, D.Y.; Vinje, J.; Jiang, X.; Gregoricus, N.; Frenck, R.W.; et al. Serological Correlates of Protection against a GII.4 Norovirus. Clin. Vaccine Immunol. 2015, 22, 923–929. [Google Scholar] [CrossRef]
- Bernstein, D.I.; Atmar, R.L.; Lyon, G.M.; Treanor, J.J.; Chen, W.H.; Jiang, X.; Vinje, J.; Gregoricus, N.; Frenck, R.W.; Moe, C.L.; et al. Norovirus Vaccine Against Experimental Human GII.4 Virus Illness: A Challenge Study in Healthy Adults. J. Infect. Dis. 2015, 211, 870–878. [Google Scholar] [CrossRef]
- Tamminen, K.; Lappalainen, S.; Huhti, L.; Vesikari, T.; Blazevic, V. Trivalent Combination Vaccine Induces Broad Heterologous Immune Responses to Norovirus and Rotavirus in Mice. PLoS ONE 2013, 8, e70409. [Google Scholar] [CrossRef]
- Heinimäki, S.; Malm, M.; Vesikari, T.; Blazevic, V. Parenterally Administered Norovirus GII.4 Virus-Like Particle Vaccine Formulated with Aluminum Hydroxide or Monophosphoryl Lipid A Adjuvants Induces Systemic but Not Mucosal Immune Responses in Mice. J. Immunol. Res. 2018, 2018, 3487095. [Google Scholar] [CrossRef]
- Ball, J.P.; Springer, M.J.; Ni, Y.; Finger-Baker, I.; Martínez, J.; Hahn, J.; Suber, J.F.; DiMarco, A.V.; Talton, J.D.; Cobb, R.R. Intranasal delivery of a bivalent norovirus vaccine formulated in an in situ gelling dry powder. PLoS ONE 2017, 12, e0177310. [Google Scholar] [CrossRef]
- Kim, L.; Liebowitz, D.; Lin, K.; Kasparek, K.; Pasetti, M.F.; Garg, S.J.; Gottlieb, K.; Trager, G.; Tucker, S.N. Safety and immunogenicity of an oral tablet norovirus vaccine, a phase I randomized, placebo-controlled trial. JCI Insight 2018, 3, 3. [Google Scholar] [CrossRef] [PubMed]
- Leroux-Roels, G.; Cramer, J.P.; Mendelman, P.M.; Sherwood, J.; Clemens, R.; Aerssens, A.; De Coster, I.; Borkowski, A.; Baehner, F.; Van Damme, P. Safety and Immunogenicity of Different Formulations of Norovirus Vaccine Candidate in Healthy Adults: A Randomized, Controlled, Double-Blind Clinical Trial. J. Infect. Dis. 2018, 217, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Treanor, J.; Sherwood, J.; Cramer, J.P.; Bouveret, N.L.C.; Lin, S.; Baehner, F.; Borkowski, A. A phase 2 study of the bivalent VLP norovirus vaccine candidate in older adults; impact of MPL adjuvant or a second dose. Vaccine 2020, 38, 5842–5850. [Google Scholar] [CrossRef] [PubMed]
- Ball, J.M.; Graham, D.Y.; Opekun, A.R.; Gilger, M.A.; Guerrero, R.A.; Estes, M.K. Recombinant Norwalk virus–like particles given orally to volunteers: Phase I study. Gastroenterology 1999, 117, 40–48. [Google Scholar] [CrossRef]
- El-Kamary, S.S.; Pasetti, M.F.; Mendelman, P.M.; Frey, S.E.; Bernstein, D.I.; Treanor, J.J.; Ferreira, J.; Chen, W.H.; Sublett, R.; Richardson, C.; et al. Adjuvanted Intranasal Norwalk Virus-Like Particle Vaccine Elicits Antibodies and Antibody-Secreting Cells That Express Homing Receptors for Mucosal and Peripheral Lymphoid Tissues. J. Infect. Dis. 2010, 202, 1649–1658. [Google Scholar] [CrossRef]
- Parra, G.I.; Bok, K.; Taylor, R.; Haynes, J.R.; Sosnovtsev, S.V.; Richardson, C.; Green, K.Y. Immunogenicity and specificity of norovirus Consensus GII.4 virus-like particles in monovalent and bivalent vaccine formulations. Vaccine 2012, 30, 3580–3586. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, K.; Wahid, R.; Richardson, C.; Bargatze, R.F.; El-Kamary, S.S.; Sztein, M.B.; Pasetti, M.F. Intranasal vaccination with an adjuvanted Norwalk virus-like particle vaccine elicits antigen-specific B memory responses in human adult volunteers. Clin. Immunol. 2012, 144, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Czakó, R.; Atmar, R.L.; Opekun, A.R.; Gilger, M.A.; Graham, D.Y.; Estes, M.K. Serum Hemagglutination Inhibition Activity Correlates with Protection from Gastroenteritis in Persons Infected with Norwalk Virus. Clin. Vaccine Immunol. 2012, 19, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Atmar, R.L.; Ettayebi, K.; Ayyar, B.V.; Neill, F.H.; Braun, R.P.; Ramani, S.; Estes, M.K. Comparison of Microneutralization and Histo-Blood Group Antigen–Blocking Assays for Functional Norovirus Antibody Detection. J. Infect. Dis. 2019, 221, 739–743. [Google Scholar] [CrossRef]
- Sherwood, J.; Mendelman, P.M.; Lloyd, E.; Liu, M.; Boslego, J.; Borkowski, A.; Jackson, A.; Faix, D. Efficacy of an intramuscular bivalent norovirus GI.1/GII.4 virus-like particle vaccine candidate in healthy US adults. Vaccine 2020, 38, 6442–6449. [Google Scholar] [CrossRef]
- Lindesmith, L.C.; Ferris, M.T.; Mullan, C.; Ferreira, J.; Debbink, K.; Swanstrom, J.; Richardson, C.; Goodwin, R.R.; Baehner, F.; Mendelman, P.M.; et al. Broad Blockade Antibody Responses in Human Volunteers after Immunization with a Multivalent Norovirus VLP Candidate Vaccine: Immunological Analyses from a Phase I Clinical Trial. PLoS Med. 2015, 12, e1001807. [Google Scholar] [CrossRef]
- Blazevic, V.; Malm, M.; Salminen, M.; Oikarinen, S.; Hyöty, H.; Veijola, R.; Vesikari, T. Multiple consecutive norovirus infections in the first 2 years of life. Eur. J. Nucl. Med. Mol. Imaging 2015, 174, 1679–1683. [Google Scholar] [CrossRef]
- Chhabra, P.; De Graaf, M.; Parra, G.I.; Chan, M.C.-W.; Green, K.; Martella, V.; Wang, Q.; White, P.A.; Katayama, K.; Vennema, H.; et al. Updated classification of norovirus genogroups and genotypes. J. Gen. Virol. 2019, 100, 1393–1406. [Google Scholar] [CrossRef]
- Kroneman, A.; Vega, E.; Vennema, H.; Vinje, J.; White, P.; Hansman, G.; Green, K.; Martella, V.; Katayama, K.; Koopmans, M. Proposal for a unified norovirus nomenclature and genotyping. Arch. Virol. 2013, 158, 2059–2068. [Google Scholar] [CrossRef]
- Kapikian, A.Z.; Wyatt, R.G.; Dolin, R.; Thornhill, T.S.; Kalica, A.R.; Chanock, R.M. Visualization by Immune Electron Microscopy of a 27-nm Particle Associated with Acute Infectious Nonbacterial Gastroenteritis. J. Virol. 1972, 10, 1075–1081. [Google Scholar] [CrossRef]
- Chan, M.C.-W.; Hu, Y.; Chen, H.; Podkolzin, A.T.; Zaytseva, E.V.; Komano, J.; Sakon, N.; Poovorawan, Y.; Vongpunsawad, S.; Thanusuwannasak, T.; et al. Global Spread of Norovirus GII.17 Kawasaki 308, 2014–2016. Emerg. Infect. Dis. 2017, 23, 1350–1354. [Google Scholar] [CrossRef] [PubMed]
- van Beek, J.; de Graaf, M.; Al-Hello, H.; Allen, D.J.; Ambert-Balay, K.; Botteldoorn, N.; Brytting, M.; Buesa, J.; Cabrerizo, M.; Chan, M.C.-W.; et al. Molecular surveillance of norovirus, 2005–2016: An epidemiological analysis of data collected from the NoroNet network. Lancet Infect. Dis. 2018, 18, 545–553. [Google Scholar] [CrossRef]
- Tohma, K.; Lepore, C.J.; Siltz, L.; Parra, G.I. Phylogenetic Analyses Suggest that Factors Other Than the Capsid Protein Play a Role in the Epidemic Potential of GII.2 Norovirus. mSphere 2017, 2, e00187-17. [Google Scholar] [CrossRef] [PubMed]
- Siebenga, J.; Vennema, H.; Zheng, D.; Vinjé, J.; Lee, B.E.; Pang, X.; Ho, E.C.M.; Lim, W.; Choudekar, A.; Broor, S.; et al. Norovirus Illness Is a Global Problem: Emergence and Spread of Norovirus GII.4 Variants, 2001–2007. J. Infect. Dis. 2009, 200, 802–812. [Google Scholar] [CrossRef] [PubMed]
- Noel, J.S.; Fankhauser, R.L.; Ando, T.; Monroe, S.; Glass, R.I. Identification of a Distinct Common Strain of “Norwalk-like Viruses” Having a Global Distribution. J. Infect. Dis. 1999, 179, 1334–1344. [Google Scholar] [CrossRef] [PubMed]
- White, P.A.; Hansman, G.S.; Li, A.; Dable, J.; Isaacs, M.; Ferson, M.; McIver, C.J.; Rawlinson, W.D. Norwalk-like virus 95/96-US strain is a major cause of gastroenteritis outbreaks in Australia. J. Med. Virol. 2002, 68, 113–118. [Google Scholar] [CrossRef]
- Lopman, B.; Vennema, H.; Kohli, E.; Pothier, P.; Sanchez, A.; Negredo, A.; Buesa, J.; Schreier, E.; Gray, J.; Gallimore, C.; et al. Increase in viral gastroenteritis outbreaks in Europe and epidemic spread of new norovirus variant. Lancet 2004, 363, 682–688. [Google Scholar] [CrossRef]
- Widdowson, M.; Cramer, E.H.; Hadley, L.; Bresee, J.S.; Beard, R.S.; Bulens, S.N.; Charles, M.; Chege, W.; Isakbaeva, E.; Wright, J.G.; et al. Outbreaks of Acute Gastroenteritis on Cruise Ships and on Land: Identification of a Predominant Circulating Strain of Norovirus—United States, 2002. J. Infect. Dis. 2004, 190, 27–36. [Google Scholar] [CrossRef]
- Bull, R.; Tu, E.T.V.; McIver, C.J.; Rawlinson, W.; White, P.A. Emergence of a New Norovirus Genotype II.4 Variant Associated with Global Outbreaks of Gastroenteritis. J. Clin. Microbiol. 2006, 44, 327–333. [Google Scholar] [CrossRef]
- Eden, J.-S.; Bull, R.A.; Tu, E.; McIver, C.J.; Lyon, M.J.; Marshall, J.A.; Smith, D.W.; Musto, J.; Rawlinson, W.D.; White, P.A. Norovirus GII.4 variant 2006b caused epidemics of acute gastroenteritis in Australia during 2007 and 2008. J. Clin. Virol. 2010, 49, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Yen, C.; Wikswo, M.E.; Lopman, B.A.; Vinje, J.; Parashar, U.D.; Hall, A.J. Impact of an Emergent Norovirus Variant in 2009 on Norovirus Outbreak Activity in the United States. Clin. Infect. Dis. 2011, 53, 568–571. [Google Scholar] [CrossRef] [PubMed]
- Van Beek, J.; Ambert-Balay, K.; Botteldoorn, N.; Eden, J.S.; Fonager, J.; Hewitt, J.; Iritani, N.; Kroneman, A.; Vennema, H.; Vinjé, J.; et al. Indications for worldwide increased norovirus activity associated with emergence of a new variant of genotype II.4, late 2012. Eurosurveillance 2013, 18, 8–9. [Google Scholar] [CrossRef] [PubMed]
- Lindesmith, L.C.; Beltramello, M.; Donaldson, E.F.; Corti, D.; Swanstrom, J.; Debbink, K.; Lanzavecchia, A.; Baric, R.S. Immunogenetic Mechanisms Driving Norovirus GII.4 Antigenic Variation. PLoS Pathog. 2012, 8, e1002705. [Google Scholar] [CrossRef] [PubMed]
- Bull, R.; Hansman, G.; Clancy, L.E.; Tanaka, M.M.; Rawlinson, W.; White, P.A. Norovirus Recombination in ORF1/ORF2 Overlap. Emerg. Infect. Dis. 2005, 11, 1079–1085. [Google Scholar] [CrossRef]
- Bull, R.; Mark, M.; UNSW Faculty of Science Biotechnology & Biomolecular Sciences Mark Tanaka; White, P. Norovirus recombination. J. Gen. Virol. 2007, 88, 3347–3359. [Google Scholar] [CrossRef] [PubMed]
- Supadej, K.; Khamrin, P.; Kumthip, K.; Kochjan, P.; Yodmeeklin, A.; Ushijima, H.; Maneekarn, N. Wide variety of recombinant strains of norovirus GII in pediatric patients hospitalized with acute gastroenteritis in Thailand during 2005 to 2015. Infect. Genet. Evol. 2017, 52, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Lindesmith, L.C.; Donaldson, E.F.; LoBue, A.D.; Cannon, J.L.; Zheng, D.-P.; Vinje, J.; Baric, R.S. Mechanisms of GII.4 Norovirus Persistence in Human Populations. PLoS Med. 2008, 5, e31. [Google Scholar] [CrossRef]
- Rohayem, J.; Münch, J.; Rethwilm, A. Evidence of Recombination in the Norovirus Capsid Gene. J. Virol. 2005, 79, 4977–4990. [Google Scholar] [CrossRef]
- Lam, T.T.-Y.; Zhu, H.; Smith, D.K.; Guan, Y.; Holmes, E.; Pybus, O. The recombinant origin of emerging human norovirus GII.4/2008: Intra-genotypic exchange of the capsid P2 domain. J. Gen. Virol. 2012, 93, 817–822. [Google Scholar] [CrossRef]
- Eden, J.-S.; Tanaka, M.M.; Boni, M.F.; Rawlinson, W.D.; White, P.A. Recombination within the Pandemic Norovirus GII.4 Lineage. J. Virol. 2013, 87, 6270–6282. [Google Scholar] [CrossRef] [PubMed]
- Mauroy, A.; Scipioni, A.; Mathijs, E.; Ziant, D.; Daube, G.; Thiry, E. Genetic and evolutionary perspectives on genogroup III, genotype 2 bovine noroviruses. Arch. Virol. 2014, 159, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, D.; Hutzenthaler, M.; Seebach, J.; Panning, M.; Umgelter, A.; Menzel, H.; Protzer, U.; Metzler, D. Norovirus GII.4 and GII.7 capsid sequences undergo positive selection in chronically infected patients. Infect. Genet. Evol. 2012, 12, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, S.M.; Lopman, B.A.; Hall, A.J.; Parashar, U.D.; Lee, B.Y. The potential economic value of a human norovirus vaccine for the United States. Vaccine 2012, 30, 7097–7104. [Google Scholar] [CrossRef]
- Simmons, K.; Gambhir, M.; Leon, J.; Lopman, B. Duration of Immunity to Norovirus Gastroenteritis. Emerg. Infect. Dis. 2013, 19, 1260–1267. [Google Scholar] [CrossRef] [PubMed]
- Thorne, L.G.; Goodfellow, I. Norovirus gene expression and replication. J. Gen. Virol. 2014, 95, 278–291. [Google Scholar] [CrossRef]
- Tan, M.; Hegde, R.S.; Jiang, X. The P Domain of Norovirus Capsid Protein Forms Dimer and Binds to Histo-Blood Group Antigen Receptors. J. Virol. 2004, 78, 6233–6242. [Google Scholar] [CrossRef]
- Choi, J.-M.; Hutson, A.M.; Estes, M.K.; Prasad, B.V.V. Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus. Proc. Natl. Acad. Sci. USA 2008, 105, 9175–9180. [Google Scholar] [CrossRef]
- Tan, M.; Jiang, X. Norovirus Gastroenteritis, Carbohydrate Receptors, and Animal Models. PLoS Pathog. 2010, 6, e1000983. [Google Scholar] [CrossRef] [PubMed]
- Glass, P.J.; White, L.J.; Ball, J.M.; Leparc-Goffart, I.; Hardy, M.E.; Estes, M.K. Norwalk Virus Open Reading Frame 3 Encodes a Minor Structural Protein. J. Virol. 2000, 74, 6581–6591. [Google Scholar] [CrossRef]
- Green, K. Caliciviridae: The noroviruses. In Fields Virology, 6th ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 582–608. [Google Scholar]
- Viskovska, M.A.; Zhao, B.; Shanker, S.; Choi, J.-M.; Deng, L.; Song, Y.; Palzkill, T.; Hu, L.; Estes, M.K.; Prasad, B.V.V. GII.4 Norovirus Protease Shows pH-Sensitive Proteolysis with a Unique Arg-His Pairing in the Catalytic Site. J. Virol. 2019, 93, 93. [Google Scholar] [CrossRef]
- Muzzarelli, K.M.; Kuiper, B.D.; Spellmon, N.; Brunzelle, J.S.; Hackett, J.; Amblard, F.; Zhou, S.; Liu, P.; Kovari, I.A.; Yang, Z.; et al. Structural and Antiviral Studies of the Human Norovirus GII.4 Protease. Biochemistry 2019, 58, 900–907. [Google Scholar] [CrossRef]
- Zeitler, C.E.; Estes, M.K.; Prasad, B.V.V. X-ray Crystallographic Structure of the Norwalk Virus Protease at 1.5-Å Resolution. J. Virol. 2006, 80, 5050–5058. [Google Scholar] [CrossRef]
- Nakamura, K.; Someya, Y.; Kumasaka, T.; Ueno, G.; Yamamoto, M.; Sato, T.; Takeda, N.; Miyamura, T.; Tanaka, N. A Norovirus Protease Structure Provides Insights into Active and Substrate Binding Site Integrity. J. Virol. 2005, 79, 13685–13693. [Google Scholar] [CrossRef] [PubMed]
- Hussey, R.J.; Coates, L.; Gill, R.S.; Erskine, P.T.; Coker, S.-F.; Mitchell, E.; Cooper, J.B.; Wood, S.; Broadbridge, R.; Clarke, I.N.; et al. A Structural Study of Norovirus 3C Protease Specificity: Binding of a Designed Active Site-Directed Peptide Inhibitor. Biochemistry 2011, 50, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Leen, E.; Baeza, G.; Curry, S. Structure of a Murine Norovirus NS6 Protease-Product Complex Revealed by Adventitious Crystallisation. PLoS ONE 2012, 7, e38723. [Google Scholar] [CrossRef]
- Someya, Y.; Takeda, N.; Miyamura, T. Identification of Active-Site Amino Acid Residues in the Chiba Virus 3C-Like Protease. J. Virol. 2002, 76, 5949–5958. [Google Scholar] [CrossRef]
- Ashkenazy, H.; Abadi, S.; Martz, E.; Chay, O.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016, 44, W344–W350. [Google Scholar] [CrossRef]
- Schechter, I.; Berger, A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967, 27, 157–162. [Google Scholar] [CrossRef]
- Wirblich, C.; Sibilia, M.; Boniotti, M.B.; Rossi, C.; Thiel, H.J.; Meyers, G. 3C-like protease of rabbit hemorrhagic disease virus: Identification of cleavage sites in the ORF1 polyprotein and analysis of cleavage specificity. J. Virol. 1995, 69, 7159–7168. [Google Scholar] [CrossRef] [PubMed]
- Sosnovtseva, S.A.; Sosnovtsev, S.V.; Green, K.Y. Mapping of the Feline Calicivirus Proteinase Responsible for Autocatalytic Processing of the Nonstructural Polyprotein and Identification of a Stable Proteinase-Polymerase Precursor Protein. J. Virol. 1999, 73, 6626–6633. [Google Scholar] [CrossRef]
- Liu, B.; Clarke, I.; Lambden, P.R. Polyprotein processing in Southampton virus: Identification of 3C-like protease cleavage sites by in vitro mutagenesis. J. Virol. 1996, 70, 2605–2610. [Google Scholar] [CrossRef]
- Liu, B.L.; Viljoen, G.J.; Clarke, I.; Lambden, P.R. Identification of further proteolytic cleavage sites in the Southampton calicivirus polyprotein by expression of the viral protease in E. coli. J. Gen. Virol. 1999, 80, 291–296. [Google Scholar] [CrossRef]
- Seah, E.L.; Marshall, J.A.; Wright, P.J. Open Reading Frame 1 of the Norwalk-Like Virus Camberwell: Completion of Sequence and Expression in Mammalian Cells. J. Virol. 1999, 73, 10531–10535. [Google Scholar] [CrossRef]
- E Hardy, M.; Crone, T.J.; E Brower, J.; Ettayebi, K. Substrate specificity of the Norwalk virus 3C-like proteinase. Virus Res. 2002, 89, 29–39. [Google Scholar] [CrossRef]
- Blakeney, S.J.; Cahill, A.; A Reilly, P. Processing of Norwalk virus nonstructural proteins by a 3C-like cysteine proteinase. Virology 2003, 308, 216–224. [Google Scholar] [CrossRef]
- Muhaxhiri, Z.; Deng, L.; Shanker, S.; Sankaran, B.; Estes, M.K.; Palzkill, T.; Song, Y.; Prasad, B.V.V. Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus. J. Virol. 2013, 87, 4281–4292. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ménard, R.; Storer, A.C. Oxyanion Hole Interactions in Serine and Cysteine Proteases. Biol. Chem. Hoppe-Seyler 1992, 373, 393–400. [Google Scholar] [CrossRef]
- Dragovich, P.S.; Webber, S.E.; Babine, R.E.; Fuhrman, S.A.; Patick, A.K.; Matthews, D.A.; Lee, C.A.; Reich, S.H.; Prins, T.J.; Marakovits, J.T.; et al. Structure-Based Design, Synthesis, and Biological Evaluation of Irreversible Human Rhinovirus 3C Protease Inhibitors. 1. Michael Acceptor Structure−Activity Studies. J. Med. Chem. 1998, 41, 2806–2818. [Google Scholar] [CrossRef]
- Tiew, K.-C.; He, G.; Aravapalli, S.; Mandadapu, S.R.; Gunnam, M.R.; Alliston, K.R.; Lushington, G.; Kim, Y.; Chang, K.-O.; Groutas, W.C. Design, synthesis, and evaluation of inhibitors of Norwalk virus 3C protease. Bioorg. Med. Chem. Lett. 2011, 21, 5315–5319. [Google Scholar] [CrossRef]
- Deng, L.; Muhaxhiri, Z.; Estes, M.K.; Palzkill, T.; Prasad, B.; Song, Y. Synthesis, activity and structure–activity relationship of noroviral protease inhibitors. MedChemComm 2013, 4, 1354–1359. [Google Scholar] [CrossRef]
- Amblard, F.; Zhou, S.; Liu, P.; Yoon, J.; Cox, B.; Muzzarelli, K.; Kuiper, B.D.; Kovari, L.C.; Schinazi, R.F. Synthesis and antiviral evaluation of novel peptidomimetics as norovirus protease inhibitors. Bioorg. Med. Chem. Lett. 2018, 28, 2165–2170. [Google Scholar] [CrossRef]
- Kankanamalage, A.C.G.; Kim, Y.; Weerawarna, P.; Uy, R.A.Z.; Damalanka, V.C.; Mandadapu, S.R.; Alliston, K.R.; Mehzabeen, N.; Battaile, K.; Lovell, S.; et al. Structure-Guided Design and Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease. Structure–Activity Relationships and Biochemical, X-ray Crystallographic, Cell-Based, and In Vivo Studies. J. Med. Chem. 2015, 58, 3144–3155. [Google Scholar] [CrossRef]
- Mandadapu, S.R.; Weerawarna, P.; Gunnam, M.R.; Alliston, K.R.; Lushington, G.H.; Kim, Y.; Chang, K.-O.; Groutas, W.C. Potent inhibition of norovirus 3CL protease by peptidyl α-ketoamides and α-ketoheterocycles. Bioorg. Med. Chem. Lett. 2012, 22, 4820–4826. [Google Scholar] [CrossRef]
- Mandadapu, S.R.; Gunnam, M.R.; Kankanamalage, A.C.G.; Uy, R.A.Z.; Alliston, K.R.; Lushington, G.H.; Kim, Y.; Chang, K.-O.; Groutas, W.C. Potent inhibition of norovirus by dipeptidyl α-hydroxyphosphonate transition state mimics. Bioorg. Med. Chem. Lett. 2013, 23, 5941–5944. [Google Scholar] [CrossRef]
- Patel, D.V.; Rielly-Gauvin, K.; Ryono, D.E.; Free, C.A.; Rogers, W.L.; Smith, S.A.; DeForrest, J.M.; Oehl, R.S.; Petrillo, E.W., Jr. Alpha-Hydroxy Phosphinyl-Based Inhibitors of Human Renin. J. Med. Chem. 1995, 38, 4557–4569. [Google Scholar] [CrossRef]
- Mandadapu, S.R.; Gunnam, M.R.; Tiew, K.-C.; Uy, R.A.Z.; Prior, A.M.; Alliston, K.R.; Hua, D.H.; Kim, Y.; Chang, K.-O.; Groutas, W.C. Inhibition of norovirus 3CL protease by bisulfite adducts of transition state inhibitors. Bioorg. Med. Chem. Lett. 2013, 23, 62–65. [Google Scholar] [CrossRef]
- Kankanamalage, A.C.G.; Kim, Y.; Rathnayake, A.D.; Alliston, K.R.; Butler, M.M.; Cardinale, S.C.; Bowlin, T.L.; Groutas, W.C.; Chang, K.-O. Design, Synthesis, and Evaluation of Novel Prodrugs of Transition State Inhibitors of Norovirus 3CL Protease. J. Med. Chem. 2017, 60, 6239–6248. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lovell, S.; Tiew, K.-C.; Mandadapu, S.R.; Alliston, K.R.; Battaile, K.; Groutas, W.C.; Chang, K.-O. Broad-Spectrum Antivirals against 3C or 3C-Like Proteases of Picornaviruses, Noroviruses, and Coronaviruses. J. Virol. 2012, 86, 11754–11762. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, D.; Kim, Y.; Lovell, S.; Prakash, O.; Groutas, W.C.; Chang, K.-O. Structural and inhibitor studies of norovirus 3C-like proteases. Virus Res. 2013, 178, 437–444. [Google Scholar] [CrossRef]
- McGeary, R.P.; Fairlie, D.P. Macrocyclic peptidomimetics: Potential for drug development. Curr. Opin. Drug Discov. Dev. 1998, 1, 208–217. [Google Scholar]
- Mandadapu, S.R.; Weerawarna, P.; Prior, A.M.; Uy, R.A.Z.; Aravapalli, S.; Alliston, K.R.; Lushington, G.; Kim, Y.; Hua, D.H.; Chang, K.-O.; et al. Macrocyclic inhibitors of 3C and 3C-like proteases of picornavirus, norovirus, and coronavirus. Bioorg. Med. Chem. Lett. 2013, 23, 3709–3712. [Google Scholar] [CrossRef]
- Damalanka, V.C.; Kim, Y.; Alliston, K.R.; Weerawarna, P.M.; Kankanamalage, A.C.G.; Lushington, G.H.; Mehzabeen, N.; Battaile, K.P.; Lovell, S.; Chang, K.-O.; et al. Oxadiazole-Based Cell Permeable Macrocyclic Transition State Inhibitors of Norovirus 3CL Protease. J. Med. Chem. 2016, 59, 1899–1913. [Google Scholar] [CrossRef]
- Weerawarna, P.; Kim, Y.; Kankanamalage, A.C.G.; Damalanka, V.C.; Lushington, G.; Alliston, K.R.; Mehzabeen, N.; Battaile, K.; Lovell, S.; Chang, K.-O.; et al. Structure-based design and synthesis of triazole-based macrocyclic inhibitors of norovirus protease: Structural, biochemical, spectroscopic, and antiviral studies. Eur. J. Med. Chem. 2016, 119, 300–318. [Google Scholar] [CrossRef] [PubMed]
- Damalanka, V.C.; Kim, Y.; Kankanamalage, A.C.G.; Lushington, G.; Mehzabeen, N.; Battaile, K.; Lovell, S.; Chang, K.-O.; Groutas, W.C. Design, synthesis, and evaluation of a novel series of macrocyclic inhibitors of norovirus 3CL protease. Eur. J. Med. Chem. 2017, 127, 41–61. [Google Scholar] [CrossRef]
- Kankanamalage, A.C.G.; Weerawarna, P.M.; Rathnayake, A.D.; Kim, Y.; Mehzabeen, N.; Battaile, K.P.; Lovell, S.; Chang, K.; Groutas, W.C. Putative structural rearrangements associated with the interaction of macrocyclic inhibitors with norovirus 3CL protease. Proteins Struct. Funct. Bioinform. 2019, 87, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.-O.; Takahashi, D.; Prakash, O.; Kim, Y. Characterization and inhibition of norovirus proteases of genogroups I and II using a fluorescence resonance energy transfer assay. Virology 2012, 423, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Hernandez, M.J.; Pal, A.; Gyan, K.E.; Charbonneau, M.-E.; Showalter, H.D.; Donato, N.J.; O’Riordan, M.; Wobus, C.E. Chemical Derivatives of a Small Molecule Deubiquitinase Inhibitor Have Antiviral Activity against Several RNA Viruses. PLoS ONE 2014, 9, e94491. [Google Scholar] [CrossRef] [PubMed]
- Perry, J.W.; Ahmed, M.; Chang, K.-O.; Donato, N.J.; Showalter, H.D.; Wobus, C.E. Antiviral Activity of a Small Molecule Deubiquitinase Inhibitor Occurs via Induction of the Unfolded Protein Response. PLoS Pathog. 2012, 8, e1002783. [Google Scholar] [CrossRef]
- Rocha-Pereira, J.; Nascimento, M.S.J.; Ma, Q.; Hilgenfeld, R.; Neyts, J.; Jochmans, D. The Enterovirus Protease Inhibitor Rupintrivir Exerts Cross-Genotypic Anti-Norovirus Activity and Clears Cells from the Norovirus Replicon. Antimicrob. Agents Chemother. 2014, 58, 4675–4681. [Google Scholar] [CrossRef]
- Kitano, M.; Hosmillo, M.; Emmott, E.; Lu, J.; Goodfellow, I. Selection and Characterization of Rupintrivir-Resistant Norwalk Virus Replicon Cells In Vitro. Antimicrob. Agents Chemother. 2018, 62, 62. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, K.; Matsushima, Y.; Nagasawa, K.; Aso, J.; Saraya, T.; Yoshihara, K.; Murakami, K.; Motoya, T.; Ryo, A.; Kuroda, M.; et al. Molecular Evolution of the Protease Region in Norovirus Genogroup II. Front. Microbiol. 2020, 10, 2991. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Douangamath, A.; Song, W.; Coker, A.R.; Chan, A.E.; Wood, S.P.; Cooper, J.B.; Resnick, E.; London, N.; von Delft, F. In crystallo-screening for discovery of human norovirus 3C-like protease inhibitors. J. Struct. Biol. X 2020, 4, 100031. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, P.; May, J.; Uhm, S.; Yon, C.; Korba, B. RNA binding by human Norovirus 3C-like proteases inhibits proteaseactivity. Virology 2013, 438, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Wobus, C.; Karst, S.M.; Thackray, L.B.; Chang, K.-O.; Sosnovtsev, S.V.; Belliot, G.; Krug, A.; Mackenzie, J.; Green, K.Y.; Virgin, H.W. Replication of Norovirus in Cell Culture Reveals a Tropism for Dendritic Cells and Macrophages. PLoS Biol. 2004, 2, e432. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.K.; Grau, K.R.; Costantini, V.; Kolawole, A.; De Graaf, M.; Freiden, P.; Graves, C.; Koopmans, M.; Wallet, S.M.; Tibbetts, S.A.; et al. Human norovirus culture in B cells. Nat. Protoc. 2015, 10, 1939–1947. [Google Scholar] [CrossRef]
- Ettayebi, K.; Crawford, S.E.; Murakami, K.; Broughman, J.R.; Karandikar, U.; Tenge, V.; Neill, F.H.; Blutt, S.E.; Zeng, X.-L.; Qu, L.; et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 2016, 353, 1387–1393. [Google Scholar] [CrossRef]
- Van Dycke, J.; Ny, A.; Conceição-Neto, N.; Maes, J.; Hosmillo, M.; Cuvry, A.; Goodfellow, I.; Nogueira, T.C.; Verbeken, E.; Matthijnssens, J.; et al. A robust human norovirus replication model in zebrafish larvae. PLoS Pathog. 2019, 15, e1008009. [Google Scholar] [CrossRef]
- Van Dycke, J.; Cuvry, A.; Knickmann, J.; Ny, A.; Rakers, S.; Taube, S.; Witte, P.D.; Neyts, J.; Rocha-Pereir, J. Infection of zebrafish larvae with human norovirus and evaluation of the in vivo efficacy of small-molecule inhibitors. Nat. Protoc. 2021, 16, 1830–1849. [Google Scholar] [CrossRef]
- Van Dycke, J.; Dai, W.; Stylianidou, Z.; Li, J.; Cuvry, A.; Roux, E.; Li, B.; Rymenants, J.; Bervoets, L.; Witte, P.D.; et al. A Novel Class of Norovirus Inhibitors Targeting the Viral Protease with Potent Antiviral Activity In Vitro and In Vivo. Viruses 2021, 13, 1852. [Google Scholar] [CrossRef]
- Karandikar, U.C.; Crawford, S.E.; Ajami, N.J.; Murakami, K.; Kou, B.; Ettayebi, K.; Papanicolaou, G.; Jongwutiwes, U.; Perales, M.-A.; Shia, J.; et al. Detection of human norovirus in intestinal biopsies from immunocompromised transplant patients. J. Gen. Virol. 2016, 97, 2291–2300. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.R.; Gilmour, K.; Breuer, J. Norovirus Infections Occur in B-Cell–Deficient Patients: Table 1. Clin. Infect. Dis. 2016, 62, 1136–1138. [Google Scholar] [CrossRef] [PubMed]
- Rolfes, M.; Sriaroon, P.; Saldaña, B.D.; Dvorak, C.; Chapdelaine, H.; Ferdman, R.; Chen, K.; Jolles, S.; Patel, N.; Kim, Y.; et al. Chronic norovirus infection in primary immune deficiency disorders: An international case series. Diagn. Microbiol. Infect. Dis. 2019, 93, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Wobus, C.; Cunha, J.; Elftman, M.; Kolawole, A. Animal Models of Norovirus Infection. In Perspectives in Medical Virology; Elsevier BV: Amsterdam, The Netherlands, 2016; pp. 397–422. [Google Scholar]
- Taube, S.; Kolawole, A.; Höhne, M.; Wilkinson, J.E.; Handley, S.A.; Perry, J.W.; Thackray, L.B.; Akkina, R.; Wobus, C.E. A Mouse Model for Human Norovirus. mBio 2013, 4, 4. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, B.; Hu, L.; Song, Y.; Patil, K.; Ramani, S.; Atmar, R.L.; Estes, M.K.; Prasad, B.V.V. Norovirus Protease Structure and Antivirals Development. Viruses 2021, 13, 2069. https://doi.org/10.3390/v13102069
Zhao B, Hu L, Song Y, Patil K, Ramani S, Atmar RL, Estes MK, Prasad BVV. Norovirus Protease Structure and Antivirals Development. Viruses. 2021; 13(10):2069. https://doi.org/10.3390/v13102069
Chicago/Turabian StyleZhao, Boyang, Liya Hu, Yongcheng Song, Ketki Patil, Sasirekha Ramani, Robert L. Atmar, Mary K. Estes, and B. V. Venkataram Prasad. 2021. "Norovirus Protease Structure and Antivirals Development" Viruses 13, no. 10: 2069. https://doi.org/10.3390/v13102069
APA StyleZhao, B., Hu, L., Song, Y., Patil, K., Ramani, S., Atmar, R. L., Estes, M. K., & Prasad, B. V. V. (2021). Norovirus Protease Structure and Antivirals Development. Viruses, 13(10), 2069. https://doi.org/10.3390/v13102069





