Abstract
Since the beginning of the 20th century, bacteriophages (phages), i.e., viruses that infect bacteria, have been used as antimicrobial agents for treating various infections. Phage preparations targeting a number of bacterial pathogens are still in use in the post-Soviet states and are experiencing a revival in the Western world. However, phages have never been used to treat diseases caused by Bacteroides fragilis, the leading agent cultured in anaerobic abscesses and postoperative peritonitis. Enterotoxin-producing strains of B. fragilis have been associated with the development of inflammatory diarrhea and colorectal carcinoma. In this study, we evaluated the molecular biosafety and antimicrobial properties of novel phage species vB_BfrS_VA7 (VA7) lysate, as well as its impact on cytokine IL-8 production in an enterotoxigenic B. fragilis (ETBF)-infected colonic epithelial cell (CEC) culture model. Compared to untreated infected cells, the addition of phage VA7 to ETBF-infected CECs led to significantly reduced bacterial counts and IL-8 levels. This in vitro study confirms the potential of phage VA7 as an antibacterial agent for use in prophylaxis or in the treatment of B. fragilis infections and associated colorectal carcinoma.
1. Introduction
Bacteriophages (phages), i.e., viruses that infect bacterial cells, were discovered at the beginning of the 20th century and were immediately used as therapeutic agents to fight bacterial infections. Phages against Escherichia coli, Shigella dysenteriae and Vibrio cholerae were among the first known antimicrobial agents [1]. There have been examples of phage therapy being used effectively to fight various acute and chronic infections [2]. However, we found no such indications in the scientific literature of phage-based treatments of Bacteroides fragilis infections including internal organ abscesses, postoperative peritonitis, sepsis, inflammatory diarrhea or colorectal carcinoma (CRC). Species within the genus Bacteroides, such as B. fragilis, B. thetaiotaomicron and B. vulgatus, are well-known residents of the natural microbiome of the human and mammalian gastrointestinal (GI) tract mucosa [3]. In these niches, they are responsible for the digestion of cellulose and the production of short-chain fatty acids, while also playing a key role in the maturation of the immune system [4]. Regardless of the numerous beneficial properties, B. fragilis also has a pathogenic nature, which is primarily associated with its spread outside the GI lumen following disease, gut perforation, abdominal surgery or trauma. Dislocation of the Bacteroides species from the intestinal wall may provoke an inflammatory response and abscess formation in the distant regions of their dissemination, which can be complicated by sepsis [5,6]. Anaerobic abscesses are usually polymicrobial, but one of the most commonly cultured agents is B. fragilis, which is suggested to be the most virulent representative of the genus [7]. More specifically, enterotoxigenic B. fragilis (ETBF) is associated with inflammatory diarrhea and identified as a risk factor for CRC [8,9,10]. Although antibiotics (e.g., broad-spectrum β-lactams and metronidazole) are generally effective [4], recent studies have shown increased isolation of multidrug-resistant B. fragilis strains from the normal microbiome of the human intestines [11].
Increased virulence of B. fragilis strains is generally associated with the presence of the bft gene in chromosomal pathogenicity island BFT PAI. The bft gene codes for fragilysin, a zinc–metalloprotease enterotoxin. Three variants or isoforms of the gene, named bft-1, bft-2 and bft-3, have been identified and sequenced, in which bft-1 was the most prevalent form in humans [12]. This B. fragilis toxin (BFT) interacts with epithelial-tight junctions of colonic epithelial cells (CECs) by binding to yet-unidentified receptors [13]. The outcome is the cleavage of intercellular E-cadherin, leading to increased permeability of the epithelial barrier and enhanced activation of the Wnt/β catenin and NF-κB cell-signaling pathways in CECs [14]. In mice, these pathways are shown to induce chemokines to recruit polymorphonuclear immature myeloid cells, with parallel triggering of ETBF-mediated distal colon tumorigenesis [15]. In addition, BFT has been associated with in vitro production of reactive oxygen species and DNA damage—two events linked to carcinogenesis [16]. The B. fragilis toxin is also shown to cause increased secretion of IL-8 in human intestinal epithelial cell cultures, which may be a provoking factor for the development of inflammatory diarrhea [14,17]. Cytokine IL-8 is also highly expressed during tumor angiogenesis, leading to the progression of cancer formation [18], and has been found to increase rapidly in ETBF-infected patients [9].
In general, CRC gut microbiota exhibit a compositional shift (dysbiosis), which includes the presence of B. fragilis, compared with the microbiota of healthy persons [19]. In 2018, Hannigan and colleagues observed that CRC-associated viromes consisted primarily of temperate phages. They concluded that these phage communities were associated with colorectal cancer and potentially impacted cancer progression by modulating the bacterial host communities towards CRC-associated gut microbiota dysbiosis [20]. Conversely, phages are increasingly considered a promising therapeutic tool against pathogenic gastrointestinal bacteria for their potential to revert dysbiosis of the gut microbiota [21].
In Georgia, sterile bacterial phage lysates, rather than purified phage preparations, are used in oral and topical phage therapies. Since the present study serves to yield preliminary information in view of possible future experimental phage therapies in Georgia, the use of sterile phage lysates was warranted. Our study aimed to assess the potential of a B. fragilis phage vB_BfrS_VA7 (VA7) lysate, previously identified as the best candidate for in vitro cell culture experiments [22], in preventing or treating inflammatory ETBF infections and, ultimately, in the prevention of CRC. Previous studies revealed a strictly virulent nature of phage VA7 attributed to fragilis-specific phages with a siphovirus morphology (Figure 1). In this preliminary study, we decided to focus on two features of this phage lysate: (a) its antimicrobial properties and (b) its impact on CEC cytokine IL-8 production in an ETBF-infected CEC model.
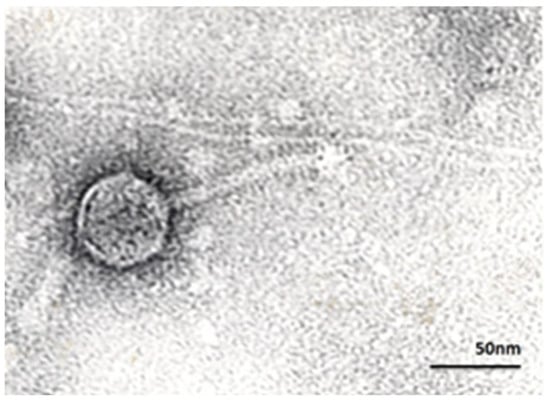
Figure 1.
Transmission electron micrograph of B. fragilis-specific phage VA7, exhibiting a siphovirus morphology with an icosahedral head of 60 nm and a tail of 100 nm. Magnification × 250,000.
2. Materials and Methods
2.1. Identification of ETBF Strains Using PCR
PCR was used to screen thirty B. fragilis isolates for the presence of the bft gene to help select an adequate ETBF strain for further use in the in vitro studies. Twenty-two B. fragilis isolates were obtained from fecal samples of patients with intra-abdominal infections, peritonitis and abscesses [22]. Eight B. fragilis strains were obtained from the strain collection at the Laboratory for Bacteriology Research (Ghent University, Ghent, Belgium). DNA was extracted from the bacterial strains using the alkaline lysis method. Briefly, approximately half of a large freshly grown B. fragilis colony was suspended in 20 µL alkaline lysis buffer (0.25% SDS, 0.05 N NaOH) and incubated for 15 min at 95 °C. After incubation, samples were centrifuged at 6000× g for 5 s and 180 µL of ultrapure water was added to each tube. Samples were centrifuged at the same speed for 5 min to remove the debris. The supernatants were stored for at least 30 min at −20 °C until analysis. PCR-based identification of the bft gene was performed according to Aitchison et al. [23]. Forward (5′–3′) GGATACATCAGCTGGGTTGTAG and reverse (5′–3′) GCGAACTCGGTTTATGCAGTGCGAAC primers (Integrated DNA Technologies, Leuven, Belgium) were selected to amplify all three subtypes of the bft gene, generating a 296 bp PCR product [20]. PCR master mix was prepared using the FastStart™ High Fidelity PCR System (Roche, Basel, Switzerland), according to the manufacturer’s instructions. PCR cycling conditions were as follows: 15 min of incubation at 95 °C, followed by 35 cycles of denaturation at 95 °C for 30 s, 30 s annealing at 55 °C and 30 s of extension at 72 °C, with a final 2 min extension phase at 72 °C.
2.2. Selection and Propagation of an ETBF Host Strain for Phage VA7
A spot test assay was performed to study the activity of phage VA7 against the identified ETBF strains. Bacterial strains were grown in enriched Brain Heart Infusion (BHI) broth (Liofilchem, Roseto degli Abruzzi, Italy). When reaching the exponential growth phase (18 to 24 h after inoculation), bacterial streaks were drawn on Petri dishes with BHI solid medium, using 10 μL loops. Drops of the phage lysates (10 µL) with a titer of 108 PFU/mL were spotted on the air-dried bacterial streaks. The plates were incubated at 37 °C for 18–24 h, after which the visible lytic plaques were evaluated [22]. The productive infectivity of the phages (efficiency of plating (EOP)) was also studied [22]. Determination of the EOP allowed us to define whether phage VA7 was able to replicate in the ETBF strains [24]. One milliliter of 109, 108, 107, 106 and 105 PFU/mL dilutions of each phage was added to 200 μL of 108 CFU/mL of bacteria. After an incubation of up to 10 min at room temperature, 2 mL of semi-solid BHI medium was added, mixed on a shaker and plated on the BHI agar plates. Following overnight incubation, the number of PFUs was counted for each phage dilution. The EOP of each strain was calculated by dividing the number of PFUs formed on the target bacterial strain by the number of PFUs generated by the non-ETBF reference strain A7 [24].
Phage VA7 lysate solutions were prepared by collecting the top layer of double-layer agar plates containing 105 PFU/mL of phages pre-incubated for 24 h with the host bacterial strain A7. The collected suspension was centrifuged at 6000× g for 30 min at 4 °C. The supernatant was filtered through 0.45 µm filters and stored at 4 °C until use. Before applying phages to the cell culture, the lysates were filtered using 0.22 µm filters. The titer of the filtered solution was determined using the double-layer agar method and plated on the BHI solid medium to evaluate the sterility of the phage stocks. The stock phage VA7 lysates produced for the genome sequencing and CEC experiments had a titer of 1010 PFU/mL.
2.3. DNA Sequencing and Analysis of VA7 Genome
Phage VA7 DNA was extracted from a high-titer phage stock, as described previously [25]. Subsequently, its genome was sequenced using an Illumina MiSeq device (Illumina, San Diego, CA, USA) at the VIB Nucleomics Core (Leuven, Belgium). After sequencing, the raw reads were trimmed (Trimmomatic) and assembled (SPAdes) in one contig [26,27]. Using MEGA X [28], the assembled phage genome was aligned to the closest characterized phage as identified by BLASTn [29] and a ViPTree [30] analysis (Bacteroides phage B124-14; NC_016770). The phage’s taxonomy was further delineated using VIRIDIC [31], which calculates the virus intergenomic distance. Next, the VA7 genome was annotated using RASTtk [32] and manually curated by BLASTp. tRNAs were identified with tRNAscan-SE v2.0. [33,34]. Finally, the phage genome was visualized using EasyFig [34].
2.4. One-Step Growth Curve
The phage VA7 one-step growth curve was evaluated to determine the burst size of the infected host cells. The experiment was accomplished according to Kropinski [35]. Bacterial strain E3 was grown in 5 mL of BHI broth for 18 h at 37 °C in a 10% CO2 atmosphere. An exponential growth phase E3 culture was diluted in BHI broth (enriched with 1 mM of CaCl2) to reach 1 × 107 CFU/mL as a final concentration. The phage VA7 lysate was added to the bacterial culture to attain a multiplicity of infection (MOI) of 0.01 (i.e., the final phage titer was equal to 1 × 105 PFU/mL). The phage–bacterial culture was incubated in a water bath at 37 °C for 10 min to achieve maximal phage to host cell adsorption. Afterwards, 0.1 mL of this mixture was taken to make ten-fold dilutions of up to 1 × 101 PFU/mL. At the same time, 0.01 μL of CHCl3 was added to 1 mL of the 1 × 102 PFU/mL dilution to serve as an adsorption control, which was stored on ice till the end of the experiment. The 1 × 103, 1 v 102 and 1 × 101 PFU/mL dilutions of the phage-bacteria mixture were incubated at 37 °C in a water bath for 60 min. Every five to ten minutes 0.1 mL from each dilution was taken to be mixed with 0.2 mL of E3 bacterial culture and 0.6% BHI overlay agar, and poured on a 1.5% BHI solid agar medium. A volume of 0.1 mL of adsorption control was also plated using the same double agar layer method. The plates were incubated for 24 h at 37 °C in a 10% CO2 atmosphere. The number of the infected cell and the burst size were determined as described by Kropinski [35].
2.5. Effect of Phage VA7 Lysate on ETBF-Infected CEC Cultures
To evaluate the effect of phage VA7 lysate on ETBF-infected CEC cultures, we adapted the model developed by Khan Mirzaei et al. [36]. Colon epithelial cell (CEC) culture line HCT 116, kindly provided by the laboratory of Klinikum rechts der Isar, Technical University of Munich, Germany, was grown in 6-well plates using 10% FCS/DMEM medium (Sigma Aldrich, St. Louis, MA, USA), without addition of antibiotics. After reaching >95% of confluence, CECs were washed twice with PBS buffer and the selected ETBF strain E3 with a final titer of 108 CFU/mL in sterile 10% FCS/DMEM medium was added to each well in a final volume of 3 mL. Blank controls consisted of 3 mL of sterile 10% FCS/DMEM medium without bacteria. For the bacteria to adhere, the CEC cultures were incubated for 3 h at 37 °C in a 5% CO2 atmosphere. After incubation, the CEC cultures were washed twice with 1 mL of PBS buffer to remove unattached bacterial cells. Subsequently, 3 mL of phage VA7 lysate (phage stock diluted 1000-fold in 10% FCS/DMEM medium to a final titer of 107 PFU/mL) was applied to the CEC cultures with the already-adhered bacterial cells at an MOI of 0.1. After incubation for another 3 h, the medium was collected and stored at −80 °C for later measurement of IL-8 levels. To remove the CECs from the bottom of the wells, 1 mL of 0.1% of Tween 20 diluted in PBS was added, followed by incubation for 5 min at 37 °C in a 5% CO2 environment. Subsequently, the detached cells were diluted ten-fold in the BHI liquid medium and 100 μL was plated on BHI solid medium for bacterial counts. A sandwich ELISA kit was used to measure IL-8 (Abcam, Cambridge, UK) levels, according to the manufacturer’s instructions. Figure 2 summarizes the experimental setup.
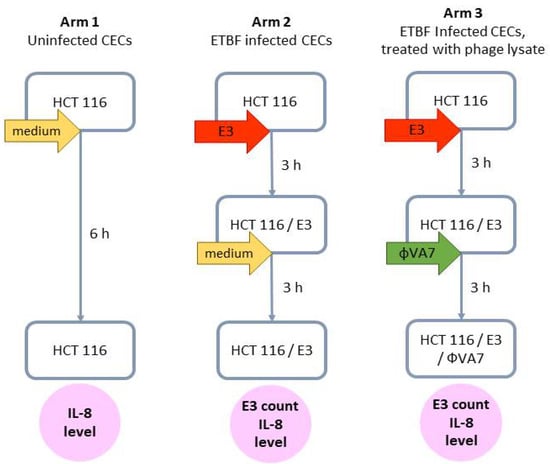
Figure 2.
Scheme of the experimental design to evaluate the effect of phage VA7 lysate on enterotoxigenic B. fragilis (ETBF) infection and IL-8 production in colonic epithelial cell (CEC) cultures. For each arm, two distinct experiments (biological replicates) were performed, and for each of these, four samples (technical replicates) were analyzed. In other words, each data point was the result of eight analyses.
2.6. Statistical Analysis
The Kruskal–Wallis test was used to evaluate differences in B. fragilis cell counts in the various experimental groups, and the one-way analysis of variance (ANOVA), as well as post hoc (Tukey HSD) and Levene’s tests, were run to evaluate differences in IL-8 release levels. The results are presented as mean values (of eight analyses) with error bars representing the 95% confidence intervals (CIs) of the means. We regarded statistical differences to be significant when p < 0.05.
3. Results and Discussion
Phage VA7 was previously identified as a potential candidate for phage therapy [22]. To evaluate its molecular biosafety, its genome sequence was determined and analyzed. The 47,095 bp double-stranded DNA genome of VA7 has a GC content of 38.53% and shows nucleotide similarity to Bacteroides phages B124-14 (96.65% sequence identity, 84% query coverage) [37], Barc2635 (94.80% sequence identity, 87% query coverage), vB_BfrS_23 (94.66% sequence identity, 85% coverage) and B40-8 (94.58% sequence identity, 74% query coverage) [38], all of which are currently unclassified Siphoviridae members. A proteome analysis using ViPTree confirmed this result. Next, the virus intergenomic distance between the most related phages and VA7 was calculated (Supplementary Figure S1), showing that Bacteroides phages VA7, B124-14, Barc2635, vB_BfrS_23 and B40-8 form five novel species within one novel unclassified Siphoviridae genus according to the International Committee for the Taxonomy of Viruses (ICTV) guidelines [39,40].
Next, the VA7 genome was annotated, revealing 68 coding sequences and no predicted tRNAs (Figure 3). The annotated genome was submitted to NCBI and is available through Genbank accession number MW916539.1. No lysogeny-related proteins are encoded on the VA7 genome, indicating its strictly lytic character. Moreover, no known virulence- or antibiotic resistance-associated genes were identified, suggesting that phage VA7 could be considered as a suitable candidate for phage therapy purposes, even though the majority of the coding sequences (72%) remain of unknown function.
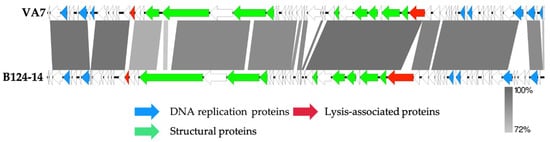
Figure 3.
Genome map of the sequenced Bacteroides phage VA7 and comparison using a BLASTn analysis (greyscale) to the closest related phage B124-14. Each arrow represents a coding sequence. In red, genes encoding packaging and lysis-associated proteins are displayed, while green shows the structural proteins and blue the DNA- and metabolism-associated proteins (adapted from EasyFig).
The phage VA7 one-step growth curve showed a latent period of about 20 min and a burst size at 30 min of about three hundred particles per infected bacterial cell (Figure 4).
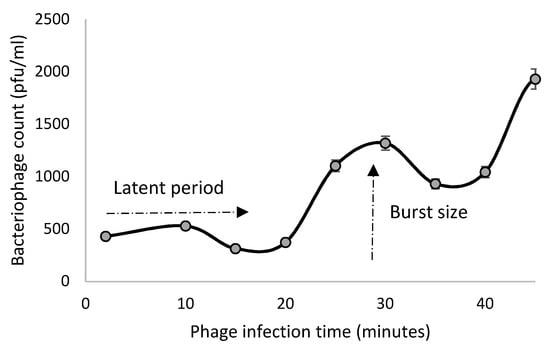
Figure 4.
One-step growth curve of phage VA7 in bacterial host A7. The evolution of phage lytic activity (PFU/mL) over time (min) is shown. The data points represent the mean of three experiments with the error bars.
To identify the enterotoxigenic B. fragilis strains in our previously described collection [22], the thirty available strains were analyzed for the presence of the bft gene by a PCR assay. This led to the amplification of a bft gene fragment with a correct size (296 bp) in four out of the thirty isolates: E1, E3, S10 and 33.
Among these, ETBF strain E3 was shown to be the most suitable host for phage VA7, being susceptible to the phage in a spot test assay and exhibiting an EOP of 0.1, which is indicative of an efficient production of new VA7 virions (productive infection) in the E3 strain. The virulence potential of strain E3 towards CECs was evaluated using a cytotoxicity assay (Supplementary Figure S2). Although ETBF strain S10 had an EOP similar to E3, the latter produced clearer lytic plaques. Phage VA7 showed lytic activity on ETBF strains E1 and 33, but no propagation was observed [22]. Therefore, the E3 strain showing susceptibility to phage VA7 was selected as a test strain in the cell culture experiments, as well as for the propagation of phage lysates used in the cell culture experiments.
Phage VA7 lysate was then evaluated for its antibacterial properties on ETBF-infected CEC cultures. Here, we observed a statistically significant (p < 0.05) decrease in bacterial cell counts in the phage-treated arm compared to the untreated infected cell culture arm. The mean bacterial cell count in the untreated ETBF (strain E3)-infected arm was 5 × 104 CFU/mL, while the mean bacterial count in the phage VA7 lysate-treated arm was about 2 log units lower (4.2 × 102 CFU/mL) (Figure 5a).
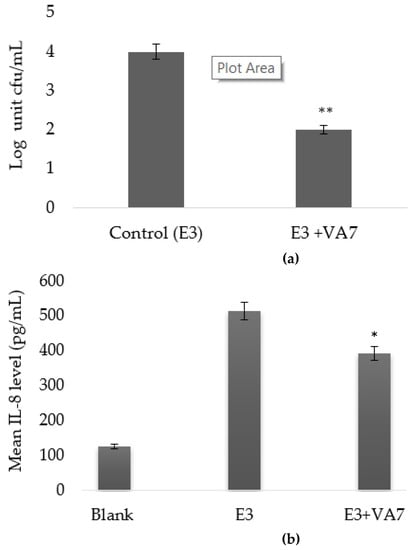
Figure 5.
Bactericidal effect (a) and IL-8 modulating effect (b) of VA7 lysate on HCT116 colonic epithelial cell (CEC) cultures infected with enterotoxigenic B. fragilis (ETBF) strain E3. Bacterial counts and IL-8 levels are presented as mean values with error bars representing the 95% confidence intervals (CIs) of the means. A base-10 logarithmic scale is used for the Y-axis of graph a. A statistically significant difference is indicated as * p < 0.05 and ** p < 0.01.
The immunomodulating effect of VA7 was also investigated in the CEC cultures. A statistically significant difference in IL-8 concentrations was observed between the phage VA7 lysate-treated arm and the untreated arm. With a mean level of 391.99 pg/mL, IL-8 production was significantly lower (p < 0.001) in the phage arm as compared to the untreated arm (514.74 pg/mL) (Figure 5b).
To the best of our knowledge, there are no reports in the scientific literature of the application of phages for the treatment of B. fragilis infections or the in vitro assessment of their therapeutic potential. Previously well-studied B. fragilis-specific phages HSP-40 and GB-124-14 were used only for the monitoring of fecal contamination in different water environments [41,42].
A previous study identified phage VA7 as a potential candidate for application against ETBF infections within in vitro or animal models, or in human phage therapy [22]. In the current study, we sequenced the genome of VA7, further supporting that it is a strictly virulent phage and that there are no counter-indications for its use in human phage therapy.
We also evaluated the antimicrobial properties of phage VA7 lysate and its impact on CEC cytokine IL-8 production in an ETBF (strain E3)-infected CEC model. Similar studies were performed on the phages of other pathogens such as E. coli, Pseudomonas aeruginosa or Staphylococcus aureus [43,44]. We observed that the addition of phage VA7 lysate to E3-infected CECs led to significantly reduced bacterial counts and lower IL-8 levels compared to untreated infected cells. These lower IL-8 levels could be explained by a potential decrease in BFT production, which, in turn, is expected to be linked to the reduction of bacterial counts [45]. Previous studies showed that a reduction in the bacterial load by cefoxitin led to the regression and disappearance of ETBF-associated neoplastic formations in the colon of mice [46]. A decrease in IL-17A, which is a pro-inflammatory cytokine released by Th-17 lymphocytes and found to be highly involved in cancer progression, was also observed [47]. By the activation of NF-κB and p42/44MAPK pathways, IL-17A was shown to stimulate the production of cytokine IL-8 [47]. IL-8 is involved in the angiogenesis of developing tumors. We speculate that directly, via BTF release, or indirectly through IL-17A stimulation, IL-8 levels are increased in the presence of ETBF and that both cytokines’ levels can be reduced through the elimination of the pathogenic cells. Even though our results are based on in vitro experiments, we suggest that the reduction in bacterial count caused by VA7 may prevent or even reverse the formation of colorectal adenocarcinoma caused by ETBF.
In the country of Georgia, phage preparations for oral and local administration are mainly produced and distributed in their lysate forms. Subsequently, we deliberately neglected to purify the phages before using them in the present model. The goal was to evaluate the antibacterial activity and CEC IL-8 response of the phage formulations that would ultimately be applied in future human prophylaxis or therapy applications in Georgia. However, the tested VA7 phage lysates undeniably contained BTF due to the production of the phage lysate in ETBF strain E3. Even though this BFT level is bound to be very low, as the lysates were diluted 1000-fold (from 1010 to 107 PFU/mL) in cell culture medium before addition to the present in vitro model, it is bound to have generated a certain bias.
We acknowledge that these are preliminary results. Future studies to evaluate the impact of phage VA7 on the induction of other relevant chemokines such as IL-17 and NF-κB, and the determination of CEC viability, are envisioned. Relevant biophysical characteristics of phage VA7, such as its stability in the human body or various solutions, and its penetration and distribution ability in relevant bodily tissues, are also of interest. These future pre-clinical studies will also include the evaluation of purified phage VA7 preparations.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/v13102044/s1: Figure S1. intergenomic similarity between Bacteroides phage VA7 and the most related phage genomes (generated by VIRIDIC), Figure S2. cytotoxicity assay using colon cancer cells (CECs), adapted from Pantosi et al.
Author Contributions
Conceptualization, N.B., K.M., E.K., I.A. and N.C.; data curation, N.B. and E.K.; methodology, N.B., N.C., K.M., E.K., M.M., N.G. and I.C.; validation, N.C., M.M. and M.V.; formal analysis, I.A., N.B., K.M., E.K., N.C., C.L. and J.W.; investigation, N.B., N.G., K.M., E.K., M.M., I.A., I.C., C.L. and J.W.; resources, N.C., M.V., I.A. and R.L.; writing—original draft preparation, N.B. and J.W.; writing—review and editing, N.C., M.M., M.V., J.-P.P., E.K., K.M., I.A., C.L., J.W. and R.L.; visualization, N.B., M.M., K.M., J.-P.P. and J.W.; supervision, N.C.; project administration, N.C. and N.B.; funding acquisition, N.C., I.A. and N.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Shota Rustaveli National Foundation of Georgia, grant numbers FR/235/7-250/14 and IG 15/1/16; and by the Eliava Foundation, grant number 1557/2. CL is supported by a PhD fellowship from FWO Vlaanderen (1S64720N). MM is supported by the Royal Higher Institute for Defense, grant number HFM/19-12. The funders had no role in the design of the study, the collection, analyses, or interpretation of data, the writing of the manuscript or the decision to publish the results.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article or supplementary material. The annotated genome was submitted to NCBI and is available through Genbank accession number MW916539.1.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Jamal, M.; Bukhari, S.M.A.U.; Andleeb, S.; Ali, M.; Raza, S.; Nawaz, M.A.; Hussain, T.; Rahman, S.U.; Shah, S.S.A. Bacteriophages: An overview of the control strategies against multiple bacterial infections in different fields. J. Basic Microbiol. 2019, 59, 123–133. [Google Scholar] [CrossRef]
- Chanishvili, N. Bacteriophages as therapeutic and prophylactic means: Summary of the Soviet and post-Soviet experiences. Curr. Drug Deliv. 2016, 13, 309–323. [Google Scholar] [CrossRef]
- Wexler, H.M. Bacteroides: The good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 2007, 20, 593–621. [Google Scholar] [CrossRef] [Green Version]
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef] [Green Version]
- Tzianabos, A.O.; Onderdonk, A.B.; Smith, R.S.; Kasper, D.L. Structure-function relationships for polysaccharide-induced intra-abdominal abscesses. Infect. Immun. 1994, 62, 3590–3593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwong, T.N.Y.; Wang, X.; Nakatsu, G.; Chow, T.C.; Tipoe, T.; Dai, R.Z.W.; Tsoi, K.K.K.; Wong, M.C.S.; Tse, G.; Chan, M.T.V.; et al. Association between bacteremia from specific microbes and subsequent diagnosis of colorectal cancer. Gastroenterology 2018, 155, 383–390. [Google Scholar] [CrossRef]
- Polk, B.F.; Kasper, D.L. Bacteroides fragilis subspecies in clinical isolates. Ann. Intern. Med. 1977, 86, 569–571. [Google Scholar] [CrossRef] [PubMed]
- Toprak, N.U.; Yagci, A.; Gulluoglu, B.M.; Akin, M.L.; Demirkalem, P.; Celenk, T.; Soyletir, G. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin. Microbiol. Infect. 2006, 12, 782–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sears, C.L.; Islam, S.; Saha, A.; Arjumand, M.; Alam, N.H.; Faruque, A.S.; Salam, M.A.; Shin, J.; Hecht, D.; Weintraub, A.; et al. Association of enterotoxigenic Bacteroides fragilis infection with inflammatory diarrhea. Clin. Infect. Dis. 2008, 47, 797–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boleij, A.; Hechenbleikner, E.M.; Goodwin, A.C.; Badani, R.; Stein, E.M.; Lazarev, M.G.; Ellis, B.; Carroll, K.C.; Albesiano, E.; Wick, E.C.; et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin. Infect. Dis. 2015, 60, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Sóki, J.; Wybo, I.; Hajdú, E.; Toprak, N.U.; Jeverica, S.; Stingu, C.S.; Tierney, D.; Perry, J.D.; Matuz, M.; Urbán, E.; et al. A Europe-wide assessment of antibiotic resistance rates in Bacteroides and Parabacteroides isolates from intestinal microbiota of healthy subjects. Anaerobe 2020, 62, 102182. [Google Scholar] [CrossRef] [PubMed]
- Sears, C.L. Enterotoxigenic Bacteroides fragilis: A rogue among symbiotes. Clin. Microbiol. Rev. 2009, 22, 349–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, S.; Shin, J.; Zhang, G.; Cohen, M.; Franco, A.; Sears, C.L. The Bacteroides fragilis toxin binds to a specific intestinal epithelial cell receptor. Infect. Immun. 2006, 74, 5382–5390. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Powell, J.; Mathioudakis, N.; Kane, S.; Fernandez, E.; Sears, C.L. Bacteroides fragilis enterotoxin induces intestinal epithelial cell secretion of interleukin-8 through mitogen-activated protein kinases and a tyrosine kinase-regulated nuclear factor-kappaB pathway. Infect. Immun. 2004, 72, 5832–5839. [Google Scholar] [CrossRef] [Green Version]
- Chung, L.; Orberg, E.T.; Geis, A.L.; Chan, J.L.; Fu, K.; DeStefano Shields, C.E.; Dejea, C.M.; Fathi, P.; Chen, J.; Finard, B.B.; et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe 2018, 23, 421. [Google Scholar] [CrossRef]
- Goodwin, A.C.; Destefano Shields, C.E.; Wu, S.; Huso, D.L.; Wu, X.; Murray-Stewart, T.R.; Hacker-Prietz, A.; Rabizadeh, S.; Woster, P.M.; Sears, C.L.; et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 15354–15359. [Google Scholar] [CrossRef] [Green Version]
- Jeon, J.I.; Ko, S.H.; Kim, J.M. Intestinal epithelial cells exposed to Bacteroides fragilis enterotoxin regulate NF-κB activation and inflammatory responses through β-catenin expression. Infect. Immun. 2019, 87, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, Y.C.; Yang, Z.B.; Cheng, X.S.; Fang, X.B.; Shen, T.; Xia, C.F.; Liu, P.; Qian, H.H.; Sun, B.; Yin, Z.F.; et al. CXCL8, overexpressed in colorectal cancer, enhances the resistance of colorectal cancer cells to anoikis. Cancer Lett. 2015, 361, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.H.; Yu, J. Gut microbiota in colorectal cancer: Mechanisms of action and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 690–704. [Google Scholar] [CrossRef]
- Hannigan, G.D.; Duhaime, M.B.; Ruffin, M.T., 4th; Koumpouras, C.C.; Schloss, P.D. Diagnostic Potential and Interactive Dynamics of the Colorectal Cancer Virome. mBio 2018, 9, e02248-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutiérrez, B.; Domingo-Calap, P. Phage Therapy in Gastrointestinal Diseases. Microorganisms 2020, 8, 1420. [Google Scholar] [CrossRef]
- Bakuradze, N.; Makalatia, K.; Merabishvili, M.; Togoshvili, L.; Chanishvili, N. Selection of active phages against B. fragilis for further study of therapeutic perspectives. Georgian Med. News 2018, 285, 111–116. [Google Scholar]
- Aitchison, A.; Frizelle, F.A.; Keenan, J.I. PCR detection of the Bacteroides fragilis enterotoxin gene relies on robust primer design. J. Clin. Microbiol. 2016, 54, 239–240. [Google Scholar] [CrossRef] [Green Version]
- Mirzaei, M.K.; Nilsson, A.S. Correction: Isolation of phages for phage therapy: A comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE 2015, 10, e0127606. [Google Scholar] [CrossRef] [Green Version]
- Makalatia, K.; Kakabadze, E.; Wagemans, J.; Grdzelishvili, N.; Bakuradze, N.; Natroshvili, G.; Macharashvili, N.; Sedrakyan, A.; Arakelova, K.; Ktsoyan, Z.; et al. Characterization of Salmonella Isolates from Various Geographical Regions of the Caucasus and Their Susceptibility to Bacteriophages. Viruses 2020, 12, 1418. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. ViPTree: The viral proteomic tree server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef] [PubMed]
- Moraru, C.; Varsani, A.; Kropinski, A.M. VIRIDIC—A Novel Tool to Calculate the Intergenomic Similarities of Prokaryote-Infecting Viruses. Viruses 2020, 12, 1268. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.J.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [Green Version]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Search and Contextual Analysis of Transfer RNA Genes. Nucl. Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Kropinski, A.M. Practical Advice on the One-Step Growth Curve. Methods Mol. Biol. 2018, 1681, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Khan Mirzaei, M.; Haileselassie, Y.; Navis, M.; Cooper, C.; Sverremark-Ekström, E.; Nilsson, A.S. Morphologically distinct Escherichia coli bacteriophages differ in their efficacy and ability to stimulate cytokine release in vitro. Front. Microbiol. 2016, 7, 437. [Google Scholar]
- Ogilvie, L.; Caplin, J.; Dedi, C.; Diston, D.; Cheek, E.; Bowler, L.; Taylor, H.; Ebdon, J.; Jones, B.V. Comparative (Meta)genomic Analysis and Ecological Profiling of Human Gut-Specific Bacteriophage wB124-14. PLoS ONE 2012, 7, e35053. [Google Scholar] [CrossRef] [Green Version]
- Hawkins, S.A.; Layton, A.C.; Ripp, S.; Williams, D.; Sayler, G.S. Genome sequence of the Bacteroides fragilis phage ATCC 51477-B1. Virol. J. 2008, 5, 97. [Google Scholar] [CrossRef] [Green Version]
- Turner, D.; Kropinski, A.; Adriaenssens, E.M. A Roadmap for Genome-Based Phage Taxonomy. Viruses 2021, 13, 506. [Google Scholar] [CrossRef]
- Tartera, C.; Jofre, J. Bacteriophages active against Bacteroides fragilis in sewage-polluted waters. Appl. Environ. Microbiol. 1987, 53, 1632–1637. [Google Scholar] [CrossRef] [Green Version]
- McMinn, B.R.; Korajkic, A.; Ashbolt, N.J. Evaluation of Bacteroides fragilis GB-124 bacteriophages as novel human-associated faecal indicators in the United States. Lett. Appl. Microbiol. 2014, 59, 115–121. [Google Scholar] [CrossRef]
- Van Belleghem, J.D.; Clement, F.; Merabishvili, M.; Lavigne, R.; Vaneechoutte, M. Pro- and anti-inflammatory responses of peripheral blood mononuclear cells induced by Staphylococcus aureus and Pseudomonas aeruginosa phages. Sci. Rep. 2017, 7, 8004. [Google Scholar] [CrossRef] [Green Version]
- Alemayehu, D.; Casey, P.G.; McAuliffe, O.; Guinane, C.M.; Martin, J.G.; Shanahan, F.; Coffey, A.; Ross, R.P.; Hill, C. Bacteriophages φMR299-2 and φNH-4 can eliminate Pseudomonas aeruginosa in the murine lung and on cystic fibrosis lung airway cells. mBio 2012, 3, e00029-12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metz, P.; Tjan, M.J.H.; Wu, S.; Pervaiz, M.; Hermans, S.; Shettigar, A.; Sears, C.L.; Ritschel, T.; Dutilh, B.E.; Boleij, A. Drug discovery and repurposing inhibits a major gut pathogen-derived oncogenic toxin. Front. Cell. Infect. Microbiol. 2019, 9, 364. [Google Scholar] [CrossRef] [PubMed]
- DeStefano Shields, C.E.; van Meerbeke, S.W.; Housseau, F.; Wang, H.; Huso, D.L.; Casero, R.A.; O’Hagan, H.M.; Sears, C.L. Reduction of murine colon tumorigenesis driven by enterotoxigenic Bacteroides fragilis using cefoxitin treatment. J. Infect. Dis. 2016, 214, 122–129. [Google Scholar] [CrossRef] [Green Version]
- Amatya, N.; Garg, A.V.; Gaffen, S.L. IL-17 signaling: The Yin and the Yang. Trends Immunol. 2017, 38, 310–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, M.; Brummer, G.; Acevedo, D.; Cheng, N. Cytokine regulation of metastasis and tumorigenicity. Adv. Cancer Res. 2016, 132, 265–367. [Google Scholar] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).