The Mouse Papillomavirus Epigenetic Signature Is Characterised by DNA Hypermethylation after Lesion Regression
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Immune Suppression and Group Descriptions
2.3. Ethics
2.4. MmuPV1 Virus and Infection Protocol
2.5. Lesion Monitoring and Harvesting
2.6. Reduced Representation Bisulphite Sequencing Library Preparation
2.7. DNA Methylation Analysis
2.8. Group Comparisons
2.9. Analysis of Viral Reads and Pathway Analysis
2.10. Statistical Analyses
3. Results
3.1. Immune-Suppressed BALB/c Mice Develop and Maintain Active MmuPV1 Infection
3.2. Genome-Scale DNA Methylation Identifies Extensive Hypermethylation in Regressed Lesions
3.3. Genes in the Core Promoter Region Are Skewed Extensively towards Hypermethylation in Regressed Lesions
3.4. Hypermethylated Genes in Regressed Lesions Are Enriched for Cellular Senescence and Cancer Related Pathways and for CTCF and RNA Polymerase II Binding
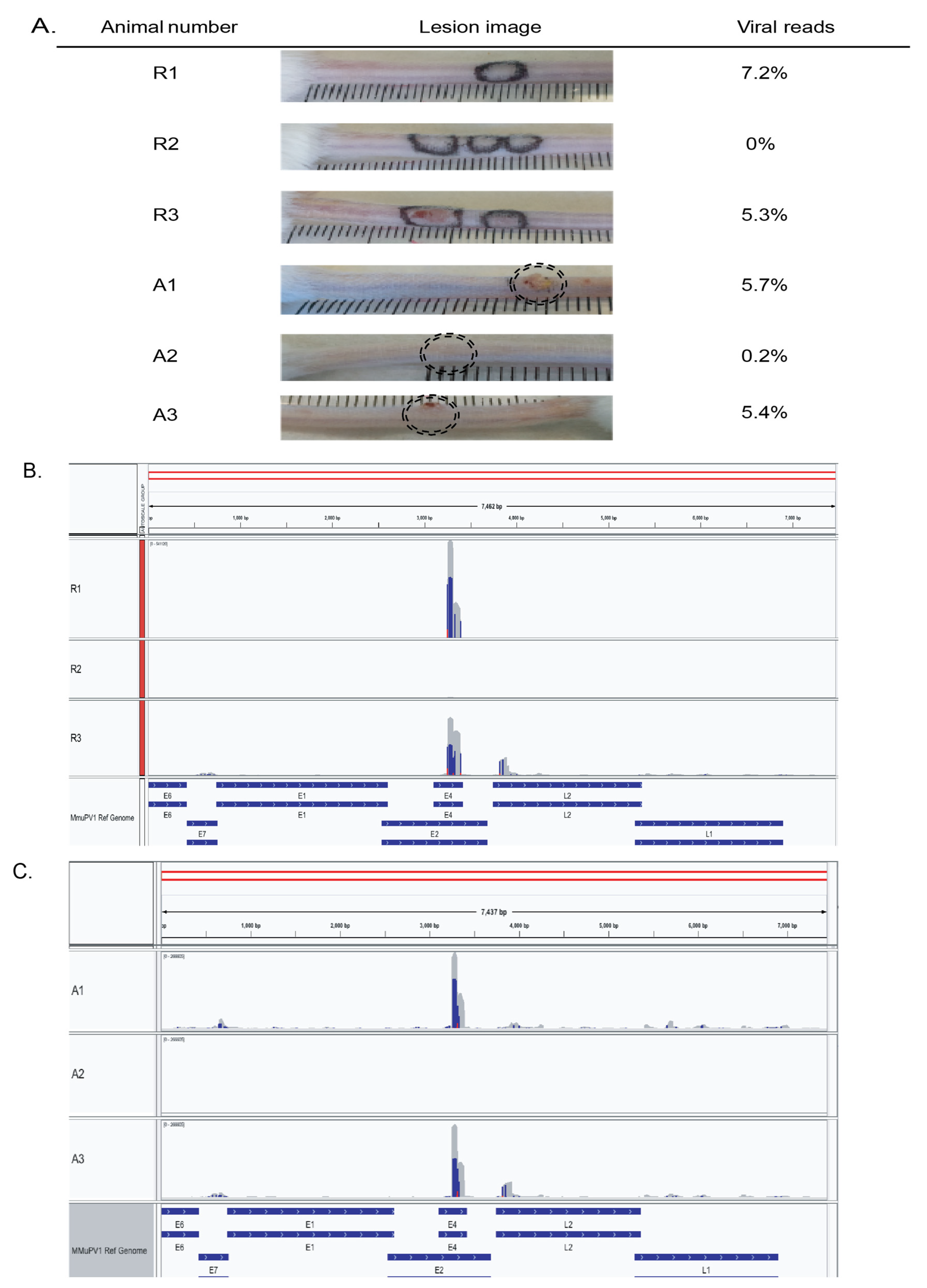
3.5. MmuPV1 Viral Reads Are Highly Concentrated in the E4 Portion of the Genome
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- de Martel, C.; Plummer, M.; Vignat, J.; Franceschi, S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int. J. Cancer 2017, 141, 664–670. [Google Scholar] [CrossRef] [PubMed]
- Antonsson, A.; Forslund, O.; Ekberg, H.; Sterner, G.; Hansson, B.G. The ubiquity and impressive genomic diversity of human skin papillomaviruses suggest a commensalic nature of these viruses. J. Virol. 2000, 74, 11636–11641. [Google Scholar] [CrossRef]
- De Jong, S.J.; Imahorn, E.; Itin, P.; Uitto, J.; Orth, G.; Jouanguy, E.; Casanova, J.-L.; Burger, B. Epidermodysplasia verruciformis: Inborn errors of immunity to human beta-papillomaviruses. Front. Microbiol. 2018, 9, 1222. [Google Scholar] [CrossRef] [PubMed]
- Mittal, A.; Colegio, O.R. Skin Cancers in Organ Transplant Recipients. Am. J. Transplant. 2017, 17, 2509–2530. [Google Scholar] [CrossRef] [PubMed]
- Bavinck, J.N.B.; Plasmeijer, E.I.; Feltkamp, M.C.W. β-Papillomavirus Infection and Skin Cancer. J. Investig. Dermatol. 2008, 128, 1355–1358. [Google Scholar] [CrossRef] [PubMed]
- Hufbauer, M.; Akgül, B. Molecular Mechanisms of Human Papillomavirus Induced Skin Carcinogenesis. Viruses 2017, 9, 187. [Google Scholar] [CrossRef]
- Karagas, M.R.; Nelson, H.; Sehr, P.; Waterboer, T.; Stukel, T.; Andrew, A.; Green, A.C.; Bavinck, J.N.B.; Perry, A.; Spencer, S.; et al. Human papillomavirus infection and incidence of squamous cell and basal cell carcinomas of the skin. J. Natl. Cancer Inst. 2006, 98, 389–395. [Google Scholar] [CrossRef]
- Tommasino, M. HPV and skin carcinogenesis. Papillomavirus Res. 2019, 7, 129–131. [Google Scholar] [CrossRef]
- Underbrink, M.P.; Howie, H.L.; Bedard, K.M.; Koop, J.I.; Galloway, D.A. E6 proteins from multiple human betapapillomavirus types degrade Bak and protect keratinocytes from apoptosis after UVB irradiation. J. Virol. 2008, 82, 10408–10417. [Google Scholar] [CrossRef]
- Li, L.; Jiang, M.; Feng, Q.; Kiviat, N.B.; Stern, J.E.; Hawes, S.; Cherne, S.; Lu, H. Aberrant methylation changes detected in cutaneous squamous cell carcinoma of immunocompetent individuals. Cell Biochem. Biophys. 2015, 72, 599–604. [Google Scholar] [CrossRef]
- Hervás-Marín, D.; Higgins, F.; Sanmartín, O.; López-Guerrero, J.A.; Bañó, M.C.; Igual, J.C.; Quilis, I.; Sandoval, J. Genome wide DNA methylation profiling identifies specific epigenetic features in high-risk cutaneous squamous cell carcinoma. PLoS ONE 2019, 14, e0223341. [Google Scholar] [CrossRef]
- Rodríguez-Paredes, M.; Bormann, F.; Raddatz, G.; Gutekunst, J.; Lucena-Porcel, C.; Köhler, F.; Wurzer, E.; Schmidt, K.; Gallinat, S.; Wenck, H.; et al. Methylation profiling identifies two subclasses of squamous cell carcinoma related to distinct cells of origin. Nat. Commun. 2018, 9, 577. [Google Scholar] [CrossRef]
- Kuss-Duerkop, S.K.; Westrich, J.A.; Pyeon, D. DNA Tumor Virus Regulation of Host DNA Methylation and Its Implications for Immune Evasion and Oncogenesis. Viruses 2018, 10, 82. [Google Scholar] [CrossRef]
- Durzynska, J.; Lesniewicz, K.; Poreba, E. Human papillomaviruses in epigenetic regulations. Mutat. Res./Rev. Mutat. Res. 2017, 772, 36–50. [Google Scholar] [CrossRef] [PubMed]
- Ingle, A.; Ghim, S.; Joh, J.; Chepkoech, I.; Bennett Jenson, A.; Sundberg, J.P. Novel laboratory mouse papillomavirus (MusPV) infection. Vet. Pathol. 2011, 48, 500–505. [Google Scholar] [CrossRef] [PubMed]
- Joh, J.; Jenson, A.B.; King, W.; Proctor, M.; Ingle, A.; Sundberg, J.P.; Ghim, S.-J. Genomic analysis of the first laboratory-mouse papillomavirus. J. Gen. Virol. 2011, 92, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J.; Egawa, N.; Griffin, H.; Kranjec, C.; Murakami, I. Human papillomavirus molecular biology and disease association. Rev. Med. Virol. 2015, 25, 2–23. [Google Scholar] [CrossRef]
- Hu, J.; Cladel, N.M.; Budgeon, L.R.; Balogh, K.K.; Christensen, N.D. The Mouse Papillomavirus Infection Model. Viruses 2017, 9, 246. [Google Scholar] [CrossRef]
- Spurgeon, M.E.; Uberoi, A.; McGregor, S.M.; Wei, T.; Ward-Shaw, E.; Lambert, P.F. A novel in vivo infection model to study papillomavirus-mediated disease of the female reproductive tract. MBio 2019, 10, e00180-19. [Google Scholar] [CrossRef]
- Uberoi, A.; Yoshida, S.; Frazer, I.H.; Pitot, H.C.; Lambert, P.F. Role of Ultraviolet Radiation in Papillomavirus-Induced Disease. PLoS Pathog. 2016, 12, e1005664. [Google Scholar] [CrossRef]
- Yu, L.; Majerciak, V.; Xue, X.-Y.; Uberoi, A.; Lobanov, A.; Chen, X.; Cam, M.; Hughes, S.H.; Lambert, P.F.; Zheng, Z.-M. Mouse papillomavirus type 1 (MmuPV1) DNA is frequently integrated in benign tumors by microhomology-mediated end-joining. PLoS Pathog. 2021, 17, e1009812. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Budgeon, L.R.; Cladel, N.M.; Balogh, K.; Myers, R.; Cooper, T.K.; Christensen, N.D. Tracking vaginal, anal and oral infection in a mouse papillomavirus infection model. J. Gen. Virol. 2015, 96, 3554–3565. [Google Scholar] [CrossRef] [PubMed]
- Handisurya, A.; Day, P.M.; Thompson, C.D.; Buck, C.B.; Pang, Y.-Y.S.; Lowy, D.R.; Schiller, J.T. Characterization of Mus musculus papillomavirus 1 infection in situ reveals an unusual pattern of late gene expression and capsid protein localization. J. Virol. 2013, 87, 13214–13225. [Google Scholar] [CrossRef]
- Lichti, U.; Anders, J.; Yuspa, S.H. Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat. Protoc. 2008, 3, 799–810. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Rodger, E.J.; Stockwell, P.A.; Weeks, R.J.; Morison, I.M. Technical considerations for reduced representation bisulfite sequencing with multiplexed libraries. J. Biomed. Biotechnol. 2012, 2012, 741542. [Google Scholar] [CrossRef]
- Chatterjee, A.; Rodger, E.J.; Stockwell, P.A.; Le Mée, G.; Morison, I.M. Generating Multiple Base-Resolution DNA Methylomes Using Reduced Representation Bisulfite Sequencing. In Oral Biology: Molecular Techniques and Applications; Seymour, G.J., Cullinan, M.P., Heng, N.C.K., Eds.; Springer: New York, NY, USA, 2017; pp. 279–298. [Google Scholar]
- Ludgate, J.L.; Wright, J.; Stockwell, P.A.; Morison, I.M.; Eccles, M.R.; Chatterjee, A. A streamlined method for analysing genome-wide DNA methylation patterns from low amounts of FFPE DNA. BMC Med Genom. 2017, 10, 54. [Google Scholar] [CrossRef]
- Stockwell, P.A.; Chatterjee, A.; Rodger, E.J.; Morison, I.M. DMAP: Differential methylation analysis package for RRBS and WGBS data. Bioinformatics 2014, 30, 1814–1822. [Google Scholar] [CrossRef]
- Chatterjee, A.; Stockwell, P.A.; Rodger, E.J.; Duncan, E.J.; Parry, M.F.; Weeks, R.J.; Morison, I.M. Genome-wide DNA methylation map of human neutrophils reveals widespread inter-individual epigenetic variation. Sci. Rep. 2015, 5, 17328. [Google Scholar] [CrossRef]
- Chatterjee, A.; Stockwell, P.A.; Rodger, E.J.; Morison, I.M. Genome-scale DNA methylome and transcriptome profiling of human neutrophils. Sci. Data 2016, 3, 160019. [Google Scholar] [CrossRef]
- Krueger, F.; Andrews, S.R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011, 27, 1571–1572. [Google Scholar] [CrossRef]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’Ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Cross, S.H.; Bird, A.P. CpG islands and genes. Curr. Opin. Genet. Dev. 1995, 5, 309–314. [Google Scholar] [CrossRef]
- Xi, Y.; Li, W. BSMAP: Whole genome bisulfite sequence MAPping program. BMC Bioinform. 2009, 10, 232. [Google Scholar] [CrossRef] [PubMed]
- Chujan, S.; Kitkumthorn, N.; Siriangkul, S.; Mutirangura, A. CCNA1 promoter methylation: A potential marker for grading Papanicolaou smear cervical squamous intraepithelial lesions. Asian Pac. J. Cancer Prev. 2014, 15, 7971–7975. [Google Scholar] [CrossRef] [PubMed]
- Teschendorff, A.E.; Liu, X.; Caren, H.; Pollard, S.M.; Beck, S.; Widschwendter, M.; Chen, L. The dynamics of DNA methylation covariation patterns in carcinogenesis. PLoS Comput. Biol. 2014, 10, e1003709. [Google Scholar] [CrossRef]
- Biktasova, A.; Hajek, M.; Sewell, A.; Gary, C.; Bellinger, G.; Deshpande, H.A.; Bhatia, A.; Burtness, B.; Judson, B.; Mehra, S.; et al. Demethylation Therapy as a Targeted Treatment for Human Papillomavirus-Associated Head and Neck Cancer. Clin. Cancer Res. 2017, 23, 7276–7287. [Google Scholar] [CrossRef]
- Gozuacik, D.; Kimchi, A. DAPk protein family and cancer. Autophagy 2006, 2, 74–79. [Google Scholar] [CrossRef]
- Yanatatsaneejit, P.; Chalertpet, K.; Sukbhattee, J.; Nuchcharoen, I.; Phumcharoen, P.; Mutirangura, A. Promoter methylation of tumor suppressor genes induced by human papillomavirus in cervical cancer. Oncol. Lett. 2020, 20, 955–961. [Google Scholar] [CrossRef]
- Lang, F.; Li, X.; Vladimirova, O.; Hu, B.; Chen, G.; Xiao, Y.; Singh, V.; Lu, D.; Li, L.; Han, H.; et al. CTCF interacts with the lytic HSV-1 genome to promote viral transcription. Sci. Rep. 2017, 7, 39861. [Google Scholar] [CrossRef]
- Pfeifer, G.P. Mechanisms of UV-induced mutations and skin cancer. Genome Instab. Dis. 2020, 1, 99–113. [Google Scholar] [CrossRef]
- Young, L.; Listgarten, J.; Trotter, M.; Andrew, S.; Tron, V. Evidence that dysregulated DNA mismatch repair characterizes human nonmelanoma skin cancer. Br. J. Dermatol. 2008, 158, 59–69. [Google Scholar] [CrossRef]
- Young, L.C.; Hays, J.B.; Tron, V.A.; Andrew, S.E. DNA mismatch repair proteins: Potential guardians against genomic instability and tumorigenesis induced by ultraviolet photoproducts. J. Investig. Dermatol. 2003, 121, 435–440. [Google Scholar] [CrossRef][Green Version]
- Lewis, C.J.; Stevenson, A.; Fear, M.W.; Wood, F.M. A review of epigenetic regulation in wound healing: Implications for the future of wound care. Wound Repair Regen 2020, 28, 710–718. [Google Scholar] [CrossRef]
- Mitra, S.; Lauss, M.; Cabrita, R.; Choi, J.; Zhang, T.; Isaksson, K.; Olsson, H.; Ingvar, C.; Carneiro, A.; Staaf, J.; et al. Analysis of DNA methylation patterns in the tumor immune microenvironment of metastatic melanoma. Mol. Oncol. 2020, 14, 933–950. [Google Scholar] [CrossRef] [PubMed]
- Handisurya, A.; Day, P.M.; Thompson, C.D.; Bonelli, M.; Lowy, D.R.; Schiller, J.T. Strain-Specific Properties and T Cells Regulate the Susceptibility to Papilloma Induction by Mus musculus Papillomavirus 1. PLoS Pathog. 2014, 10, e1004314. [Google Scholar] [CrossRef] [PubMed]
- Fraga, M.; Herranz, M.; Espada, J.; Ballestar, E.; Paz, M.F.; Ropero, S.; Erkek, E.; Bozdogan, O.; Peinado, H.; Niveleau, A.; et al. A mouse skin multistage carcinogenesis model reflects the aberrant DNA methylation patterns of human tumors. Cancer Res. 2004, 64, 5527–5534. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Xu, X. Reduction in DNA methyltransferases and alteration of DNA methylation pattern associate with mouse skin ageing. Exp. Dermatol. 2014, 23, 357–359. [Google Scholar] [CrossRef]
- Wei, T.; Buehler, D.; Ward-Shaw, E.; Lambert, P.F. An Infection-Based Murine Model for Papillomavirus-Associated Head and Neck Cancer. mBio 2020, 11, e00908-20. [Google Scholar] [CrossRef]
- Maufort, J.P.; Williams, S.M.; Pitot, H.C.; Lambert, P.F. Human papillomavirus 16 E5 oncogene contributes to two stages of skin carcinogenesis. Cancer Res. 2007, 67, 6106–6112. [Google Scholar] [CrossRef]
- Xue, X.-Y.; Majerciak, V.; Uberoi, A.; Kim, B.-H.; Gotte, D.; Chen, X.; Cam, M.; Lambert, P.F.; Zheng, Z.-M. The full transcription map of mouse papillomavirus type 1 (MmuPV1) in mouse wart tissues. PLoS Pathog. 2017, 13, e1006715. [Google Scholar] [CrossRef] [PubMed]
- Maglennon, G.A.; McIntosh, P.; Doorbar, J. Persistence of viral DNA in the epithelial basal layer suggests a model for papillomavirus latency following immune regression. Virology 2011, 414, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Middleton, K.; Peh, W.; Southern, S.; Griffin, H.; Sotlar, K.; Nakahara, T.; El-Sherif, A.; Morris, L.; Seth, R.; Hibma, M.; et al. Organization of human papillomavirus productive cycle during neoplastic progression provides a basis for selection of diagnostic markers. J. Virol. 2003, 77, 10186–10201. [Google Scholar] [CrossRef]
- Peh, W.L.; Middleton, K.; Christensen, N.; Nicholls, P.; Egawa, K.; Sotlar, K.; Brandsma, J.; Percival, A.; Lewis, J.; Liu, W.J.; et al. Life Cycle Heterogeneity in Animal Models of Human Papillomavirus-Associated Disease. J. Virol. 2002, 76, 10401. [Google Scholar] [CrossRef]
- Kremer, W.W.; Vink, F.J.; van Zummeren, M.; Dreyer, G.; Rozendaal, L.; Doorbar, J.; Bleeker, M.C.G.; Meijer, C.J.L.M. Characterization of cervical biopsies of women with HIV and HPV co-infection using p16ink4a, ki-67 and HPV E4 immunohistochemistry and DNA methylation. Mod. Pathol. 2020, 33, 1968–1978. [Google Scholar] [CrossRef] [PubMed]
- Galván, S.C.; Martínez-Salazar, M.; Galván, V.M.; Méndez, R.; Díaz-Contreras, G.T.; Alvarado-Hermida, M.; Alcántara-Silva, R.; García-Carrancá, A. Analysis of CpG methylation sites and CGI among human papillomavirus DNA genomes. BMC Genom. 2011, 12, 580. [Google Scholar] [CrossRef] [PubMed]
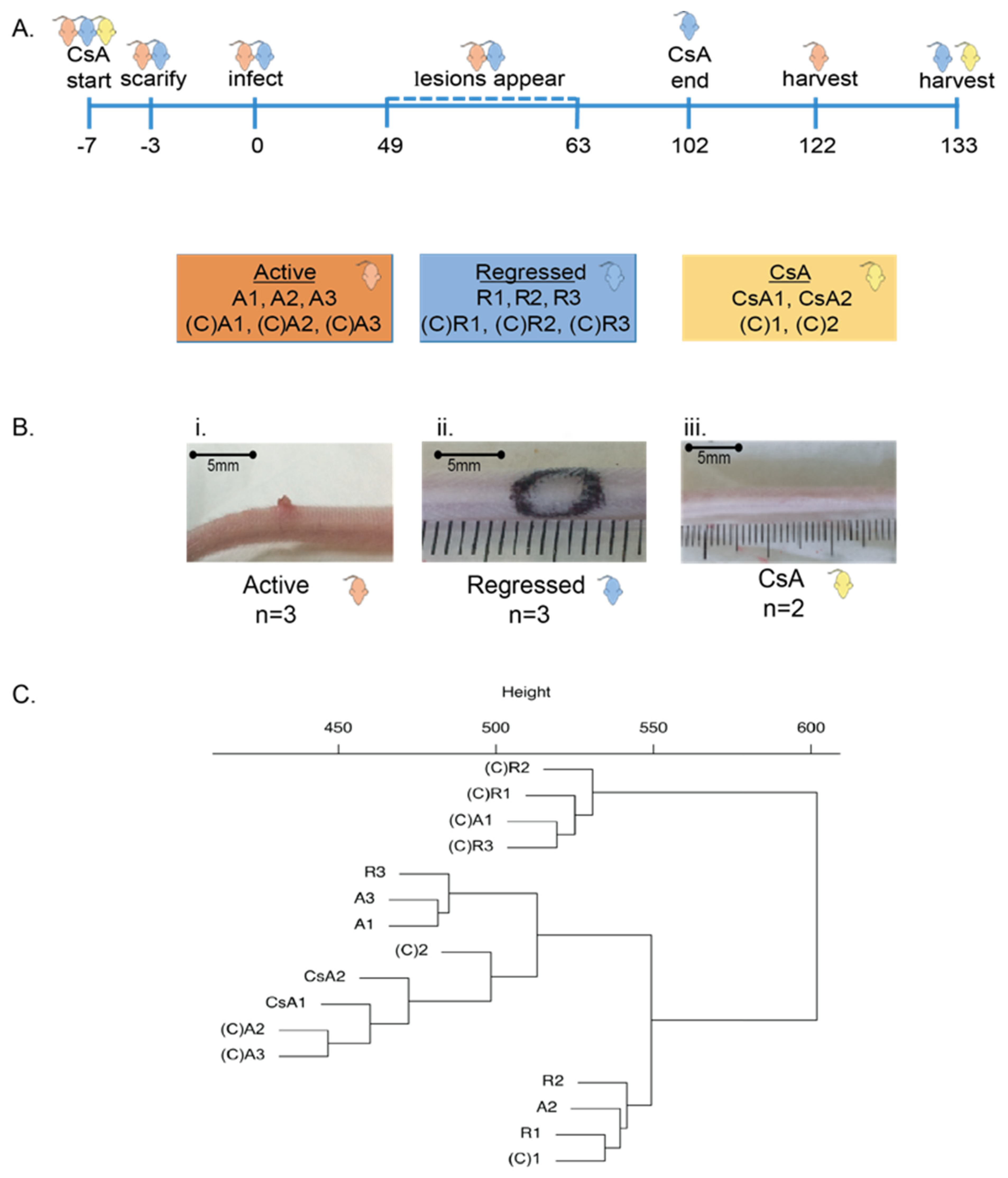
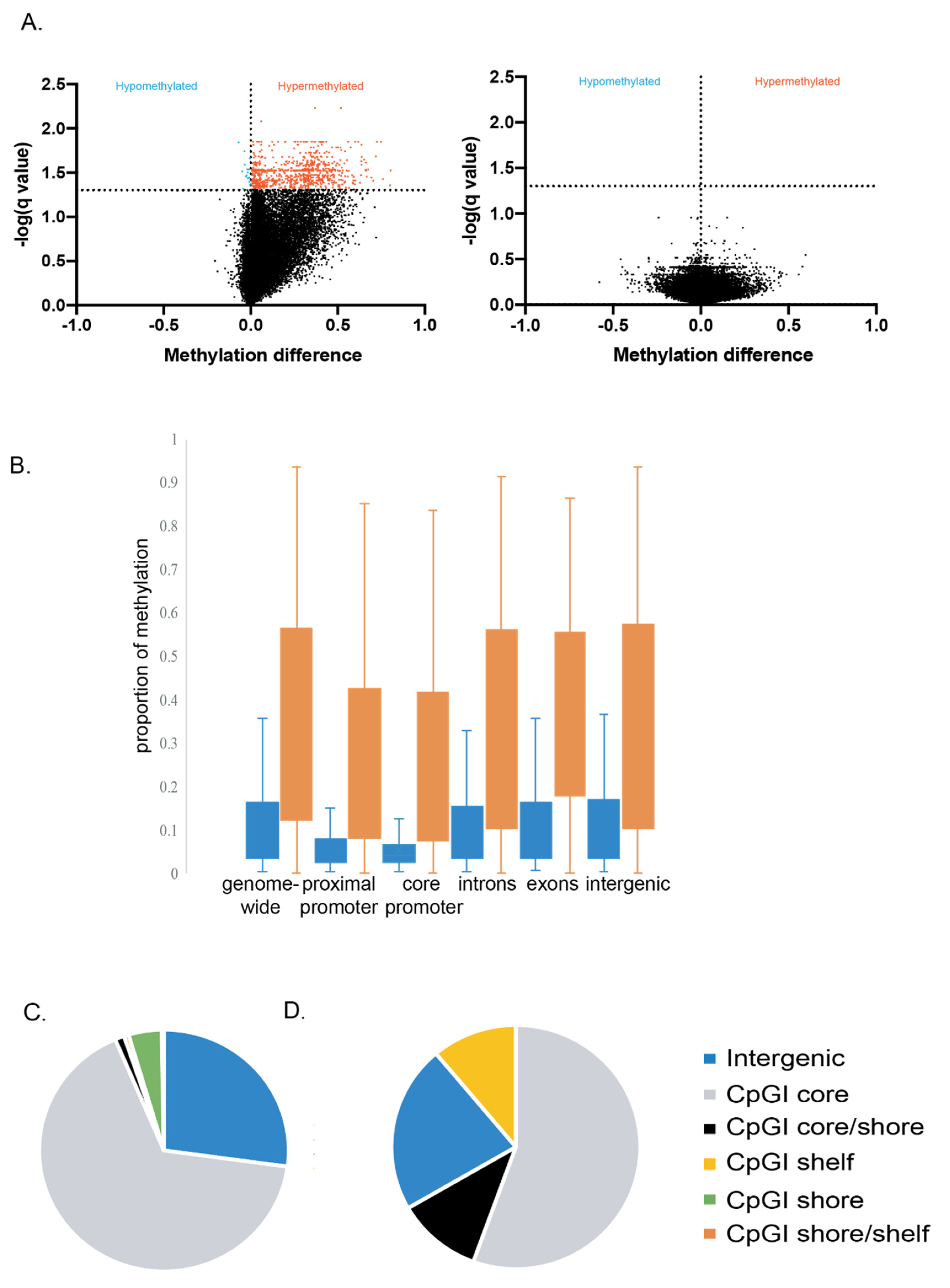
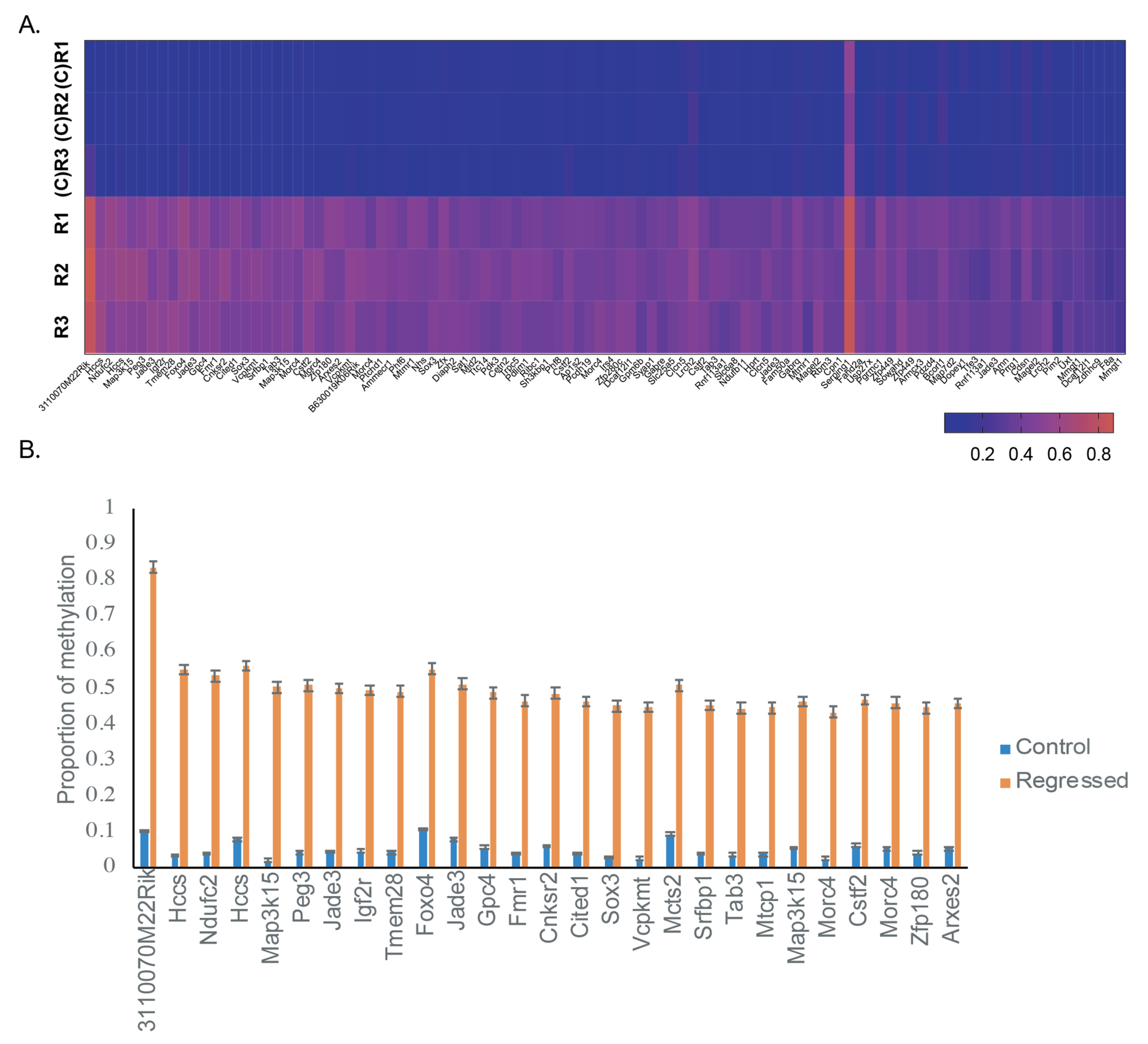
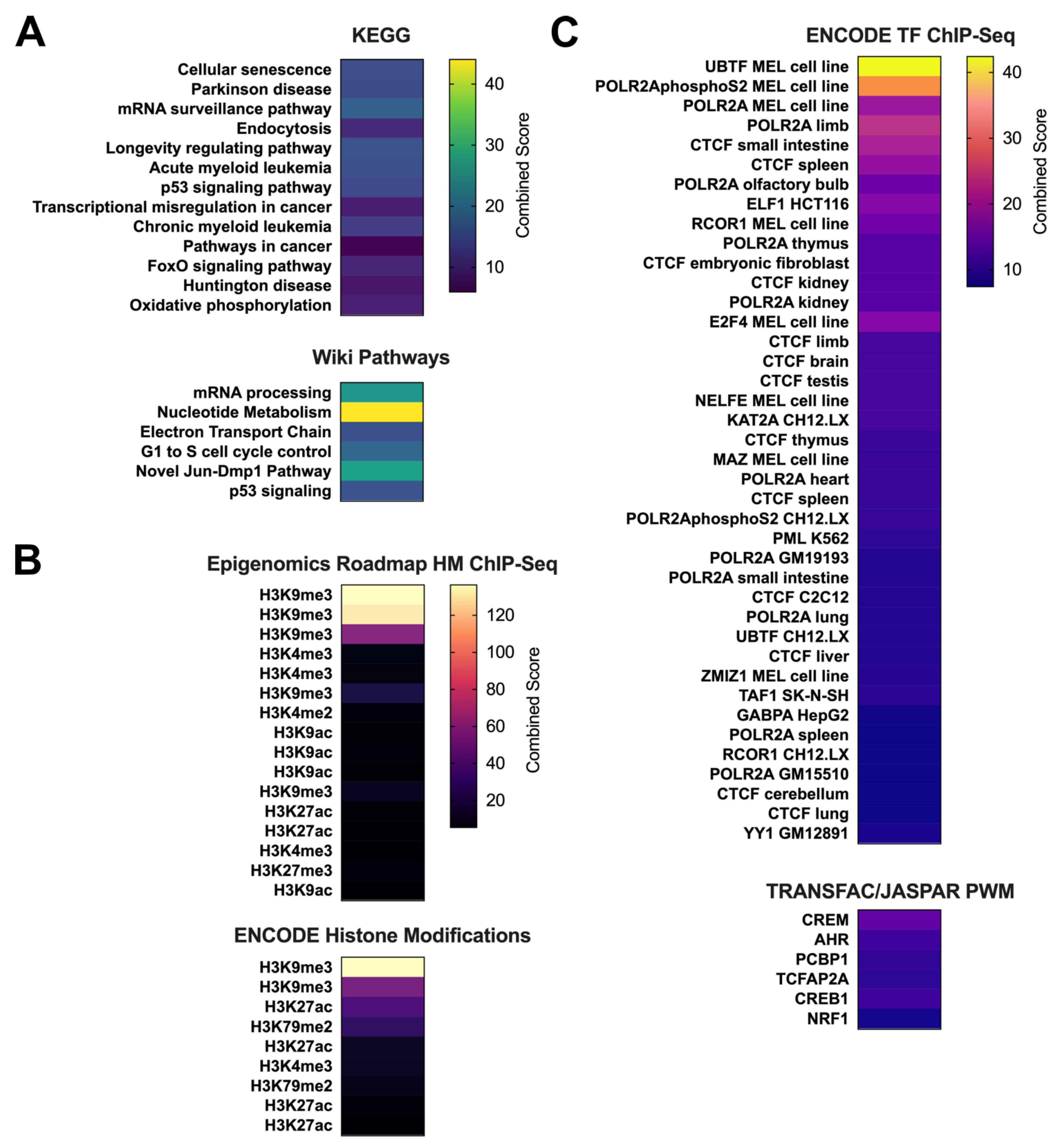
| Comparison Groups | Individual | No. of Unique Alignments | Mapping Efficiency | No. of DMFs at 5% FDR |
|---|---|---|---|---|
| Regressed | R1 | 6,942,498 | 44.3% | 834 |
| R2 | 7,861,430 | 48.6% | ||
| R3 | 7,240,969 | 47.0% | ||
| Regressed Control | (C)R1 | 11,964,235 | 50.6% | |
| (C)R2 | 12,197,807 | 49.8% | ||
| (C)R3 | 9,824,427 | 39.5% | ||
| Actively increasing lesions | A1 | 7,449,228 | 49.6% | 0 |
| A2 | 10,712,796 | 33.7% | ||
| A3 | 6,442,790 | 49.5% | ||
| Active Control | (C)A1 | 13,217,138 | 38.3% | |
| (C)A2 | 9,198,760 | 49.8% | ||
| (C)A3 | 10,374,183 | 51.9% | ||
| No CsA control | (C)1 | 6,434,780 | 47.8% | 0 |
| (C)2 | 6,853,519 | 49.1% | ||
| CsA | CsA1 | 9,428,602 | 49.5% | |
| CsA2 | 12,426,447 | 52.7% | ||
| Non-scarified | CsA1, CsA2 | 0 | ||
| Scarified | (C)A1, (C)A2, (C)A3 | |||
| Active | A1, A2, A3 | 0 | ||
| Regressed | R1, R2, R3 |
| Group | Sample | Total Reads | Viral Reads | % Viral Reads | % in E4 | Infection Status |
|---|---|---|---|---|---|---|
| Regressed lesions | R1 | 15,674,849 | 1,129,427 | 7.2 | 74 | Regressed lesion |
| R2 | 16,166,560 | 5871 | 0 | 0 | Regressed lesion | |
| R3 | 15,392,780 | 819,114 | 5.3 | 59.9 | Regressed lesion | |
| Regressed Control | (C)R1 | 23,666,799 | 3713 | 0 | 0 | Mock-infected |
| (C)R2 | 24,496,535 | 2868 | 0 | 0 | Mock-infected | |
| (C)R3 | 24,877,419 | 4143 | 0 | 0 | Mock-infected | |
| Actively increasing lesions | A1 | 15,020,374 | 849,677 | 5.7 | 52.1 | Active lesion |
| A2 | 31,835,719 | 48,521 | 0.2 | 11.4 | Active lesion | |
| A3 | 13,025,548 | 708,976 | 5.4 | 56 | Active lesion | |
| Active Control | (C)A1 | 34,512,945 | 6246 | 0 | 0 | Mock-infected |
| (C)A2 | 18,479,427 | 2534 | 0 | 0 | Mock-infected | |
| (C)A3 | 19,996,210 | 2475 | 0 | 0 | Mock-infected | |
| CsA | CsA1 | 19,044,226 | 1271 | 0 | 0 | CsA/No infection |
| CsA2 | 23,584,967 | 1829 | 0 | 0 | CsA/No infection | |
| No CsA Control | (C)1 | 13,470,444 | 1106 | 0 | 0 | No CsA/Not inf. |
| (C)2 | 13,946,497 | 1282 | 0 | 0 | No CsA/Not inf. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tschirley, A.M.; Stockwell, P.A.; Rodger, E.J.; Eltherington, O.; Morison, I.M.; Christensen, N.; Chatterjee, A.; Hibma, M. The Mouse Papillomavirus Epigenetic Signature Is Characterised by DNA Hypermethylation after Lesion Regression. Viruses 2021, 13, 2045. https://doi.org/10.3390/v13102045
Tschirley AM, Stockwell PA, Rodger EJ, Eltherington O, Morison IM, Christensen N, Chatterjee A, Hibma M. The Mouse Papillomavirus Epigenetic Signature Is Characterised by DNA Hypermethylation after Lesion Regression. Viruses. 2021; 13(10):2045. https://doi.org/10.3390/v13102045
Chicago/Turabian StyleTschirley, Allison M., Peter A. Stockwell, Euan J. Rodger, Oliver Eltherington, Ian M. Morison, Neil Christensen, Aniruddha Chatterjee, and Merilyn Hibma. 2021. "The Mouse Papillomavirus Epigenetic Signature Is Characterised by DNA Hypermethylation after Lesion Regression" Viruses 13, no. 10: 2045. https://doi.org/10.3390/v13102045
APA StyleTschirley, A. M., Stockwell, P. A., Rodger, E. J., Eltherington, O., Morison, I. M., Christensen, N., Chatterjee, A., & Hibma, M. (2021). The Mouse Papillomavirus Epigenetic Signature Is Characterised by DNA Hypermethylation after Lesion Regression. Viruses, 13(10), 2045. https://doi.org/10.3390/v13102045







