Molecular Characterization of Hovenia Dulcis-Associated Virus 1 (HDaV1) and 2 (HDaV2): New Tentative Species within the Order Picornavirales
Abstract
1. Introduction
2. Material and Methods
2.1. Plant Material
2.2. Enrichment of Viral Particles
2.3. High Throughput Sequencing and Analysis
2.4. RNA Extraction and Virus Detection by RT-PCR
2.5. 3′ RACE
2.6. Phylogenetic Analyses
2.7. Nucleotide Composition Analysis (NCA)
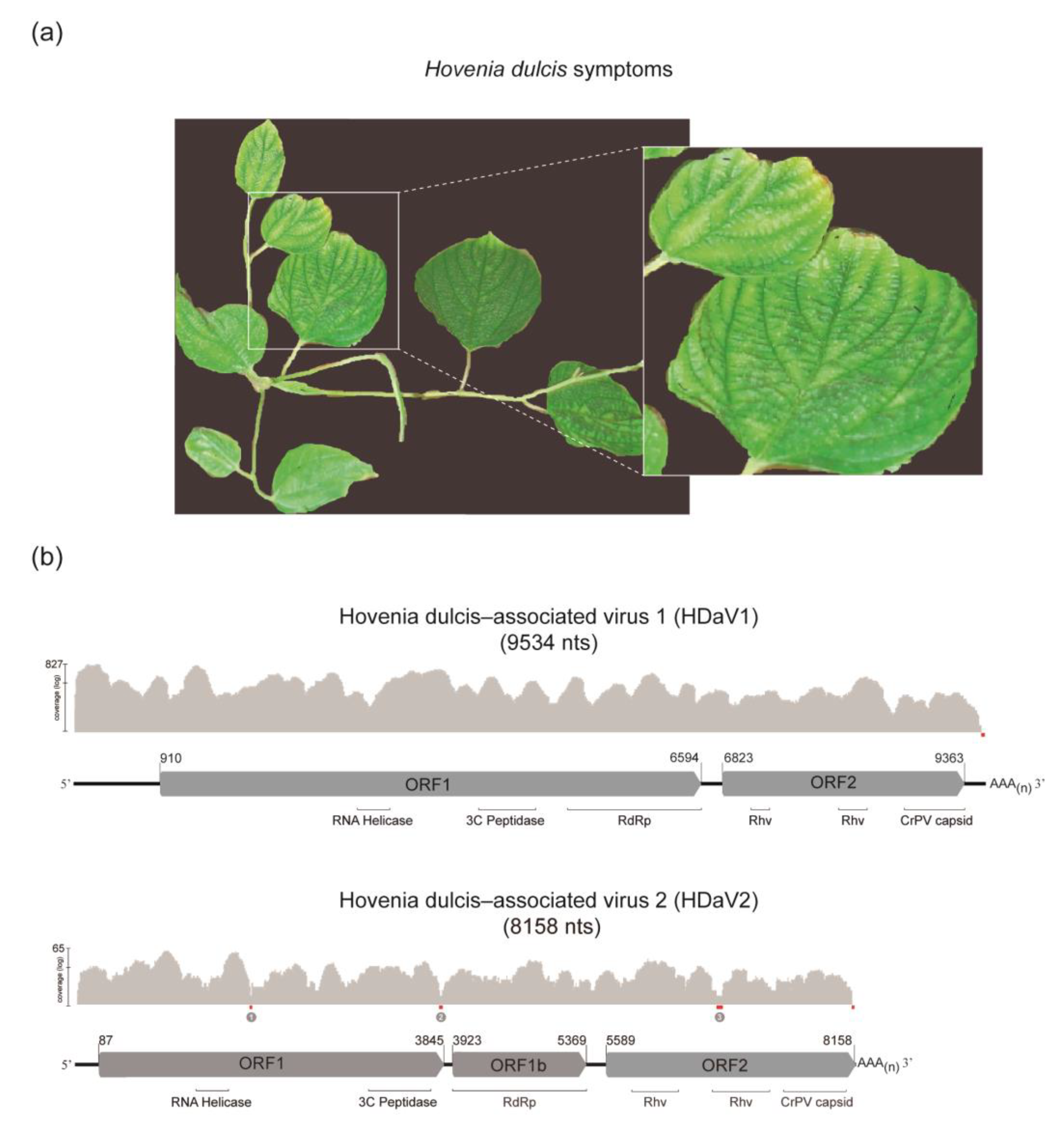
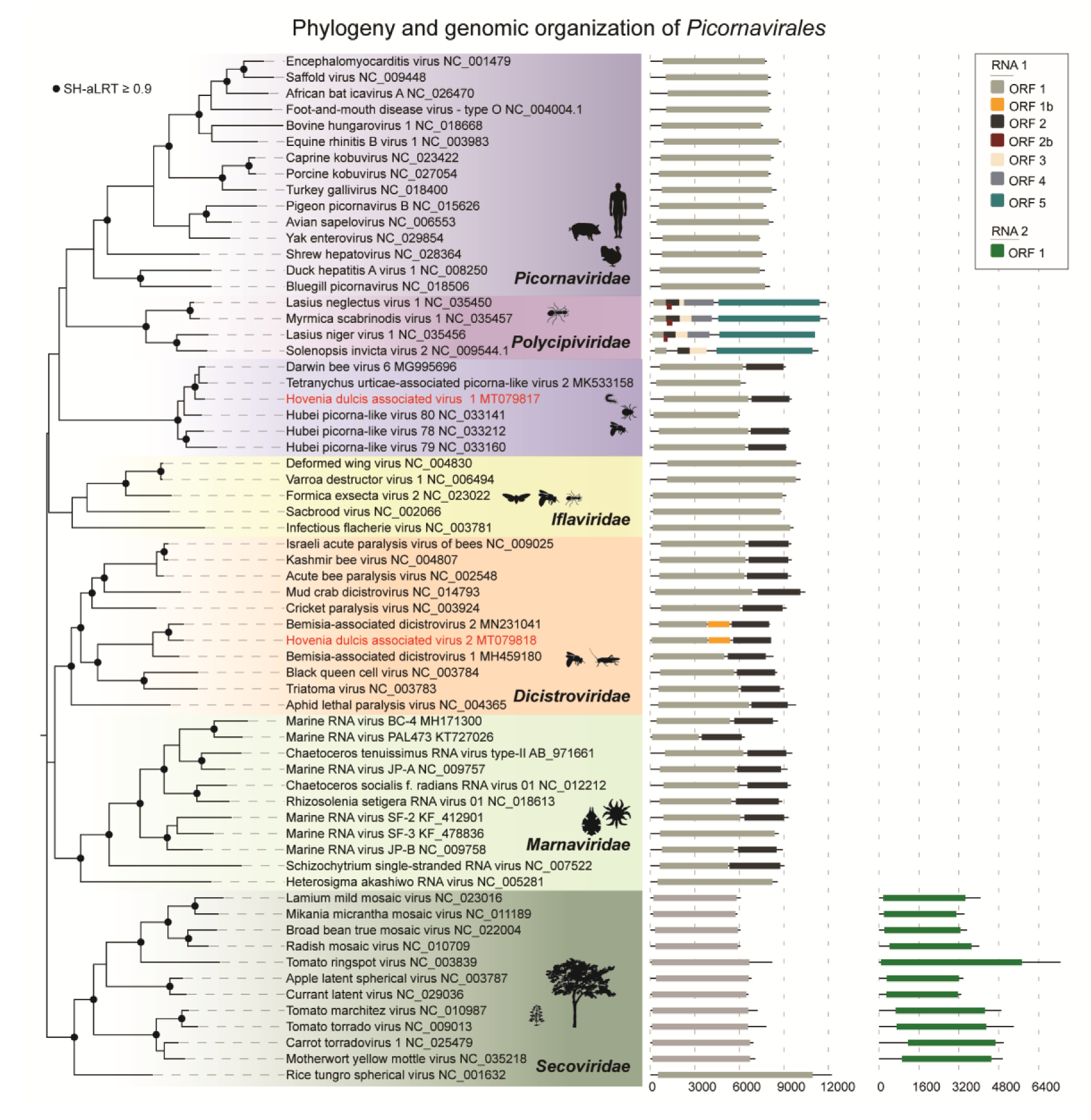
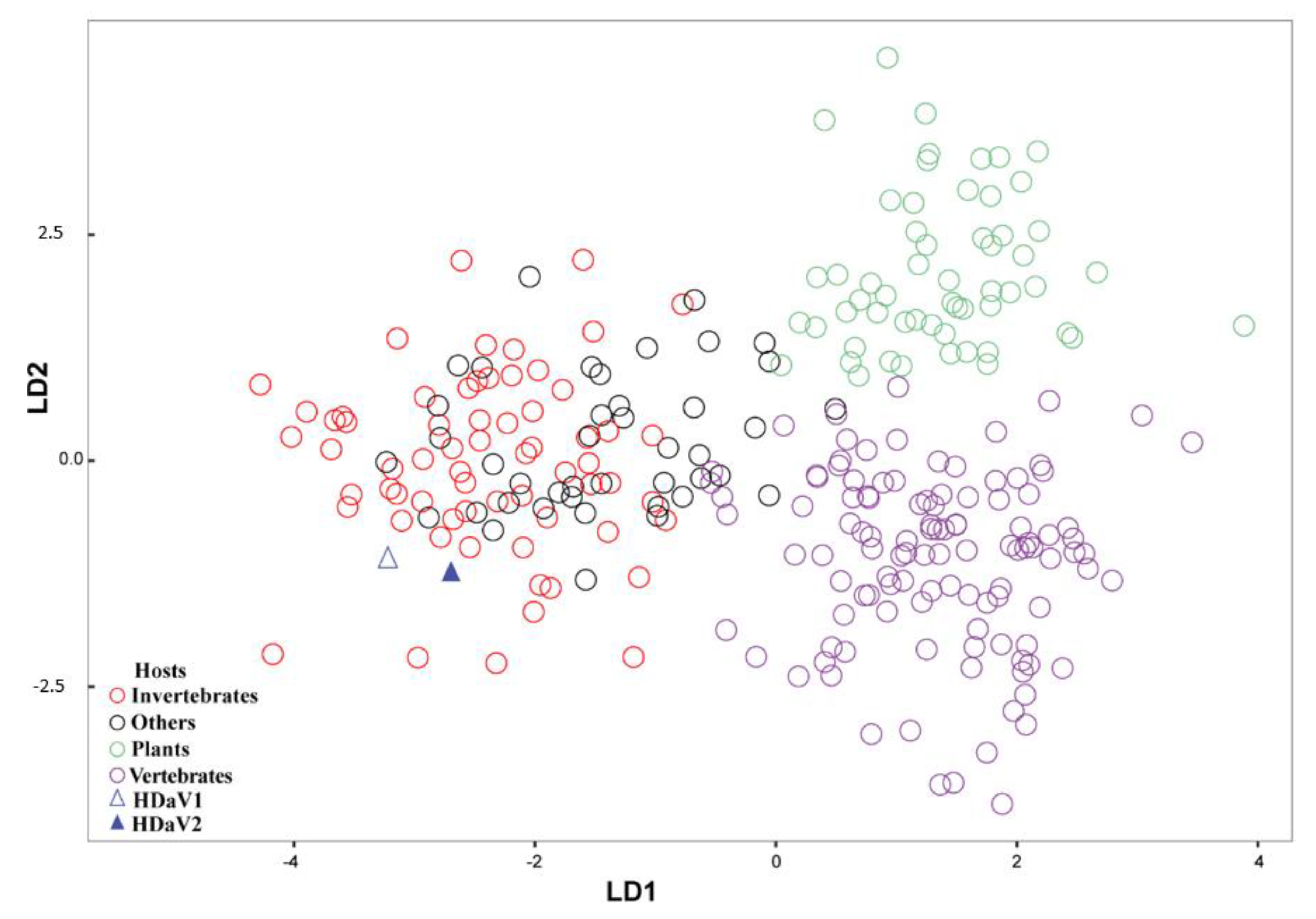
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Keenan, R.J.; Reams, G.A.; Achard, F.; de Freitas, J.V.; Grainger, A.; Lindquist, E. Dynamics of global forest area: Results from the FAO Global Forest Resources Assessment 2015. For. Ecol. Manag. 2015, 352, 9–20. [Google Scholar] [CrossRef]
- Bełka, M.; Gonthier, P.; Nicolotti, G. 2013: Infectious Forest Diseases; Gonthier, P., Nicolotti, G., Eds.; CABI: Wallingford, UK; Boston, MA, USA, 2015; p. 641. [Google Scholar]
- Nienhaus, F.; Castello, J.D. Viruses in forest trees. Annu. Rev. Phytopathol. 1989, 27, 165–186. [Google Scholar] [CrossRef]
- Büttner, C.; Von Bargen, S.; Bandte, M.; Mühlbach, H.-P. Forest diseases caused by viruses. In Infectious Forest Diseases; CABI: Wallingford, UK, 2013; pp. 50–75. [Google Scholar]
- Lin, M.T.; Kitajima, E.W.; Costa, C.L. Association of cassia mild mosaic virus with dieback of Cassia macranthera in central Brazil. Plant Dis. 1980, 64, 587–589. [Google Scholar] [CrossRef]
- Beserra, J.E.A., Jr.; de Carvalho, M.G.; Barguil, B.M.; Zerbini, F.M. Partial genome sequence of a Potyvirus and of a virus in the order Tymovirales found in Senna macranthera in Brazil. Trop. Plant Pathol. 2012, 36, 116–120. [Google Scholar] [CrossRef]
- Lin, M.T.; Kitajima, E.W.; Cupertino, F.P.; Costa, C.L. Properties of a possible carlavirus isolated from a cerrado native plant, Cassia sylvestris. Plant Dis. 1979, 63, 501–505. [Google Scholar]
- Gama, M.; Kitagima, E.W.; Avila, A.C.; Lim, M.T. Um Carlavirus em seringueira (Hevea brasiliensis). Fitopatol. Bras. 1983, 8, 621. [Google Scholar]
- Nicolini, C.; Pio-Ribeiro, G.; Andrade, G.P.; Melo, F.L.; Oliveira, V.C.; Guimarães, F.C.; Resende, R.O.; Kitajima, E.W.; Rezende, J.A.M.; Nagata, T. A distinct tymovirus infecting Cassia hoffmannseggii in Brazil. Virus Genes 2012, 45, 190–194. [Google Scholar] [CrossRef]
- Morozova, O.; Marra, M.A. Applications of next-generation sequencing technologies in functional genomics. Genomics 2008, 92, 255–264. [Google Scholar] [CrossRef]
- De Bruijn, F.J. Handbook of Molecular Microbial Ecology II: Metagenomics in Different Habitats; John Wiley & Sons: Hoboken, NJ, USA, 2011; pp. 15–24. [Google Scholar]
- Prabha, K.; Baranwal, V.K.; Jain, R.K. Applications of next generation high throughput sequencing technologies in characterization, discovery and molecular interaction of plant viruses. Indian J. Virol. 2013, 24, 157–165. [Google Scholar] [CrossRef]
- Barba, M.; Czosnek, H.; Hadidi, A. Historical perspective, development and applications of next-generation sequencing in plant virology. Viruses 2014, 6, 106–136. [Google Scholar] [CrossRef]
- Roossinck, M.J.; Martin, D.P.; Roumagnac, P. Plant virus metagenomics: Advances in virus discovery. Phytopathology 2015, 105, 716–727. [Google Scholar] [CrossRef] [PubMed]
- Melcher, U.; Muthukumar, V.; Wiley, G.B.; Min, B.E.; Palmer, M.W.; Verchot-Lubicz, J.; Ali, A.; Nelson, R.S.; Roe, B.A.; Thapa, V.; et al. Evidence for novel viruses by analysis of nucleic acids in virus-like particle fractions from Ambrosia psilostachya. J. Virol. Methods 2008, 152, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Pardina, P.E.R.; Bejerman, N.; Luque, A.V.; Di Feo, L. Complete nucleotide sequence of an Argentinean isolate of sweet potato virus G. Virus Genes 2012, 45, 593–595. [Google Scholar] [CrossRef] [PubMed]
- Candresse, T.; Filloux, D.; Muhire, B.; Julian, C.; Galzi, S.; Fort, G.; Bernardo, P.; Daugrois, J.-H.; Fernandez, E.; Martin, D.P. Appearances can be deceptive: Revealing a hidden viral infection with deep sequencing in a plant quarantine context. PLoS ONE 2014, 9, e102945. [Google Scholar] [CrossRef] [PubMed]
- Sheveleva, A.; Kudryavtseva, A.; Speranskaya, A.; Belenikin, M.; Melnikova, N.; Chirkov, S. Complete genome sequence of a novel Plum pox virus strain W isolate determined by 454 pyrosequencing. Virus Genes 2013, 47, 385–388. [Google Scholar] [CrossRef]
- Rott, M.; Xiang, Y.; Boyes, I.; Belton, M.; Saeed, H.; Kesanakurti, P.; Hayes, S.; Lawrence, T.; Birch, C.; Bhagwat, B.; et al. Application of next generation sequencing for diagnostic testing of tree fruit viruses and viroids. Plant Dis. 2017, 101, 1489–1499. [Google Scholar] [CrossRef]
- Maliogka, V.I.; Minafra, A.; Saldarelli, P.; Ruiz-García, A.B.; Glasa, M.; Katis, N.; Olmos, A. Recent advances on detection and characterization of fruit tree viruses using high-throughput sequencing technologies. Viruses 2018, 10, 436. [Google Scholar] [CrossRef]
- Fajardo, T.V.M.; Silva, F.N.; Eiras, M.; Nickel, O. High-throughput sequencing applied for the identification of viruses infecting grapevines in Brazil and genetic variability analysis. Trop. Plant Pathol. 2017, 42, 250–260. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Gruber, A.R.; Bernhart, S.H.; Lorenz, R. The ViennaRNA web services. In RNA Bioinformatics; Springer: Berlin/Heidelberg, Germany, 2015; pp. 307–326. [Google Scholar]
- Lorenz, A.R.; Bernhart, S.H.; Neubock, R.; Hofacker, I.L. The vienna RNA websuite. Nucleic Acids Res. 2008, 36 (Suppl. 2), W70–W74. [Google Scholar]
- Lorenz, R.; Bernhart, S.H.; Zu Siederdissen, C.H.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. Vienna RNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Verwoerd, T.C.; Dekker, B.M.; Hoekema, A. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 1989, 17, 2362. [Google Scholar] [CrossRef] [PubMed]
- Silva, K.N.; Nicolini, C.; Silva, M.S.; Fernandes, C.D.; Nagata, T.; Resende, R.O. First report of Johnsongrass mosaic virus (JGMV) infecting Pennisetum purpureum in Brazil. Plant Dis. 2013, 97, 1003. [Google Scholar] [CrossRef]
- Chen, J.; Adams, M.J. A universal PCR primer to detect members of the Potyviridae and its use to examine the taxonomic status of several members of the family. Arch. Virol. 2001, 146, 757–766. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003, 1, 2–3. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Kapoor, A.; Simmonds, P.; Lipkin, W.I.; Zaidi, S.; Delwart, E. Use of nucleotide composition analysis to infer hosts for three novel picorna-like viruses. J. Virol. 2010, 84, 10322–10328. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P. SSE: A nucleotide and amino acid sequence analysis platform. BMC Res. Notes 2012, 5, 50. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Roberts, J.M.K.; Anderson, D.L.; Durr, P.A. Metagenomic analysis of Varroa-free Australian honey bees (Apis mellifera) shows a diverse Picornavirales virome. J. Gen. Virol. 2018, 99, 818–826. [Google Scholar] [CrossRef] [PubMed]
- Bourgarel, M.; Noël, V.; Pfukenyi, D.; Michaux, J.; André, A.; Becquart, P.; Cerqueira, F.; Barrachina, C.; Boué, V.; Talignani, L.; et al. Next-generation sequencing on insectivorous bat guano: An accurate tool to identify arthropod viruses of potential agricultural concern. Viruses 2019, 11, 1102. [Google Scholar] [CrossRef] [PubMed]
- Bonning, B.C.; Miller, W.A. Dicistroviruses. Annu. Rev. Entomol. 2010, 55, 129–150. [Google Scholar] [CrossRef]
- Nakasu, E.Y.T.; Hedil, M.; Nagata, T.; Michereff-Filho, M.; Lucena, V.S.; Inoue-Nagata, A.K. Complete genome sequence and phylogenetic analysis of a novel dicistrovirus associated with the whitefly Bemisia tabaci. Virus Res. 2019, 260, 49–52. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.D.; Tian, J.H.; Chen, L.J.; Chen, X.; Li, C.X.; Qin, X.C.; Li, J.; Cao, J.P.; Eden, J.S.; et al. Redefining the invertebrate RNA virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef]
- François, S.; Mutuel, D.; Duncan, A.B.; Rodrigues, L.R.; Danzelle, C.; Lefevre, S.; Santos, I.; Frayssinet, M.; Fernandez, E.; Filloux, D.; et al. A new prevalent densovirus discovered in Acari. Insight from metagenomics in viral communities associated with two-spotted mite (Tetranychus urticae) populations. Viruses 2019, 11, 233. [Google Scholar] [CrossRef]
- Olendraite, I.; Lukhovitskaya, N.I.; Porter, S.D.; Valles, S.M.; Firth, A.E. Polycipiviridae: A proposed new family of polycistronic picorna-like RNA viruses. J. Gen. Virol. 2017, 98, 2368–2378. [Google Scholar] [CrossRef]
- Le Gall, O.; Christian, P.; Fauquet, C.M.; King, A.M.Q.; Knowles, N.J.; Nakashima, N.; Stanway, G.; Gorbalenya, A.E. Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T= 3 virion architecture. Arch. Virol. 2008, 153, 715. [Google Scholar] [CrossRef]
- Lang, A.S.; Vlok, M.; Suttle, C.A. Assigning 4 new and 2 unassigned genera to the family Marnaviridae. Int. Comm. Taxon. Viruses ICTV 2018, 2, 1–6. [Google Scholar]
- King, A.M.Q.; Lefkowitz, E.; Adams, M.J.; Carstens, E.B. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Gildow, F.E.; D’arcy, C.J. Cytopathology and experimental host range of Rhopalosiphum padi virus, a small isometric RNA virus infecting cereal grain aphids. J. Invertebr. Pathol. 1990, 55, 245–257. [Google Scholar] [CrossRef]
- Li, J.L.; Cornman, R.S.; Evans, J.D.; Pettis, J.S.; Zhao, Y.; Murphy, C.; Peng, W.J.; Wu, J.; Hamilton, M.; Boncristiani, H.F.; et al. Systemic spread and propagation of a plant-pathogenic virus in European honeybees, Apis mellifera. MBio Am. Soc. Microbiol. 2014, 5, e00898-13. [Google Scholar] [CrossRef] [PubMed]
- Flenniken, M.L. Honey bee-infecting plant virus with implications on honey bee colony health. MBio 2014, 5, e00877-14. [Google Scholar] [CrossRef] [PubMed]
- Galbraith, D.A.; Fuller, Z.L.; Ray, A.M.; Brockmann, A.; Frazier, M.; Gikungu, M.W.; Martinez, J.F.I.; Kapheim, K.M.; Kerby, J.T.; Kocher, S.D.; et al. Investigating the viral ecology of global bee communities with high-throughput metagenomics. Sci. Rep. 2018, 8, 8879. [Google Scholar] [CrossRef]
- Jiwaji, M.; Matcher, G.F.; de Bruyn, M.-M.; Awando, J.A.; Moodley, H.; Waterworth, D.; Jarvie, R.A.; Dorrington, R.A. Providence virus: An animal virus that replicates in plants or a plant virus that infects and replicates in animal cells? PLoS ONE 2019, 14, e0217494. [Google Scholar] [CrossRef]
- Regelin, Z.T. Translation and Replication of Rhopalosiphum Padi Virus RNA in a Plant Cellular Environment. Master of Science Thesis, Digital Repository Iowa State University, Ames, Iowa, 30 April 2010. [Google Scholar]
- Ma, Y.; Marais, A.; Lefebvre, M.; Theil, S.; Svanella-Dumas, L.; Faure, C.; Candresse, T. Phytovirome analysis of wild plant populations: Comparison of double-stranded RNA and virion-associated nucleic acid metagenomic approaches. J. Virol. 2019, 94, e01462-19. [Google Scholar] [CrossRef]
| Primer Name | Primer Sequence 5′–3′ | AT 1 (°C) | Amplicon Size (bp) | Application/Target Genomic Regions |
|---|---|---|---|---|
| HDaV1_7626_F 2 | AGTCACTGGTGCGTTAGGTG | 57 | 993 | Detection/Capsid |
| HDaV1_8618_R 3 | GTAAGCATACCTCCACGCGA | |||
| HDaV2_6983_F | GAATGAACTGCGTGCTACAC | 59 | 757 | Detection/Capsid |
| HDaV2_7739_R | CCGGGGGAAAACAGCAGT | |||
| C1622_254_F 4 | TTAATGGGGTTGCAGGGCTT | 60 | 519 | Detection/RdRp |
| C1622_772_R 4 | TCATGACTCCTATGCGCCAC | |||
| C1177_207_F 5 | GTGTCGTTTGTATCGCAGGC | 59 | 674 | Detection/RdRp |
| C1177_880_R 5 | CGCGCTCATAGCCAAACAAA | |||
| C_1797_31_F 6 | ATTGAAAACGCGACCTGCAC | 59 | 571 | Detection/RdRp |
| C1797_601_R 6 | GCGGGATAAGCTCACCAAGT | |||
| HDaV2_1630_F | TGCAAGAGTACCAGGAACAGAATAAT | 54 | 608 | Low coverage region 1/ORF1 |
| HDaV2_2236_R | GCAAGGCCATGATACATGACCA | |||
| HDaV2_3431_F | AGAAAGTGTTTACTATGTAGCACCAACT | 59 | 549 | Low coverage region 2/ORF1b |
| HDaV2_3981_R | CTATTCCTTGGCAGGCTTGACG | |||
| HDaV2_6422_F | GTCTGCTCCTGATGCTAATCCG | 58 | 540 | Low coverage region 3 |
| HDaV2_6961_R | GCTGGGACATCATCAAGGGAAC | |||
| Oligod50TM4 | GTTTTCCCAGTCACGACTTAATTAA(T)50 | 65 | − | Race cDNA |
| M4 | GTTTTCCCAGTCACGACT | 56 | − | Race 3′ PCR |
| HDaV1_9041_F | CCTCAGAAGTTTTCGAGACTGC | 56 | − | Race 3′ PCR |
| HDaV2_7256_F | ACCTCACAAATATACTGTTGGTGAGG | 60 | − | Race 3′ PCR |
| HDaV2_7518_F | CCTGAACTTGGTATATTGGATGTTCCC | 60 | − | Race 3′ PCR |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nery, F.M.B.; Melo, F.L.; Boiteux, L.S.; Ribeiro, S.G.; Resende, R.O.; Orílio, A.F.; Batista, J.G.; Lima, M.F.; Pereira-Carvalho, R.C. Molecular Characterization of Hovenia Dulcis-Associated Virus 1 (HDaV1) and 2 (HDaV2): New Tentative Species within the Order Picornavirales. Viruses 2020, 12, 950. https://doi.org/10.3390/v12090950
Nery FMB, Melo FL, Boiteux LS, Ribeiro SG, Resende RO, Orílio AF, Batista JG, Lima MF, Pereira-Carvalho RC. Molecular Characterization of Hovenia Dulcis-Associated Virus 1 (HDaV1) and 2 (HDaV2): New Tentative Species within the Order Picornavirales. Viruses. 2020; 12(9):950. https://doi.org/10.3390/v12090950
Chicago/Turabian StyleNery, Flávia M. B., Fernando L. Melo, Leonardo S. Boiteux, Simone G. Ribeiro, Renato O. Resende, Anelise F. Orílio, Josiane G. Batista, Mirtes F. Lima, and Rita C. Pereira-Carvalho. 2020. "Molecular Characterization of Hovenia Dulcis-Associated Virus 1 (HDaV1) and 2 (HDaV2): New Tentative Species within the Order Picornavirales" Viruses 12, no. 9: 950. https://doi.org/10.3390/v12090950
APA StyleNery, F. M. B., Melo, F. L., Boiteux, L. S., Ribeiro, S. G., Resende, R. O., Orílio, A. F., Batista, J. G., Lima, M. F., & Pereira-Carvalho, R. C. (2020). Molecular Characterization of Hovenia Dulcis-Associated Virus 1 (HDaV1) and 2 (HDaV2): New Tentative Species within the Order Picornavirales. Viruses, 12(9), 950. https://doi.org/10.3390/v12090950






