Abstract
Fast and reliable detection of bacterial pathogens in clinical samples, contaminated food products, and water supplies can drastically improve clinical outcomes and reduce the socio-economic impact of disease. As natural predators of bacteria, bacteriophages (phages) have evolved to bind their hosts with unparalleled specificity and to rapidly deliver and replicate their viral genome. Not surprisingly, phages and phage-encoded proteins have been used to develop a vast repertoire of diagnostic assays, many of which outperform conventional culture-based and molecular detection methods. While intact phages or phage-encoded affinity proteins can be used to capture bacteria, most phage-inspired detection systems harness viral genome delivery and amplification: to this end, suitable phages are genetically reprogrammed to deliver heterologous reporter genes, whose activity is typically detected through enzymatic substrate conversion to indicate the presence of a viable host cell. Infection with such engineered reporter phages typically leads to a rapid burst of reporter protein production that enables highly sensitive detection. In this review, we highlight recent advances in infection-based detection methods, present guidelines for reporter phage construction, outline technical aspects of reporter phage engineering, and discuss some of the advantages and pitfalls of phage-based pathogen detection. Recent improvements in reporter phage construction and engineering further substantiate the potential of these highly evolved nanomachines as rapid and inexpensive detection systems to replace or complement traditional diagnostic approaches.
1. Introduction
The development of quick and reliable methods for pathogen detection and identification is critical to improve the prevention and treatment of bacterial diseases in various fields from food production to health care. While culture-based detection remains the gold standard for detection and identification of bacterial pathogens, it can be time and labor intensive, typically requiring more than 48 h to allow selective bacterial growth to ensure reliable detection [1,2]. Within the clinic, early microbial identification is important for ensuring patients receive optimal antibiotic treatment. Approximately 30–50% of patients receive ineffective antibiotic therapy because physicians must treat immediately with first-line, broad-spectrum antibiotics until the results of culture-based detection are available [3]. For example, blood culture assays require 48 to 72 h to complete [4], with fastidious organisms such as Bacillus species and HACEK (Haemophilus species, Aggregatibacter species, Cardiobacterium hominis, Eikenella corrodens, and Kingella species) organisms requiring several days to produce a conclusive result [5]. This not only affects patient survival due to the prescription of inappropriate antibiotics [6,7] but the misuse of antibiotics is a direct contributor to the global spread of antimicrobial resistant bacteria [8,9,10,11].
The increasing scale of production and global distribution of food goods, especially fresh fruit, vegetables and ready-to-eat products, makes quick and accurate microbial detection essential for ensuring circulation of high-quality and safe foods. Again, conventional culture-based methods are incommensurate with the quick turnaround time required by food producers today [12]. For example, ISO 11290-1:2017 guidelines for culture-based Listeria monocytogenes detection recommends a minimum of 24 h for colony formation on chromogenic agar, with an additional 24 h for slow-growing bacteria and an additional 96 h to enable complete morphological identification of colonies (e.g., phospholipase activity) [13]. As such, food products requiring test results prior to their release (positive release) are placed in temporary quarantine, which costs money and affects product half-life. Nevertheless, food products can still arrive on supermarket shelves without satisfactory microbial assessment, leading to costly product recalls as well as the spread of life-threatening foodborne disease outbreaks [14]. In Europe alone, over 23 million annual illnesses and 5000 deaths are associated with the consumption of contaminated food products [15]; in 2018, EU member states reported 5146 foodborne outbreaks, mostly from Salmonella-contaminated produce [16]. To meet the demands from the clinic and food producers for improved bacterial detection, a multitude of rapid diagnostic systems have been established that attempt to circumvent the need for extensive selective culturing.
While culture-based methods remain the mainstay diagnostic, clinical testing is becoming increasingly reliant on culture-independent diagnostic tests (CIDTs) such as nucleic acid amplification or ELISA-based antigen detection [17,18,19], as well as matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) and whole-genome sequencing (WGS) for detection of bacteria [20,21]. The advantage of these approaches is the potential for automation, making them reproducible and easy to use while also allowing sensitive detection of non-culturable organisms and polymicrobial infections (multiplex detection). However, the specificity of these approaches can be affected by detection of closely related, non-target species generating false-positive results [22,23]. Furthermore, nucleic acid-based methods lack the ability to differentiate between DNA from viable and dead bacterial cells, meaning they can assess microbial viability retrospectively, i.e., changes in nucleic acid levels over a given timeframe; however, they are incapable of determining viability within discrete samples, making identification of only viable (and potentially infections) bacteria extremely challenging. The adoption of CIDTs, WGS and MALDI-TOF-MS by the food industry is also a significantly bigger challenge than clinical samples due to the lower levels of bacterial contaminants typically present and the greater variability in product matrices where heterogeneous levels of fats, proteins and debris can impede bacterial detection. A robust and sensitive alternative is to implement phage-based diagnostics.
Bacteriophages (phages) are environmentally ubiquitous viruses that infect specific bacterial hosts. The ability of phages to target and kill bacteria down to the species or even strain level, has led to the resurgence of interest over the last decade in their use as therapeutics (i.e., phage therapy) against antibiotic resistant bacterial infections [24,25,26]. Phages are also used as biocontrol agents to tackle foodborne bacteria such as Listeria and Salmonella [27]. Phages have evolved highly efficient mechanisms to attach to bacteria under various harsh environments that provide a clear advantage over other affinity-based system for bacterial recognition in complex sample matrices. The ability of phage particles to bind with high affinity and specificity to their target bacteria has led to their use as affinity molecules instead of other bio-probes such as antibodies in biosensor-based assays [28,29,30] and for bacterial enrichment and detection when conjugated to magnetic nanobeads [31] (Figure 1A). A drawback of using whole-phage particles exclusively as affinity molecules is their relatively large size and (unless they are inactivated) their basal lytic activity that may destroy target bacteria before downstream detection can be completed. The alternative to whole-phage bio-probes is to use phage proteins that confer host binding (Figure 1B,C). For example, cell wall-binding domains (CBDs) of phage endolysins have been successfully used for detecting various Gram-positive bacteria, e.g., Listeria [32,33], Bacillus cereus [34], and Clostridium tyrobutyricum [35]. Phages recognize their bacterial hosts using specialized receptor-binding proteins (RBPs), identified as tail fibers and tailspikes, which initiate the attachment of the phage to specific receptors on the bacterial cell wall [36,37]. Various RBP-based detection assays have been developed for detecting Salmonella [38,39], Shigella [40], and Pseudomonas aeruginosa [41]. By combining Listeria-specific CBDs and RBPs, a glycotyping system was also recently developed for discerning different Listeria serovars [42]. In addition to using the ability of phages or phage proteins to bind a distinct host range of bacteria, many research groups have developed highly efficient reporter phage systems that exploit the rapid infection, DNA replication, and phage production kinetics within infected target cells (Figure 1D–G). This can be achieved by simply monitoring an increase in phage titer (Section 2), or by quantifying the production of phage-encoded heterologous proteins such as fluorescent proteins, luciferases, or other enzymatic reporters (Section 5). This review focuses on the design principles that govern the engineering of heterologous reporters into phages (Section 3) and how these phages can be used to detect a wide variety of clinically and industrially important bacterial pathogens.

Figure 1.
Overview of phage-based pathogen detection. Capture-based detection: The limited host ranges of phages towards a given genus, species or subspecies of bacteria make them ideal candidates for development into detection bio-probes. (A) The high binding affinity of whole-phage particles has led to their use as bio-probes in biosensors [28,29,30], or by conjugation with radioactive tracers [43] fluorophores [44], magnetic nanoparticles [31,45] or a combination of both [46] to label and enrich bacteria for detection. (B) Host specificity of phages is mediated by specialized receptor-binding proteins (RBPs) that provide equivalent binding capabilities as whole phages, but at a fraction of the size. Recently, RBPs have been applied in biosensors [47], ELISA-based assays [38,40], and for glycotyping Salmonella [39] and Listeria [32]. (C) Alternatively, cell wall-binding domains (CBDs) of phage endolysins have proven highly effective at detecting Gram-positive pathogens. CBDs have recently been used to detect B. cereus using biosensors [34] lateral flow assays [48] and magnetic enrichment-based detection [49]. In addition, Listeria-targeting CBDs were used to glycotype and identify Listeria serovars [42] and Clostridium tyrobutyricum-targeting CBDs have been employed for spore detection during cheese spoilage [35,50]. Infection-based detection: Infection of a bacterial host by lytic phage leads to rapid progeny phage production and ensuing cell lysis. (D) Released progeny phages [51,52] or bacterial cell content (e.g., ATP, DNA, RNA and bacterial proteins) provide excellent markers for downstream detection of the original bacterial host [53,54,55]. Alternatively, genetically engineered phages encoding reporters such as fluorescent proteins (E) [56,57,58], luciferases (F) [59,60,61,62] or hydrolyzing enzymes (e.g., β-galactosidase) (G) [63,64] are used. These phages express the reporter proteins during host infection to produce an amplifying fluorescent or bioluminescent signal upon the addition of a substrate. The rapid and sensitive nature of reporter phage-based systems has made them ideal tools for detecting low levels of viable, contaminating bacteria in many matrices, including foods, water and clinical samples.
2. Phage Amplification-Based Detection
After successful host cell adsorption, a phage typically injects its genome and internal proteins into the bacterium and manipulates host metabolic processes to begin production of progeny phages, which are subsequently released upon host cell lysis. The number of progeny phages produced and the time required (latent period) can vary greatly between different phages and the hosts they encounter. For instance, coliphage T4 produces 150–200 progeny phage per infected cell within 25 min under optimal laboratory conditions [65]; however, under environmental and unfavorable conditions, phage amplification (i.e., burst size and latent period) can be affected [66]. Nevertheless, phage amplification can occur quickly within a contaminated sample to generate a large number of progeny particles. An increase in phage numbers indicates the presence of a susceptible host within the sample. The change in phage titer can be determined by traditional plaque assays using suitable indicator strains [52,67,68], by physical detection of the phage particles using ELISA-based assays [69], or as discussed below, by measuring the increase in phage nucleic acid content. Sensitivity of these assays can be improved by capturing and enriching the progeny phage particles [70] by using lateral flow assays [71,72,73,74] or (magnetic) bead-based enrichment. For example, antibody-conjugated beads were used to isolate amplified MS2 phages from solution for detection of Escherichia coli [75]. In addition to detecting whole-phage particles, real-time PCR methods have also been developed for detecting the increase in phage DNA generated during infection [55,68,76]. For instance, Griffiths and colleagues coupled capture of pathogenic E. coli and Salmonella Newport from food samples by phage coated paper dipsticks with qPCR-mediated detection of progeny phage DNA [55]. Phage RNA has also been used as a biomarker. For instance, surrogate marker loci in engineered mycobacteriophages [77,78] or simply the phage’s natural RNA [54] have both been detected using qPCR methods. Methods that are based on reverse transcription of phage RNA can offer enhanced sensitivity and are less prone to false-positive results than DNA amplification-based detection systems [79].
In addition to progeny phage detection, cell lysis releases an abundance of cellular content that can be used as diagnostic markers. For instance, ATP is easily detectable after release from cells using a bioluminescence reaction with firefly luciferase [80]. This kind of bioluminescence-based sensing of ATP released from cells after infection with phage K enabled detection of Stapylococcus spp. in fluid from orthopedic artificial joints [53]. Other methods have visualized the change in redox reactions by Salmonella enterica Typhi and Paratyphi after phage infection [81], or measured the release of eDNA from phage infected E. coli [82]. Table 1 provides a detailed overview of phage amplification-based detection assays for bacterial pathogens. A downside of most phage amplification-based assays is the requirement for completion of the phage lytic cycle to allow the release of biomarkers or progeny phages for detection. In contrast, many reporter phage-based systems do not require cell lysis or completion of the phage infection cycle to detect infected cells. Host adsorption and phage genome injection followed by reporter gene expression is generally sufficient to produce a detectable signal, allowing reporter phages to circumvent the gamut of intracellular phage defense mechanisms that can inhibit various stages of the phage infection cycle [83,84].

Table 1.
Phage amplification-based detection assays published since 2016.
3. Design Rules for Reporter Phage Engineering
Compared to measuring phage amplification or released cell content, reporter proteins can be overexpressed within the bacterial host using strong promoters to produce an intense and amplified signal for instant detection upon substrate addition, providing a more sensitive diagnostic assay than relying on phage lysis alone. As detailed in Table 2, an abundance of reporter phage systems have been developed in the last four years using different engineering platforms and reporter genes. As important as the choice of engineering technique seems to be, the question of how to design the reporter phage is of paramount importance. Four principles for engineering an effective reporter phage are:

Table 2.
Reporter phage-based detection assays published since 2016.
- The phage has to be able to infect or transduce the reporter gene into the target bacteria. Therefore, phages capable of infecting a broad spectrum of representative strains for the target pathogen should be used.
- Both the reporter gene and the method of detection have to be suitable for the target bacteria, expected microbial content, and the sampling environment. Inclusion of a pre-enrichment step is highly recommended when low bacterial numbers are expected in the sample, for instance, the zero tolerance policy for common food pathogens (e.g., Salmonella, Campylobacter and E. coli) necessitates a pre-enrichment of food samples to ensure bacterial amplification to a minimal threshold for detection. In addition, as food components can affect fluorescence-based detection or physically block biosensors, detection from complex matrices should be coupled with an initial capture and enrichment step, e.g., bead-based magnetic separation.
- Identify a suitable region within the phage genome that allows for integration of a heterologous reporter gene without disrupting infectivity. Furthermore, lysogenic phages can be modified to become strictly lytic by targeted deletion of genes involved in prophage integration and maintenance. The removal of such genes makes room for additional payloads within space-limited phage genomes. For example, Kim et al. 2017 deleted whole regions unrelated to phage infectivity, such as the integrase (int), adenine methylase (am), O-acetyltransferase (oac), and anti-immunity protein (aip) genes and replaced them with the luxCDABE operon [88]. In Listeria phages A500 and A006, the lysogeny control module was removed, rendering the phages strictly lytic (Meile et al. 2020). DS6A-derived mycobacteriophages Φ2GFP12 and Φ2GFP13 were created by replacing two independent regions including the integrase gene or mazG with the L5 promoter driven mVenus reporter cassette [58]. In the E. coli phages IP008BK and IP052BK, the non-essential small outer capsid gene soc was replaced with the cytochrome c peroxidase gene ccp [89].
- Reporter gene expression should be tuned to ensure sufficient production and subsequent signal generation for detection. For instance, a heterologous strong promoter can be used or the reporter gene can be inserted into a region controlled by a strong endogenous promoter (e.g., structural genes). For instance, expression of NanoLuc from an early promoter controlling the anti-CRISPR locus of Listeria phage A006 provided rapid protein production and early detection during the latent period [90]. Still, many reporter genes are inserted downstream of the strongly expressed but rather late endogenous promoters of the major capsid gene [62,91] the endolysin gene [62], or the tail spike protein [88]. Some examples of recently used constitutive exogenous promoters are PL(L5) for mycobacteria [58,92,93,94], and PrplU [95] and phi10 T7 [96] for E. coli. In addition, the N-terminal leader sequence pelB was added to increase soluble heterologous gene expression and direct translated reporter proteins to the periplasmic space ([97,98,99]).
In addition, various reporter proteins do not require cell lysis or completion of the phage infection cycle to produce a signal. Genome injection and potentially some degree of DNA replication is therefore sufficient [58,62]. In fact, an intact phage genome may not even be required: non-replicative phage particles carrying a luciferase-containing plasmid (fittingly named Smarticles™ (Roche/Geneweave, Los Gatos, CA, USA)) simply transduce the luciferase gene into the target bacteria for detection [100]; however, this approach does remove the possibility of signal amplification from secondary phage infections.
4. Engineering Platforms Available for Reporter Phage Generation
Current approaches for engineering of phages can be assigned to three categories: (i) direct cloning; (ii) homologous recombination with or without CRISPR-Cas counter selection; and (iii) whole genome activation, a.k.a. “rebooting”.
4.1. Direct Cloning
Phages can be cloned using phage vectors (phasmids or phagemids), which are plasmids with an additional origin of replication and packaging sequence originating from a phage. This approach is quick and efficient due to the ability to insert genes using standard cloning procedures (e.g., restriction enzyme digestions and insertion into multiple cloning sites) and phasmids can be simply propagated as plasmids or lytically as phages. However, the application is currently limited to mycobacteriophages [116,117] and Gram-negative targeting phages, such as lambda, M13 and T7 [118,119,120]. Recently published fluoromycobateriophages described in Table 2 were engineered using phasmids following the same strategy: TM4 shuttle phasmids are cloned in vitro to contain the desired reporter gene (e.g., GFP or RFP), and the phasmid DNA is then packaged into phage λ particles for amplification in an intermediate E. coli host before transformation into a fast growing M. smegmatis strain mc2155 to produce the final infectious virions [56,57] (Figure 2A). A major limitation of using this system is the relatively small packaging capacity of the lambda phage capsid (~53 kb), meaning larger genomes are unsuitable for phasmid construction, especially for simultaneous insertion of reporter genes [121].
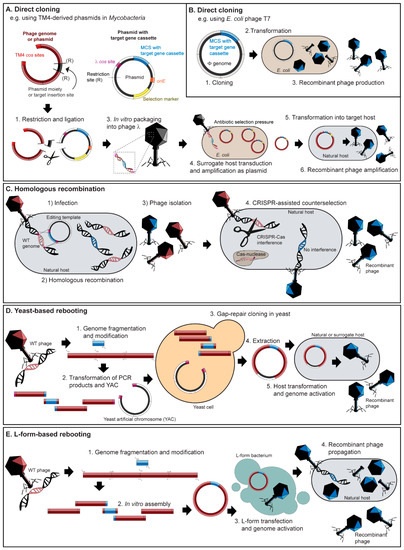
Figure 2.
Overview of engineering strategies for reporter phage generation. (A) Heterogeneous reporter genes can be inserted into dual function shuttle phasmids capable of plasmid replication in E. coli and phage replication in a target host, e.g., Mycobacterium spp. (B) Reporter genes can also be directly inserted into phage genomes, e.g., T7Select®(Novagen). (C) Homologous recombination (and recombineering with bacteriophage-encoded homologous recombination systems, such as the coliphage λ Red system) involves the swapping of a reporter gene containing homology arms featured on an editing plasmid with the homologous region located within a phage genome. Homologous recombination can be combined with CRISPR-Cas counter selection to facilitate the removal of wild-type phages to improve identification of recombinant phages. (D) PCR products of a phage genome and a reporter gene insert are transformed alongside a linearized yeast replicon fragment from a yeast artificial chromosome (YAC) into yeast cells. The phage genome is assembled in the YAC vector by gap-repair cloning. YAC–phage DNA is extracted and directly transformed into the host bacterium leading to the production of recombinant phages. (E) A synthetic genome can be in vitro assembled (e.g., using Gibson assembly) from PCR fragments featuring a reporter gene insert. The synthetic genome can be transformed into L-form bacteria leading to genome activation and the release of viable phage for subsequent propagation on the phage host.
An alternative direct cloning platform used by Nugen and colleagues is the T7Select® system (Novagen, Gauteng, South Africa), originally designed as a phage-display vector, which is capable of displaying peptides and large protein antigens (up to 1200 amino acids) on the capsid [122]. By incorporating a stop codon downstream of the reporter cassette, heterologous reporter proteins are released instead of being displayed [96] (Figure 2B). This system was used to create various T7-based systems for detecting E. coli in water and food [60,97,98,105,106]. Not only is a commercial kit available for T7 phage engineering (i.e., T7Select® (Novagen)), but as a model phage it has a genome suitable for insertion of heterologous genes and a well-characterized host range, making it an ideal phage for reporter phage-based assay development as can be observed by its frequent use over the last four years (Table 2).
4.2. Homologous Recombination Combined with CRISPR-Cas Selection
Phage engineering strategies based on homologous recombination make use of editing plasmids carrying the desired genome modifications flanked by homology regions. When host cells harboring editing plasmids are infected with wild-type phage, double homologous recombination can occur during replication of the phage genome, leading to the production of wild-type and recombinant phages (Figure 2C). Due to low recombination rates (10−10–10−4), screening can be extremely laborious and typically requires a suitable selection marker, such as antibiotic resistance in lysogens, a change in host range [118], or plaque morphology [91]. As an example, the temperate, Bacillus anthracis phage Wβ∷luxAB-2 was generated using spectinomycin resistance transduction as the selection marker. The phage carried Vibrio harveyi luxAB and the spectinomycin resistance (spc) cassette under the control of an optimized promoter (LuxAB-2) in a non-essential region of the genome that generated bioluminescence and provided spectinomycin resistance to the lysogen [101,123]. In the absence of selection markers, low recombination frequencies can be increased using the lambda red homologous recombination-mediated genetic engineering (recombineering) system [124,125,126], which is commonly used for modifying E. coli and Salmonella targeting phages such as the reporter phage ΦV10 for detecting Shiga toxin producing E. coli O157:H7 [102]. The system originates from phage lambda but can be expressed from the bacterial chromosome or a separate plasmid, making it widely applicable for engineering other phages (reviewed in detail by [127]).
Homologous recombination-based methods can be coupled with CRISPR-Cas systems to facilitate enrichment of recombinant phages by sequence-specific counter selection of wild-type phage genomes (Figure 2C). [128,129,130]. This approach has been developed for engineering Gram-negative and Gram-positive targeting phages. For instance, Jackson et al. 2016 employed the E. coli type I-E CRISPR-Cas system [128,131] to create a T7 based reporter phage carrying an alkaline phosphatase for colorimetric detection of E. coli [107]. Similarly, a Listeria ivanovii type II-A CRISPR-Cas system [132] was used for the modification of lytic Listeria phage A511 to create two variants of A511::nluc that transduce bioluminescence into Listeria spp. for detection [62]. While the use of CRISPR-Cas counter selection can greatly improve recombinant phage identification, a current bottleneck is the lack of well-characterized and programmable CRISPR-Cas systems that can be used for reporter phage engineering in different bacterial hosts.
4.3. Synthetic Genome Rebooting
Synthetic genome rebooting involves assembling a complete genome from individual PCR fragments (or synthesized DNA) for subsequent transformation into a surrogate bacterial host for phage production. One approach is to transform the genome fragments together with a yeast artificial chromosome (YAC) into Saccaromyces cervisiae featuring an efficient gap repair system to produce a full phage genome cloned into a replicative yeast plasmid [133,134] (Figure 2D). The assembled YAC–phage DNA is extracted and subsequently transformed into a bacterial host for virion production. By switching the tail fiber genes between synthetic T7-like phage genomes, this method could redirect phage host range between E. coli, Klebsiella and Yersinia hosts [134]. Recently, this approach was used to engineer E. coli phage λ-mKate, a lysogenic phage featuring a red fluorescent reporter to observe prophage induction in bacterial host cells within phagocytes [135]. A major restriction to this approach is the requirement to transform a bacterial production strain with the assembled genome phages. While this is possible for some Gram-negative bacteria, it can be challenging to transfer such large viral genomes across the thick wall of Gram-positive cells. To overcome this limitation for Gram-positive targeting phage engineering, a phage engineering method using a strain of L-form Listeria for rebooting of synthetic phage genomes was recently developed [136] (Figure 2E). L-forms are wall-deficient bacteria that retain metabolic activity and the ability of cell division [137,138,139]. Induction of L-forms occurs by prolonged subcultivation under selective pressure of cell wall targeting antibiotics in a medium that provides osmoprotective conditions, preventing the wall-deficient cells from hypotonic lysis. Using L-forms as surrogate hosts for the reactivation of synthetic, in vitro assembled genomes provides many advantages: multiple modifications can be achieved in a single step and no cloning of potentially toxic phage genes is required. Low-efficiency homologous recombination and subsequent screening can be avoided because only correctly assembled genomes and viable recombinant phages are isolated using this technology. While native phage genomes of different sizes and structures can easily be rebooted in L-forms [140], the current limitation of this approach is the assembly of large (>100 kb) genomes in vitro, using approaches such as Gibson assembly of multiple genetic fragments [141].
5. Overview of Reporter Phage Systems
5.1. Bioluminescence-Based Detection
The phenomenon of bioluminescence is found across a wide diversity of life including bacteria, fungi, insects and a variety of marine organisms [142]. Light (photon) emission occurs when a substrate is oxidized by a member of a class of enzymes called luciferases. Due to highly sensitive signal detection and ease of use, luciferase reporter phages have found broad application for bacterial detection. The ideal luciferase leads to bright and sustained light emission with low background. Preferably, the luciferase should be structurally stable in different environmental conditions. We recently compared the performance of several isogenic reporter phages encoding for different luciferases. Bacterial, cnidarian and crustacean luciferase coding sequences derived from Vibrio harveyi (luxAB), Gaussia princeps (gluc), Renilla reniformis (rluc) and Oplophorus gracilirostris (nluc) were inserted into the Listeria phage A500. The light-emitting properties of the NLuc reporter phage (A500::nluc ΔLCR) were clearly superior, indicated by a 100-fold larger increase in luminescence values compared to other reporters. Overall, the assay using the NLuc luciferase was highly sensitive and able to directly detect as few as three L. monocytogenes cells [62]. Nluc is an engineered luciferase (19 kDa) that produces a glow-type bioluminescent signal upon addition of its substrate (furimazine; signal half-life > 2 h) [143] and is widely used in reporter phage assays as detailed in Table 2. The first published reporter phage encoding NLuc is E. coli phage ΦV10 for detecting E. coli O157:H7 [102]. Another recent example is the broad host-range, nluc-containing Myovirus A511 (A511::nlucCPS) that detects a single L. monocytogenes cell in 25 g of various artificially contaminated food samples within less than 24 h. In addition to A511::nlucCPS-mediated detection, other nluc Listeria phages can be used for serovar differentiation of food isolates [62]. Furthermore, Dow et al. 2018 used acoustic separation and microfluidics to separate bacteria from blood cells and then employed the NLuc-reporter phage K1E for detection of E. coli [103]. A set of T7-based phages encoding a NLuc-carbohydrate-binding module fusion protein (NLuc-CBM) were evaluated for detection of E. coli in water and food samples [60,98,104,105]. One specific assay used cellulose-coated beads to concentrate and purify NLuc-CBM after its production from infected E. coli cells, enabling more sensitive detection down to 1 CFU/100 mL drinking water within 10 h [60].
The luxCDABE operon encodes for the luciferase (LuxA and LuxB) and the enzymes that produce its substrate (Lux C, D and E). Typically, only luxA and luxB are inserted into reporter phages and its substrate, a fatty aldehyde, is applied exogenously to the reaction solution [59]. Introducing a complete luxCDABE operon creates a substrate-independent reporter system; however, the relatively large size (approximately 6 kb) of the operon complicates phage engineering. Nevertheless, temperate E. coli phages HK620 and HK97 that use different packaging systems both proved functional upon integration of the complete luxCDABE operon and were used to detect E. coli in solution [95]. Similarly, ΦV10lux was engineered to contain the full operon for detection of enterohemorrhagic E. coli O157:H7 in food. In order to make room for the luxCDABE operon on the genome of this phage, non-essential regions were removed, which additionally converted the phage to a strictly lytic lifestyle. The operon itself was integrated behind the tailspike gene and expression driven from the endogenous phage promoter [88].
5.2. Colorimetry-Based Detection
Colorimetric signals arise from the result of enzymatic substrate conversion that can be visually interpreted. Beta-galactosidase (β-gal) is a glycoside hydrolase (lacZ) encoded in the lac operon of E. coli. T7 phages have been engineered to carry the lacZ gene, leading to β-gal expression during phage infection [63,64,87]. Upon release from the cell, the enzyme hydrolyses a colorimetric substrate for visual detection. For example, phage T7LacZ, in combination with chlorophenol red-β-d-galactopyranoside (CPRG) as a substrate for β-gal, was employed for antibiotic resistance profiling [63] and for detecting E. coli in food samples [64]. Similarly, T7 phages encoding alkaline phosphatase enabled detection of E. coli by hydrolysis of the substrate p-nitrophenyl phosphate (pNPP) to p-nitrophenol (pNP) [97,107] or upon reaction with nitro-blue tetrazolium chloride NBT and 5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt (BCIP) [60,106]. The alkaline phosphatase could be functionalized with a cellulose-specific carbohydrate-binding module (CBM) from Cellulomonas fimi and consequently captured on magnetic cellulose [97] or cellulose filters [60] for detection. The latter enabled detection of 1 CFU/100 mL of E. coli in drinking water. Furthermore, detection of enterohemorrhagic E. coli in fresh produce was recently achieved using the cytochrome c peroxidase gene ccp as a reporter in recombinant phages PP01ccp [108], IP008BK and IP052BK [89] Bacterial detection using this system is based on the oxidation of cytochrome c and associated shift in absorbance at 550 nm after release of the reporter enzyme from the cells. Overall, colorimetric assays are straightforward and cost effective, but are limited greatly by the composition of the sample matrix, for instance, colored solutions can interfere with the visual readout and the pH can have a significant effect on substrate conversion [63]. Capture and detection approaches are therefore advisable when using colorimetric reporters.
5.3. Electrochemistry-Based Detection
Electrochemical biosensors are relatively simple and cost-effective while remaining sensitive and specific. Usually, electrochemical measurements are based on the detection of electroactive species and thus are not influenced by turbidity or color of the samples. Modulation of electrical properties is a result of redox reactions occurring among analytes. In addition to the use of phages as physical bio-probes in biosensors (reviewed in [144]), reporter proteins produced during phage infection can be used as the analyte for detection. T7-based reporter phages featuring the LacZ operon produced β-galactosidase (β-gal) that is released upon cell lysis. Enzymatic activity is detected by measuring the level of 4-aminophenol (PAP) produced upon hydrolysis of the substrate 4-aminophenyl-β-d-galactopyranoside (PAPG) by β-gal. The electroactive PAP product is subsequently monitored by amperometry (detection of ions in solution) [109]. Immobilization of biomarkers on the surface of electrodes can also serve to increase the sensitivity of electrochemical biosensors. Wang et al. engineered a T7-based reporter phage featuring a gold-binding peptide fused to an alkaline phosphatase (GBPs-ALP) that is released during cell lysis and binds directly to the gold biosensor surface. The activity of GBPs-ALP-coated electrodes was subsequently measured electrochemically using linear sweep voltammetry (LSV), which enabled detection of 105 CFU/mL in drinking water after 2 h [110].
5.4. Fluorescence-Based Detection
The majority of fluorescence-based reporter phage assays developed in the last four years involve the detection of Mycobacterium and drug susceptibility testing (DST). Engineered fluoromycobacteriophages are combined with the drug of interest and the clinical isolates. Phage-mediated fluorescence only occurs in drug-resistant bacteria, which is detected by fluorescence microscopy or flow cytometry [94,112]. Due to its broad host range against several Mycobacterium species, the majority of recently employed fluoromycobacteriophages are derivatives of the temperate phage TM4 [145,146]. Additionally, these fluoromycobacteriophages are thermosensitive, thus they do not lyse their host cells at 37 °C. Thermosensitivity of the fluorophages ensures the survival and thus detection of drug-resistant hosts during DST. Recently, the TM4-derived phage Φ2GFP10 was used to detect low-frequency drug-resistant subpopulations of M. tuberculosis in vitro and in sputum from a South African TB patient [94] a second-generation fluoromycobacteriophage with optimized expression of a mCherrybomb gene in mycobacteria with improved fluorescent signal allowed shorter time to detection of M. tuberculosis [112]. mCherrybomb-Φ proved useful in a microscopy-based approach for detection of Mycobacterium spp. and determination of rifampicin resistance directly from Brazilian TB patient sputum within days. This phage was further used for evaluation of phage based DST and discrimination between M. tuberculosis complex (MTBC) and non-tuberculous mycobacteria (NTM) strains [111]. Other fluorescence-based assays for the detection of E. coli relied on the detection of the reporter with antibodies [106,115] or have been using a fluorescent substrate for the alkaline phosphatase reporter [114].
6. Future Perspective
Reporter phage-based assays combine host-specific binding with rapid intracellular phage multiplication and gene expression to provide highly sensitive detection. Nevertheless, the major limitations to practical reporter phage application are food matrix effects and the restricted host ranges of many phages that is intrinsically linked to phage resistance. The first limitation can be circumvented by coupling detection to specific capture of the target bacterium or the amplified reporter protein. Restricted host range and phage resistance are mechanistically interconnected and caused either by a lack of phage adsorption/receptor engagement or by the presence of intracellular defense mechanisms within individual strains. In therapeutic settings, this limitation is often circumvented by using multiple phages with complementary host ranges (the “phage cocktail”). With faster engineering protocols at hand, the cocktail approach could also be adopted in future reporter phage studies. In addition, synthetic biology and phage genome engineering may offer more targeted strategies to tune a limited number of well-characterized phage backbones towards the specific needs of each reporter phage application. This could be achieved by reprogramming the phage binding range through targeted RBP or baseplate engineering. Several recent studies suggest that redirecting and tuning host specificity is a viable option, at least for Sipho- and Podoviridae [134,147,148]. Counteracting intracellular defenses through phage engineering could be another viable strategy to expand the detection range of reporter phages. For example, RM systems can be counteracted through specific methylation within the phage production strains or CRISPR-Cas systems could potentially be inactivated via phage-mediated delivery of anti-CRISPR proteins. Due to the large variety and complexity of intracellular defenses, this strategy may be even more challenging and requires a detailed understanding of the underlying mechanisms. Despite these limitations, many reporter phage candidates already perform exceptionally well and will continue to offer an inexpensive and rapid alternative to culture-based and molecular diagnostics, which will be further improved through genetic engineering.
Author Contributions
Conceptualization, S.M. and M.D.; data curation, S.M.; writing—original draft preparation, S.M., M.D., and S.K.; writing—review and editing, S.M., M.D., S.K., and M.J.L.; visualization, S.M. and M.D.; funding acquisition, S.K. and M.J.L. All authors have read and agreed to the published version of the manuscript.
Funding
S.M. and S.K. are funded through the Swiss National Science Foundation, grant PZ00P3_174108.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Opota, O.; Croxatto, A.; Prod’Hom, G.; Greub, G. Blood culture-based diagnosis of bacteraemia: State of the art. Clin. Microbiol. Infect. 2015, 21, 313–322. [Google Scholar] [CrossRef]
- Cho, I.-H.; Ku, S. Current Technical Approaches for the Early Detection of Foodborne Pathogens: Challenges and Opportunities. Int. J. Mol. Sci. 2017, 18, 2078. [Google Scholar] [CrossRef] [PubMed]
- Abram, T.J.; Cherukury, H.; Ou, C.-Y.; Vu, T.; Toledano, M.; Li, Y.; Grunwald, J.T.; Toosky, M.N.; Tifrea, D.F.; Slepenkin, A.; et al. Rapid bacterial detection and antibiotic susceptibility testing in whole blood using one-step, high throughput blood digital PCR. Lab Chip 2019, 20, 477–489. [Google Scholar] [CrossRef]
- Shi, X.; Kadiyala, U.; Vanepps, J.S.; Yau, S.-T. Culture-free bacterial detection and identification from blood with rapid, phenotypic, antibiotic susceptibility testing. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, J.H.; Hindler, J.F.; Reller, L.B.; Weinstein, M.P. New Consensus Guidelines from the Clinical and Laboratory Standards Institute for Antimicrobial Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria. Clin. Infect. Dis. 2007, 44, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Luna, C.M.; Aruj, P.; Niederman, M.S.; Garzón, J.; Violi, D.; Prignoni, A.; Rios, F.; Baquero, S.; Gando, S. Appropriateness and delay to initiate therapy in ventilator-associated pneumonia. Eur. Respir. J. 2006, 27, 158–164. [Google Scholar] [CrossRef]
- Kang, C.-I.; Kim, S.-H.; Park, W.B.; Lee, K.-D.; Bin Kim, H.; Kim, E.-C.; Oh, M.-D.; Choe, K.-W. Bloodstream Infections Caused by Antibiotic-Resistant Gram-Negative Bacilli: Risk Factors for Mortality and Impact of Inappropriate Initial Antimicrobial Therapy on Outcome. Antimicrob. Agents Chemother. 2005, 49, 760–766. [Google Scholar] [CrossRef]
- Merlin, C. Reducing the Consumption of Antibiotics: Would That Be Enough to Slow Down the Dissemination of Resistances in the Downstream Environment? Front. Microbiol. 2020, 11, 33. [Google Scholar] [CrossRef] [PubMed]
- Canton, R.; Horcajada, J.P.; Oliver, A.; Garbajosa, P.R.; Vila, J. Inappropriate use of antibiotics in hospitals: The complex relationship between antibiotic use and antimicrobial resistance. Enferm. Infecc. Y Microbiol. Clínica 2013, 31, 3–11. [Google Scholar] [CrossRef]
- Ventola, C.L. The Antibiotic Resistance Crisis: Part 1: Causes and threats. P T Peer Rev. J. Formul. Manag. 2015, 40, 277–283. [Google Scholar]
- Caniça, M.; Manageiro, V.; Abriouel, H.; Moran-Gilad, J.; Franz, C.M. Antibiotic resistance in foodborne bacteria. Trends Food Sci. Technol. 2019, 84, 41–44. [Google Scholar] [CrossRef]
- Zwietering, M.; Jacxsens, L.; Membré, J.-M.; Nauta, M.; Peterz, M. Relevance of microbial finished product testing in food safety management. Food Control. 2016, 60, 31–43. [Google Scholar] [CrossRef]
- ISO. Microbiology of the Food–Horizontal Method for the Detection and Enumeration of Listeria Monocytogenes and of Listeria spp. Part 1: Detection Method, International Standard ISO 11290-1: 2017; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Soon, J.M.; Brazier, A.K.; Wallace, C.A. Determining common contributory factors in food safety incidents–A review of global outbreaks and recalls 2008–2018. Trends Food Sci. Technol. 2020, 97, 76–87. [Google Scholar] [CrossRef]
- Havelaar, A.; Kirk, M.; Torgerson, P.; Gibb, H.J.; Hald, T.; Lake, R.J.; Praet, N.; Bellinger, D.C.; De Silva, N.R.; Gargouri, N.; et al. World Health Organization Global Estimates and Regional Comparisons of the Burden of Foodborne Disease in 2010. PLoS Med. 2015, 12, e1001923. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union One Health 2018 Zoonoses Report. EFSA J. 2019, 17, e05926. [Google Scholar]
- Langley, G.; Besser, J.; Iwamoto, M.; Lessa, F.C.; Cronquist, A.; Skoff, T.H.; Chaves, S.; Boxrud, D.; Pinner, R.W.; Harrison, L.H. Effect of Culture-Independent Diagnostic Tests on Future Emerging Infections Program Surveillance. Emerg. Infect. Dis. 2015, 21, 1582–1588. [Google Scholar] [CrossRef]
- Imdad, A.; Retzer, F.; Thomas, L.S.; McMILLIAN, M.; Garman, K.; Rebeiro, P.F.; A Deppen, S.; Dunn, J.R.; Woron, A.M. Impact of Culture-Independent Diagnostic Testing on Recovery of Enteric Bacterial Infections. Clin. Infect. Dis. 2017, 66, 1892–1898. [Google Scholar] [CrossRef] [PubMed]
- Shea, S.; Kubota, K.A.; Maguire, H.; Gladbach, S.; Woron, A.; Atkinson-Dunn, R.; Couturier, M.R.; Miller, M.B. Clinical Microbiology Laboratories’ Adoption of Culture-Independent Diagnostic Tests Is a Threat to Foodborne-Disease Surveillance in the United States. J. Clin. Microbiol. 2016, 55, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Quainoo, S.; Coolen, J.P.M.; Van Hijum, S.A.F.T.; Huynen, M.A.; Melchers, W.J.G.; Van Schaik, W.; Wertheim, H.F.L. Whole-Genome Sequencing of Bacterial Pathogens: The Future of Nosocomial Outbreak Analysis. Clin. Microbiol. Rev. 2017, 30, 1015–1063. [Google Scholar] [CrossRef] [PubMed]
- Feucherolles, M.; Cauchie, H.-M.; Penny, C. MALDI-TOF Mass Spectrometry and Specific Biomarkers: Potential New Key for Swift Identification of Antimicrobial Resistance in Foodborne Pathogens. Microorganisms 2019, 7, 593. [Google Scholar] [CrossRef]
- Marder, E.P.; Cieslak, P.R.; Cronquist, A.B.; Dunn, J.; Lathrop, S.; Rabatsky-Ehr, T.; Ryan, P.; Smith, K.; Tobin-D’Angelo, M.; Vugia, D.J.; et al. Incidence and Trends of Infections with Pathogens Transmitted Commonly Through Food and the Effect of Increasing Use of Culture-Independent Diagnostic Tests on Surveillance—Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2013–2016. Morb. Mortal. Wkly. Rep. 2017, 66, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.; Sánchez-Juanes, F.; Porras-Guerra, I.; García-García, M.I.; García-Sánchez, J.; González-Buitrago, J.; Muñioz-Bellido, J.L. Microorganisms direct identification from blood culture by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 2011, 17, 546–551. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef] [PubMed]
- Altamirano, F.L.G.; Barr, J.J. Phage Therapy in the Postantibiotic Era. Clin. Microbiol. Rev. 2019, 32, 32. [Google Scholar] [CrossRef]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef]
- Strydom, A.; Witthuhn, R.C. Listeria monocytogenes: A Target for Bacteriophage Biocontrol. Compr. Rev. Food Sci. Food Saf. 2015, 14, 694–704. [Google Scholar] [CrossRef]
- Shabani, A.; Lawrence, M.; Marquette, C.A.; Mandeville, R. Carbon microarrays for the direct impedimetric detection of Bacillus anthracis using Gamma phages as probes. Analyst 2013, 138, 1434. [Google Scholar] [CrossRef]
- Bhardwaj, N.; Bhardwaj, S.K.; Mehta, J.; Mohanta, G.C.; Deep, A. Bacteriophage immobilized graphene electrodes for impedimetric sensing of bacteria (Staphylococcus arlettae). Anal. Biochem. 2016, 505, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.; He, Y.; Fan, E.; Wang, L.; Lu, S.; Fu, Z. Label-free electrochemiluminescent biosensor for rapid and sensitive detection of pseudomonas aeruginosa using phage as highly specific recognition agent. Biosens. Bioelectron. 2017, 94, 429–432. [Google Scholar] [CrossRef]
- Chen, J.; Alcaine, S.D.; Jiang, Z.; Rotello, V.; Nugen, S.R. Detection of Escherichia coliin Drinking Water Using T7 Bacteriophage-Conjugated Magnetic Probe. Anal. Chem. 2015, 87, 8977–8984. [Google Scholar] [CrossRef]
- Sumrall, E.T.; Keller, A.P.; Shen, Y.; Loessner, M.J. Structure and function of Listeria teichoic acids and their implications. Mol. Microbiol. 2020, 113, 627–637. [Google Scholar] [CrossRef] [PubMed]
- Kretzer, J.W.; Schmelcher, M.; Loessner, M.J. Ultrasensitive and Fast Diagnostics of Viable Listeria Cells by CBD Magnetic Separation Combined with A511::luxAB Detection. Viruses 2018, 10, 626. [Google Scholar] [CrossRef] [PubMed]
- Kong, M.; Sim, J.; Kang, T.; Nguyen, H.H.; Park, H.K.; Chung, B.H.; Ryu, S. A novel and highly specific phage endolysin cell wall binding domain for detection of Bacillus cereus. Eur. Biophys. J. 2015, 44, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Torres, N.; Ávila, M.; Narbad, A.; Mayer, M.J.; Garde, S. Use of fluorescent CTP1L endolysin cell wall-binding domain to study the evolution of Clostridium tyrobutyricum during cheese ripening. Food Microbiol. 2019, 78, 11–17. [Google Scholar] [CrossRef]
- Dunne, M.; Hupfeld, M.; Klumpp, J.; Loessner, M.J. Molecular Basis of Bacterial Host Interactions by Gram-Positive Targeting Bacteriophages. Viruses 2018, 10, 397. [Google Scholar] [CrossRef] [PubMed]
- Hyman, P.; Van Raaij, M.J. Bacteriophage T4 long tail fiber domains. Biophys. Rev. 2017, 10, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Denyes, J.M.; Dunne, M.; Steiner, S.; Mittelviefhaus, M.; Weiss, A.; Schmidt, H.; Klumpp, J.; Loessner, M.J. Modified Bacteriophage S16 Long Tail Fiber Proteins for Rapid and Specific Immobilization and Detection of Salmonella Cells. Appl. Environ. Microbiol. 2017, 83, e00277-17. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Rabsch, W.; Broeker, N.K.; Barbirz, S. Bacteriophage tailspike protein based assay to monitor phase variable glucosylations in Salmonella O-antigens. BMC Microbiol. 2016, 16, 207. [Google Scholar] [CrossRef] [PubMed]
- Kunstmann, S.; Scheidt, T.; Buchwald, S.; Helm, A.; Mulard, L.A.; Fruth, A.; Barbirz, S. Bacteriophage Sf6 Tailspike Protein for Detection of Shigella flexneri Pathogens. Viruses 2018, 10, 431. [Google Scholar] [CrossRef]
- He, Y.; Shi, Y.; Liu, M.; Wang, Y.; Wang, L.; Lu, S.; Fu, Z. Nonlytic Recombinant Phage Tail Fiber Protein for Specific Recognition of Pseudomonas aeruginosa. Anal. Chem. 2018, 90, 14462–14468. [Google Scholar] [CrossRef]
- Sumrall, E.T.; Röhrig, C.; Hupfeld, M.; Selvakumar, L.; Du, J.; Dunne, M.; Schmelcher, M.; Shen, Y.; Loessner, M.J. Glycotyping and specific separation of Listeria monocytogenes with a novel bacteriophage protein toolkit. Appl. Environ. Microbiol. 2020. [Google Scholar] [CrossRef]
- Cardoso, M.E.; Fernández, L.; Tejería, E.; Esperón, P.; Terán, M.; Elena, C.; Leticia, F.; Emilia, T.; Patricia, E.; Mariella, T. Evaluation of a Labelled Bacteriophage with 99mTc as a Potential Agent for Infection Diagnosis. Curr. Radiopharm. 2016, 9, 137–142. [Google Scholar] [CrossRef]
- Kim, J.; Vu, B.; Kourentzi, K.; Willson, R.C.; Conrad, J.C. Increasing Binding Efficiency via Reporter Shape and Flux in a Viral Nanoparticle Lateral-Flow Assay. ACS Appl. Mater. Interfaces 2017, 9, 6878–6884. [Google Scholar] [CrossRef]
- Zhou, Y.; Ramasamy, R.P. Isolation and separation of Listeria monocytogenes using bacteriophage P100-modified magnetic particles. Colloids Surfaces B Biointerfaces 2019, 175, 421–427. [Google Scholar] [CrossRef]
- Janczuk, M.; Richter, Ł.; Hoser, G.; Kawiak, J.; Łoś, M.; Niedziółka-Jönsson, J.; Paczesny, J.; Hołyst, R. Bacteriophage-Based Bioconjugates as a Flow Cytometry Probe for Fast Bacteria Detection. Bioconjug. Chem. 2016, 28, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.J.; Lim, W.K. Rapid label-free detection of E. coli using a novel SPR biosensor containing a fragment of tail protein from phage lambda. Prep. Biochem. Biotechnol. 2018, 48, 498–505. [Google Scholar] [CrossRef]
- Kong, M.; Shin, J.H.; Heu, S.; Park, J.-K.; Ryu, S. Lateral flow assay-based bacterial detection using engineered cell wall binding domains of a phage endolysin. Biosens. Bioelectron. 2017, 96, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Kong, M.; Lee, J.-H.; Ryu, S.; Park, S. Detection of Bacillus Cereus Using Bioluminescence Assay with Cell Wall-binding Domain Conjugated Magnetic Nanoparticles. BioChip J. 2018, 12, 287–293. [Google Scholar] [CrossRef]
- Gómez-Torres, N.; Dunne, M.; Garde, S.; Meijers, R.; Narbad, A.; Ávila, M.; Mayer, M.J. Development of a specific fluorescent phage endolysin for in situ detection of Clostridium species associated with cheese spoilage. Microb. Biotechnol. 2017, 11, 332–345. [Google Scholar] [CrossRef] [PubMed]
- Swift, B.M.; Huxley, J.N.; Plain, K.M.; Begg, D.J.; De Silva, K.; Purdie, A.C.; Whittington, R.J.; Rees, C.E.D. Evaluation of the limitations and methods to improve rapid phage-based detection of viable Mycobacterium avium subsp. paratuberculosis in the blood of experimentally infected cattle. BMC Veter Res. 2016, 12, 115. [Google Scholar] [CrossRef] [PubMed]
- Ben Said, M.; Ben Saad, M.; Achouri, F.; Bousselmi, L.; Ghrabi, A. Detection of active pathogenic bacteria under stress conditions using lytic and specific phage. Water Sci. Technol. 2019, 80, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Šuster, K.; Podgornik, A.; Cör, A. Quick bacteriophage-mediated bioluminescence assay for detecting Staphylococcus spp. in sonicate fluid of orthopaedic artificial joints. New Microbiol. 2017, 40, 190–196. [Google Scholar]
- Malagon, F.; Estrella, L.A.; Stockelman, M.G.; Hamilton, T.; Teneza-Mora, N.; Biswas, B. Phage-Mediated Molecular Detection (PMMD): A Novel Rapid Method for Phage-Specific Bacterial Detection. Viruses 2020, 12, 435. [Google Scholar] [CrossRef] [PubMed]
- Anany, H.; Brovko, L.; Eldougdoug, N.; Sohar, J.; Fenn, H.; AlAsiri, N.; Jabrane, T.; Mangin, P.; Ali, M.M.; Kannan, B.; et al. Print to detect: A rapid and ultrasensitive phage-based dipstick assay for foodborne pathogens. Anal. Bioanal. Chem. 2017, 410, 1217–1230. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.; Hartman, T.; Eisenberg, N.; O’Donnell, M.R.; Kriakov, J.; Govender, K.; Makume, M.; Thaler, D.S.; Hatfull, G.F.; Sturm, A.W.; et al. 2GFP10, a High-Intensity Fluorophage, Enables Detection and Rapid Drug Susceptibility Testing of Mycobacterium tuberculosis Directly from Sputum Samples. J. Clin. Microbiol. 2012, 50, 1362–1369. [Google Scholar] [CrossRef]
- Piuri, M.; Jacobs, W.R., Jr.; Hatfull, G.F. Fluoromycobacteriophages for Rapid, Specific, and Sensitive Antibiotic Susceptibility Testing of Mycobacterium tuberculosis. PLoS ONE 2009, 4, e4870. [Google Scholar] [CrossRef]
- Mayer, O.; Jain, P.; Weisbrod, T.R.; Biro, D.; Ho, L.; Jacobs-Sera, D.; Hatfull, G.F.; Jacobs, W.R., Jr. Fluorescent Reporter DS6A Mycobacteriophages Reveal Unique Variations in Infectibility and Phage Production in Mycobacteria. J. Bacteriol. 2016, 198, 3220–3232. [Google Scholar] [CrossRef]
- Loessner, M.J.; E Rees, C.; Stewart, G.S.; Scherer, S. Construction of luciferase reporter bacteriophage A511::luxAB for rapid and sensitive detection of viable Listeria cells. Appl. Environ. Microbiol. 1996, 62, 1133–1140. [Google Scholar] [CrossRef]
- Hinkley, T.C.; Singh, S.; Garing, S.; Le Ny, A.-L.M.; Nichols, K.P.; Peters, J.E.; Talbert, J.N.; Nugen, S.R. A phage-based assay for the rapid, quantitative, and single CFU visualization of E. coli (ECOR #13) in drinking water. Sci. Rep. 2018, 8, 14630. [Google Scholar] [CrossRef]
- Nguyen, C.; Makkar, R.; Sharp, N.J.; Page, M.A.; Molineux, I.J.; Schofield, D.A. Detection of Bacillus anthracis spores from environmental water using bioluminescent reporter phage. J. Appl. Microbiol. 2017, 123, 1184–1193. [Google Scholar] [CrossRef]
- Meile, S.; Sarbach, A.; Du, J.; Schuppler, M.; Saez, C.; Loessner, M.J.; Kilcher, S. Engineered Reporter Phages for Rapid Bioluminescence-Based Detection and Differentiation of Viable Listeria Cells. Appl. Environ. Microbiol. 2020, 86. [Google Scholar] [CrossRef]
- Chen, J.; Alcaine, S.D.; Jackson, A.A.; Rotello, V.; Nugen, S.R. Development of Engineered Bacteriophages for Escherichia coli Detection and High-Throughput Antibiotic Resistance Determination. ACS Sens. 2017, 2, 484–489. [Google Scholar] [CrossRef]
- Chen, J.; Picard, R.A.; Wang, D.; Nugen, S.R. Lyophilized Engineered Phages for Escherichia coli Detection in Food Matrices. ACS Sens. 2017, 2, 1573–1577. [Google Scholar] [CrossRef]
- Abedon, S.T.; Hyman, P.; Thomas, C. Experimental Examination of Bacteriophage Latent-Period Evolution as a Response to Bacterial Availability. Appl. Environ. Microbiol. 2003, 69, 7499–7506. [Google Scholar] [CrossRef]
- Denes, T.; Wiedmann, M. Environmental responses and phage susceptibility in foodborne pathogens: Implications for improving applications in food safety. Curr. Opin. Biotechnol. 2014, 26, 45–49. [Google Scholar] [CrossRef]
- Ben Said, M.; Ben Saad, M.; Achouri, F.; Bousselmi, L.; Ghrabi, A. The application of phage reactivation capacity to sens bacterial viability and activity after photocatalytic treatment. Environ. Technol. 2020, 1–9. [Google Scholar] [CrossRef]
- Swift, B.M.; Convery, T.W.; Rees, C.E.D. Evidence of Mycobacterium tuberculosis complex bacteraemia in intradermal skin test positive cattle detected using phage-RPA. Virulence 2016, 7, 779–788. [Google Scholar] [CrossRef]
- Stewart, L.D.; Foddai, A.; Elliott, C.; Grant, I.R. Development of a novel phage-mediated immunoassay for the rapid detection of viable Mycobacterium avium subsp paratuberculosis. J. Appl. Microbiol. 2013, 115, 808–817. [Google Scholar] [CrossRef]
- Rames, E.; Macdonald, J. The QuantiPhage assay: A novel method for the rapid colorimetric detection of coliphages using cellulose pad materials. Water Res. 2019, 149, 98–110. [Google Scholar] [CrossRef]
- Bhowmick, T.; Mirrett, S.; Reller, L.B.; Price, C.; Qi, C.; Weinstein, M.P.; Kirn, T.J. Controlled Multicenter Evaluation of a Bacteriophage-Based Method for Rapid Detection of Staphylococcus aureus in Positive Blood Cultures. J. Clin. Microbiol. 2013, 51, 1226–1230. [Google Scholar] [CrossRef]
- Sullivan, K.V.; Turner, N.N.; Roundtree, S.S.; McGowan, K.L. Rapid Detection of Methicillin-Resistant Staphylococcus aureus (MRSA) and Methicillin-Susceptible Staphylococcus aureus (MSSA) Using the KeyPath MRSA/MSSA Blood Culture Test and the BacT/ALERT System in a Pediatric Population. Arch. Pathol. Lab. Med. 2013, 137, 1103–1105. [Google Scholar] [CrossRef] [PubMed]
- Stambach, N.R.; Carr, S.A.; Cox, C.R.; Voorhees, K.J. Rapid Detection of Listeria by Bacteriophage Amplification and SERS-Lateral Flow Immunochromatography. Viruses 2015, 7, 6631–6641. [Google Scholar] [CrossRef] [PubMed]
- Cox, C.R.; Jensen, K.R.; Mondesire, R.R.; Voorhees, K.J. Rapid detection of Bacillus anthracis by γ phage amplification and lateral flow immunochromatography. J. Microbiol. Methods 2015, 118, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Mido, T.; Schaffer, E.M.; Dorsey, R.W.; Sozhamannan, S.; Hofmann, E.R. Sensitive detection of live Escherichia coli by bacteriophage amplification-coupled immunoassay on the Luminex® MAGPIX instrument. J. Microbiol. Methods 2018, 152, 143–147. [Google Scholar] [CrossRef]
- Sergueev, K.V.; Filippov, A.A.; Nikolich, M.P. Highly Sensitive Bacteriophage-Based Detection of Brucella abortus in Mixed Culture and Spiked Blood. Viruses 2017, 9, 144. [Google Scholar] [CrossRef]
- Mulvey, M.C.; Lemmon, M.; Rotter, S.; Lees, J.; Einck, L.; Nacy, C.A. Optimization of a Nucleic Acid-Based Reporter System to Detect Mycobacterium tuberculosis Antibiotic Sensitivity. Antimicrob. Agents Chemother. 2014, 59, 407–413. [Google Scholar] [CrossRef]
- Mulvey, M.C.; Sacksteder, K.A.; Einck, L.; A Nacy, C. Generation of a Novel Nucleic Acid-Based Reporter System to Detect Phenotypic Susceptibility to Antibiotics in Mycobacterium tuberculosis. mBio 2012, 3, e00312-11. [Google Scholar] [CrossRef]
- Wang, X.; Li, X.; Liu, S.; Ren, H.; Yang, M.; Ke, Y.; Huang, L.; Liu, C.; Liu, B.; Chen, Z. Ultrasensitive Detection of Bacteria by Targeting Abundant Transcripts. Sci. Rep. 2016, 6, 20393. [Google Scholar] [CrossRef]
- Marques, S.M.; Peralta, F.; Estevesdasilva, J.; Da Silva, J.C.E. Optimized chromatographic and bioluminescent methods for inorganic pyrophosphate based on its conversion to ATP by firefly luciferase. Talanta 2009, 77, 1497–1503. [Google Scholar] [CrossRef]
- Vaidya, A.; Ravindranath, S.; Annapure, U.S. Detection and differential identification of typhoidal Salmonella using bacteriophages and resazurin. 3 Biotech 2020, 10, 196–198. [Google Scholar] [CrossRef]
- Yang, X.; Wisuthiphaet, N.; Young, G.M.; Nitin, N. Rapid detection of Escherichia coli using bacteriophage-induced lysis and image analysis. PLoS ONE 2020, 15, e0233853. [Google Scholar]
- Hampton, H.G.; Watson, B.N.J.; Fineran, P.C. The arms race between bacteria and their phage foes. Nature 2020, 577, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Bernheim, A.; Sorek, R. The pan-immune system of bacteria: Antiviral defence as a community resource. Nat. Rev. Genet. 2019, 18, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Rees, J.C.; Barr, J.R. Detection of methicillin-resistant Staphylococcus aureus using phage amplification combined with matrix-assisted laser desorption/ionization mass spectrometry. Anal. Bioanal. Chem. 2016, 409, 1379–1386. [Google Scholar] [CrossRef] [PubMed]
- Tamariz, J.; Guevara, V.; Guerra, H. Rapid detection of salmonellosis due to Salmonella enterica serovar Typhimurium in Peruvian commercially bred cavies, using indigenous wild bacteriophages. Germs 2018, 8, 178–185. [Google Scholar] [CrossRef]
- Chen, J.; Jackson, A.A.; Rotello, V.; Nugen, S.R. Colorimetric Detection of Escherichia coli Based on the Enzyme-Induced Metallization of Gold Nanorods. Small 2016, 12, 2469–2475. [Google Scholar] [CrossRef]
- Kim, J.; Kim, M.; Kim, S.; Ryu, S. Sensitive detection of viable Escherichia coli O157:H7 from foods using a luciferase-reporter phage phiV10 lux. Int. J. Food Microbiol. 2017, 254, 11–17. [Google Scholar] [CrossRef]
- Hoang, H.A.; Quy, N.T.; Chi, N.V. Detection of Escherichia coli in ready-to-eat fresh vegetables using broad-host-range recombinant phages. J. Appl. Microbiol. 2018, 124, 1610–1616. [Google Scholar] [CrossRef]
- Osuna, B.A.; Karambelkar, S.; Mahendra, C.; Sarbach, A.; Johnson, M.C.; Kilcher, S.; Bondy-Denomy, J. Critical Anti-CRISPR Locus Repression by a Bi-functional Cas9 Inhibitor. Cell Host Microbe 2020. [Google Scholar] [CrossRef]
- Born, Y.; Fieseler, L.; Thöny, V.; Leimer, N.; Duffy, B.; Loessner, M.J. Engineering of Bacteriophages Y2::dpoL1-C and Y2::luxAB for Efficient Control and Rapid Detection of the Fire Blight Pathogen, Erwinia amylovora. Appl. Environ. Microbiol. 2017, 83, 83. [Google Scholar] [CrossRef]
- Jain, P.; Weinrick, B.; Kalivoda, E.J.; Yang, H.; Munsamy, V.; Vilcheze, C.; Weisbrod, T.R.; Larsen, M.H.; O’Donnell, M.R.; Pym, A.S.; et al. Dual-Reporter Mycobacteriophages (Φ 2 DRMs) Reveal Preexisting Mycobacterium tuberculosis Persistent Cells in Human Sputum. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Gu, Y.; Jiang, G.; Ma, Y.; Zhao, L.; Sun, Z.; Jain, P.; O’Donnell, M.; Larsen, M.; Jacobs, W.R., Jr.; et al. Evaluation of a High-Intensity Green Fluorescent Protein Fluorophage Method for Drug- Resistance Diagnosis in Tuberculosis for Isoniazid, Rifampin, and Streptomycin. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, M.; Larsen, M.H.; Brown, T.S.; Jain, P.; Munsamy, V.; Wolf, A.; Uccellini, L.; Karim, F.; De Oliveira, T.; Mathema, B.; et al. Early Detection of Emergent Extensively Drug-Resistant Tuberculosis by Flow Cytometry-Based Phenotyping and Whole-Genome Sequencing. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [PubMed]
- Franche, N.; Vinay, M.; Ansaldi, M. Substrate-independent luminescent phage-based biosensor to specifically detect enteric bacteria such as E. coli. Environ. Sci. Pollut. Res. 2016, 24, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Alcaine, S.D.; Pacitto, D.; Sela, D.A.; Nugen, S.R. Phage & phosphatase: A novel phage-based probe for rapid, multi-platform detection of bacteria. Analyst 2015, 140, 7629–7636. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Hinkley, T.; Nugen, S.R.; Talbert, J.N. Colorimetric detection of Escherichia coli using engineered bacteriophage and an affinity reporter system. Anal. Bioanal. Chem. 2019, 411, 7273–7279. [Google Scholar] [CrossRef]
- Hinkley, T.C.; Garing, S.; Singh, S.; Le Ny, A.-L.M.; Nichols, K.P.; E Peters, J.; Talbert, J.N.; Nugen, S.R. Reporter bacteriophage T7NLC utilizes a novel NanoLuc::CBM fusion for the ultrasensitive detection of Escherichia coli in water. Analyst 2018, 143, 4074–4082. [Google Scholar] [CrossRef]
- Pulkkinen, E.; Hinkley, T.C.; Nugen, S.R. Utilizing in vitro DNA assembly to engineer a synthetic T7 Nanoluc reporter phage for Escherichia coli detection. Integr. Boil. 2019, 11, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Syal, K.; Mo, M.; Yu, H.; Iriya, R.; Jing, W.; Guodong, S.; Wang, S.; Grys, T.E.; Haydel, S.E.; Tao, N. Current and emerging techniques for antibiotic susceptibility tests. Theranostics 2017, 7, 1795–1805. [Google Scholar] [CrossRef]
- Sharp, N.J.; Molineux, I.J.; Page, M.A.; Schofield, D.A. Rapid Detection of Viable Bacillus anthracis Spores in Environmental Samples by Using Engineered Reporter Phages. Appl. Environ. Microbiol. 2016, 82, 2380–2387. [Google Scholar] [CrossRef]
- Zhang, D.; Coronel-Aguilera, C.P.; Romero, P.L.; Perry, L.; Minocha, U.; Rosenfield, C.; Gehring, A.G.; Paoli, G.C.; Bhunia, A.K.; Applegate, B. The Use of a Novel NanoLuc -Based Reporter Phage for the Detection of Escherichia coli O157:H7. Sci. Rep. 2016, 6, 33235. [Google Scholar] [CrossRef] [PubMed]
- Dow, P.; Kotz, K.; Gruszka, S.; Holder, J.; Fiering, J. Acoustic separation in plastic microfluidics for rapid detection of bacteria in blood using engineered bacteriophage. Lab Chip 2018, 18, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Hinkley, T.C.; Garing, S.; Jain, P.; Williford, J.; Le Ny, A.-L.M.; Nichols, K.P.; Peters, J.E.; Talbert, J.N.; Nugen, S.R. A Syringe-Based Biosensor to Rapidly Detect Low Levels of Escherichia coli (ECOR13) in Drinking Water Using Engineered Bacteriophages. Sensors 2020, 20, 1953. [Google Scholar] [CrossRef]
- Kozak, S.; Alcaine, S.D. Phage-based forensic tool for spatial visualization of bacterial contaminants in cheese. J. Dairy Sci. 2020, 103, 5964–5971. [Google Scholar] [CrossRef] [PubMed]
- Alcaine, S.D.; Law, K.; Ho, S.; Kinchla, A.; Sela, D.; Nugen, S.R. Bioengineering bacteriophages to enhance the sensitivity of phage amplification-based paper fluidic detection of bacteria. Biosens. Bioelectron. 2016, 82, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.A.; Hinkley, T.C.; Talbert, J.N.; Nugen, S.R.; Sela, D.A. Genetic optimization of a bacteriophage-delivered alkaline phosphatase reporter to detect Escherichia coli. Analyst 2016, 141, 5543–5548. [Google Scholar] [CrossRef] [PubMed]
- Hoang, H.A.; Nhung, N.T. Development of a bacteriophage-based Method for Detection of Escherichia coli O157:H7 in Fresh Vegetables. Food Saf. 2018, 6, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Chen, J.; Nugen, S.R. Electrochemical Detection of Escherichia coli from Aqueous Samples Using Engineered Phages. Anal. Chem. 2017, 89, 1650–1657. [Google Scholar] [CrossRef]
- Wang, D.; Hinkley, T.C.; Chen, J.; Talbert, J.N.; Nugen, S.R. Phage based electrochemical detection of Escherichia coli in drinking water using affinity reporter probes. Analyst 2019, 144, 1345–1352. [Google Scholar] [CrossRef]
- Rondón, L.; Urdániz, E.; Latini, C.; Payaslian, F.; Matteo, M.; Sosa, E.J.; Porto, D.A.F.D.; Turjanski, A.G.; Nemirovsky, S.I.; Hatfull, G.F.; et al. Fluoromycobacteriophages Can Detect Viable Mycobacterium tuberculosis and Determine Phenotypic Rifampicin Resistance in 3–5 Days from Sputum Collection. Front. Microbiol. 2018, 9, 9. [Google Scholar] [CrossRef]
- Urdániz, E.; Rondón, L.; A Marti, M.; Hatfull, G.F.; Piuri, M. Rapid Whole-Cell Assay of Antitubercular Drugs Using Second-Generation Fluoromycobacteriophages. Antimicrob. Agents Chemother. 2016, 60, 3253–3256. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Nugen, S.R. Detection of protease and engineered phage-infected bacteria using peptide-graphene oxide nanosensors. Anal. Bioanal. Chem. 2019, 411, 2487–2492. [Google Scholar] [CrossRef]
- Wisuthiphaet, N.; Yang, X.; Young, G.M.; Nitin, N. Rapid detection of Escherichia coli in beverages using genetically engineered bacteriophage T7. AMB Express 2019, 9, 55. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Song, Y.; Luan, T.; Ma, L.; Su, L.; Wang, S.; Yan, X. Specific detection of live Escherichia coli O157:H7 using tetracysteine-tagged PP01 bacteriophage. Biosens. Bioelectron. 2016, 86, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, W.R.J. Gene Transfer in Mycobacterium tuberculosis: Shuttle Phasmids to Enlightenment. Microbiol. Spectr. 2014, 2, 3–25. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Ding, M.; Zhang, N.; Li, J. Mycobacteriophages: An important tool for the diagnosis of Mycobacterium tuberculosis (Review). Mol. Med. Rep. 2012, 12, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.P.; Cleto, S.; Sillankorva, S.; Azeredo, J.; Lu, T.K. Genetically Engineered Phages: A Review of Advances over the Last Decade. Microbiol. Mol. Boil. Rev. 2016, 80, 523–543. [Google Scholar] [CrossRef]
- Messing, J. [2] New M13 vectors for cloning. Methods Enzymol. 1983, 101, 20–78. [Google Scholar] [CrossRef] [PubMed]
- Melnikov, A.A.; Tchernov, A.P.; Fodor, I.; Bayev, A.A. Lambda phagemids and their transducing properties. Gene 1984, 28, 29–35. [Google Scholar] [CrossRef]
- Marinelli, L.J.; Hatfull, G.F.; Piuri, M. Recombineering: A powerful tool for modification of bacteriophage genomes. Bacteriophage 2012, 2, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, A.; Griffin, K.; Studier, F.W.; McCormick, M.; Berg, J.; Novy, R.; Mierendorf, R. T7Select phage display system: A powerful new protein display system based on bacteriophage T7. Innovations 1996, 6, 1–6. [Google Scholar]
- Schofield, D.; Westwater, C. Phage-mediated bioluminescent detection of Bacillus anthracis. J. Appl. Microbiol. 2009, 107, 1468–1478. [Google Scholar] [CrossRef] [PubMed]
- Ellis, H.M.; Yu, D.; Ditizio, T.; Court, D.L. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc. Natl. Acad. Sci. USA 2001, 98, 6742–6746. [Google Scholar] [CrossRef] [PubMed]
- Thomason, L.C.; Sawitzke, J.A.; Li, X.; Costantino, N.; Court, D.L. Recombineering: Genetic Engineering in Bacteria Using Homologous Recombination. Curr. Protoc. Mol. Boil. 2014, 106, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Thomason, L.; Court, D.L.; Bubunenko, M.; Costantino, N.; Wilson, H.; Datta, S.; Oppenheim, A. Recombineering: Genetic Engineering in Bacteria Using Homologous Recombination. Curr. Protoc. Mol. Boil. 2007, 78, 1–16. [Google Scholar] [CrossRef]
- Sawitzke, J.A.; Thomason, L.C.; Costantino, N.; Bubunenko, M.; Datta, S.; Court, N.L. Recombineering: In Vivo Genetic Engineering in E. coli, S. enterica, and Beyond. Methods Enzymol. 2007, 421, 171–199. [Google Scholar] [CrossRef]
- Kiro, R.; Shitrit, D.; Qimron, U. Efficient engineering of a bacteriophage genome using the type I-E CRISPR-Cas system. RNA Boil. 2014, 11, 42–44. [Google Scholar] [CrossRef]
- Martel, B.; Moineau, S. CRISPR-Cas: An efficient tool for genome engineering of virulent bacteriophages. Nucleic Acids Res. 2014, 42, 9504–9513. [Google Scholar] [CrossRef]
- Lemay, M.-L.; Tremblay, D.M.; Moineau, S. Genome Engineering of Virulent Lactococcal Phages Using CRISPR-Cas9. ACS Synth. Boil. 2017, 6, 1351–1358. [Google Scholar] [CrossRef]
- Brouns, S.J.J.; Jore, M.M.; Lundgren, M.; Westra, E.R.; Slijkhuis, R.J.H.; Snijders, A.P.L.; Dickman, M.J.; Makarova, K.S.; Koonin, E.V.; Van Der Oost, J. Small CRISPR RNAs Guide Antiviral Defense in Prokaryotes. Science 2008, 321, 960–964. [Google Scholar] [CrossRef]
- Hupfeld, M.; Trasanidou, D.; Ramazzini, L.; Klumpp, J.; Loessner, M.J.; Kilcher, S. A functional type II-A CRISPR–Cas system from Listeria enables efficient genome editing of large non-integrating bacteriophage. Nucleic Acids Res. 2018, 46, 6920–6933. [Google Scholar] [CrossRef] [PubMed]
- Jaschke, P.R.; Lieberman, E.K.; Rodriguez, J.; Sierra, A.; Endy, D. A fully decompressed synthetic bacteriophage øX174 genome assembled and archived in yeast. Virology 2012, 434, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Ando, H.; Lemire, S.; Pires, D.P.; Lu, T.K. Engineering Modular Viral Scaffolds for Targeted Bacterial Population Editing. Cell Syst. 2015, 1, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Bodner, K.; Melkonian, A.L.; Barth, A.I.; Kudo, T.; Tanouchi, Y.; Covert, M.W. Engineered Fluorescent E. coli Lysogens Allow Live-Cell Imaging of Functional Prophage Induction Triggered inside Macrophages. Cell Syst. 2020, 10, 254–264. [Google Scholar] [CrossRef]
- Kilcher, S.; Studer, P.; Muessner, C.; Klumpp, J.; Loessner, M.J. Cross-genus rebooting of custom-made, synthetic bacteriophage genomes in L-form bacteria. Proc. Natl. Acad. Sci. USA 2018, 115, 567–572. [Google Scholar] [CrossRef]
- Allan, E.J.; Hoischen, C.; Gumpert, J. Chapter 1 Bacterial L-Forms. Adv. Appl. Microbiol. 2009, 68, 1–39. [Google Scholar]
- Errington, J. L-form bacteria, cell walls and the origins of life. Open Boil. 2013, 3, 120143. [Google Scholar] [CrossRef]
- Studer, P.; Staubli, T.; Wieser, N.; Wolf, P.; Schuppler, M.; Loessner, M.J. Proliferation of Listeria monocytogenes L-form cells by formation of internal and external vesicles. Nat. Commun. 2016, 7, 13631. [Google Scholar] [CrossRef]
- Kilcher, S.; Loessner, M.J. Engineering Bacteriophages as Versatile Biologics. Trends Microbiol. 2018, 27, 355–367. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; A Hutchison, C.; O Smith, H. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Widder, E. Bioluminescence in the Ocean: Origins of Biological, Chemical, and Ecological Diversity. Science 2010, 328, 704–708. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.P.; Unch, J.; Binkowski, B.F.; Valley, M.P.; Butler, B.L.; Wood, M.G.; Otto, P.; Zimmerman, K.; Vidugiris, G.; Machleidt, T.; et al. Engineered Luciferase Reporter from a Deep Sea Shrimp Utilizing a Novel Imidazopyrazinone Substrate. ACS Chem. Boil. 2012, 7, 1848–1857. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Chau, Y.; Lee, Y.-K. Phage-based Electrochemical Sensors: A Review. Micromachines 2019, 10, 855. [Google Scholar] [CrossRef] [PubMed]
- Ford, M.; Stenström, C.; Hendrix, R.; Hatfull, G. Mycobacteriophage TM4: Genome structure and gene expression. Tuber. Lung Dis. 1998, 79, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Timme, T.L.; Brennan, P.J. Induction of Bacteriophage from Members of the Mycobacterium avium, Mycobacterium intracellulare, Mycobacterium scrofulaceum Serocomplex. J. Microbiol. 1984, 130, 2059–2066. [Google Scholar] [CrossRef]
- Dunne, M.; Rupf, B.; Tala, M.; Qabrati, X.; Ernst, P.; Shen, Y.; Sumrall, E.; Heeb, L.; Plückthun, A.; Loessner, M.J.; et al. Reprogramming Bacteriophage Host Range through Structure-Guided Design of Chimeric Receptor Binding Proteins. Cell Rep. 2019, 29, 1336–1350. [Google Scholar] [CrossRef]
- Yehl, K.; Lemire, S.; Yang, A.C.; Ando, H.; Mimee, M.; Torres, M.D.T.; De La Fuente-Nunez, C.; Lu, T.K. Engineering Phage Host-Range and Suppressing Bacterial Resistance through Phage Tail Fiber Mutagenesis. Cell 2019, 179, 459–469. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).