Abstract
Accurate pathogen detection and diagnosis is paramount in clinical success of treating patients. There are two general paradigms in pathogen detection: molecular and immuno-based, and phage-based detection is a third emerging paradigm due to its sensitivity and selectivity. Molecular detection methods look for genetic material specific for a given pathogen in a sample usually by polymerase chain reaction (PCR). Immuno-methods look at the pathogen components (antigens) by antibodies raised against that pathogen specific antigens. There are different variations and products based on these two paradigms with advantages and disadvantages. The third paradigm at least for bacterial pathogen detection entails bacteriophages specific for a given bacterium. Sensitivity and specificity are the two key parameters in any pathogen detection system. By their very nature, bacteriophages afford the best sensitivity for bacterial detection. Bacteria and bacteriophages form the predator-prey pair in the evolutionary arms race and has coevolved over time to acquire the exquisite specificity of the pair, in some instances at the strain level. This specificity has been exploited for diagnostic purposes of various pathogens of concern in clinical and other settings. Many recent reviews focus on phage-based detection and sensor technologies. In this review, we focus on a very special group of pathogens that are of concern in biodefense because of their potential misuse in bioterrorism and their extremely virulent nature and as such fall under the Centers for Disease and Prevention (CDC) Category A pathogen list. We describe the currently available phage methods that are based on the usual modalities of detection from culture, to molecular and immuno- and fluorescent methods. We further highlight the gaps and the needs for more modern technologies and sensors drawing from technologies existing for detection and surveillance of other pathogens of clinical relevance.
1. Introduction
Bacteriophages, also known as phages, are ubiquitous viruses that infect bacteria and archaea. Like their eukaryote relatives, phages can consist of either double-stranded or single-stranded DNA or RNA genomes. This diversity of phages can be seen in both their morphology and genome architecture, with genome size that ranges from ~3–500 kb. First described independently in 1915 by Frederick Twort and in 1917 by Felix d’Herelle, we now know phages are the most abundant taxon in the biosphere with an estimated 1031 viral particles. Since their discovery, phages have had an enormous impact on our understanding of genetics, molecular biology, bacterial evolution, and marine and soil ecology. Furthermore, the study of bacterial defenses against phages have led to powerful scientific tools including restriction enzymes and the revolutionary gene editing tools of the CRISPR-Cas system. The discovery of restriction enzyme system was awarded the Nobel Prize for Physiology/Medicine in 1978 and in 2020, CRISPR-Cas-based gene editing application was awarded the Nobel Prize in Chemistry.
Phages were the first to have their entire genomes sequenced and, indeed, the use of phage derived products such as T7 DNA polymerase, T4 DNA ligase, M13 vectors were critical in whole genome sequencing projects from phages to humans. Phages themselves have played an immense role in developing tools and methods for pathogen detection and identification. From the Center for Disease Control and Prevention’s (CDC) gold standard for “phage typing” bacterial isolates to the development of the phage display system for discovery of binding peptides and antibodies, researchers have found creative ways to exploit phages for novel pathogen detection platforms. There are many reviews on the use of phages for pathogen detection using different assays, platforms and biosensors [,,,,,,,,]. This review will focus on how phages have been exploited for detection of those pathogens that are of most concern due to their history and potential for use as biowarfare and bioterrorism agents.
Historically, the use of phages in bacterial detection started with phage typing schemes for strains of specific pathogens such as Escherichia coli, Klebsiella, Shigella, Salmonella, and Vibrio cholera []. Over time, more diverse and innovative approaches have been taken. The state of the art in phage-based detection of biodefense pathogens is illustrated in Figure 1 and summarized in Table 1 along with key features such as limit of detection and time to result. In addition, the whole genome sequence features of phages that infect different biodefense pathogens are provided in a Supplementary table. In the following sections, we elaborate on the detection modalities currently available for individual pathogens.
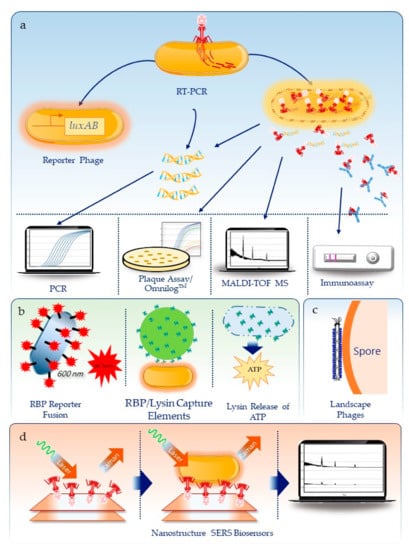
Figure 1.
Summary of approaches used to detect biothreats using bacteriophage. (a) Methods dependent on the infection and metabolic activity or lysis of the bacteria cell, including reporter phages, molecular and immuno-detection for phage RNA/DNA and proteins, respectively, detection of phage proteins by mass spectrometry, and the traditional plaque assay or real-time, high throughput detection on the Omnilog™. (b) Use of phage components, including receptor binding proteins (RBP) reporter fusions to reporter molecules like florescent proteins, use of RBPs or lysins as capture agents on Qdots or gold nanoparticles, or use of lysins to release ATP that can be measured using a luminescent enzyme assay. (c) Engineered filamentous phages for spore detection and (d) as capture agents on plasmonic nanostructures coupled with surface enhanced Raman spectroscopy (SERS) for Brucella detection.

Table 1.
Summary of phage-based methods for the detection of biodefense pathogens.
2. Use of Pathogen Specific Phages
2.1. Anthrax
Following the 2001 anthrax letter attacks in the United States, there has been a renewed emphasis on early warning and rapid detection of the deadliest pathogens on the CDC Select Agent list (https://www.selectagents.gov/SelectAgentsandToxinsList.html). Bacillus anthracis is the bacterium that causes the infectious disease anthrax and is a Gram positive spore forming pathogen. Anthrax spores can be produced at high concentrations and disseminated efficiently and have been weaponized for use in bioterrorism. For these reasons it is an ideal biological weapon of mass destruction. Left untreated for more than 24 h, anthrax is usually fatal. Diagnosis is difficult symptomatically and it was traditionally identified by clinical, microbiological, and morphological methods.
Culture-based methods: Use of phages for detection of B. anthracis vegetative cells dates back to the 1950s with the identification of a phage variant γ, which could infect and lyse encapsulated smooth forms from active infections of B. anthracis and differentiate between B. anthracis and B. cereus strains []. This assay was further refined and eventually FDA approved in 2005 for clinical diagnosis and is used by the Laboratory Response Network (LRN) [,]. Specificity of the assay is very high with very few non-B anthracis strains being susceptible to infection. With over a hundred B. anthracis strains tested, very few have been found to be resistant to phage γ. One of the disadvantages of culture-based γ phage assay is that it takes 20 h for the confirmation from visualizing clear plaques. A faster high throughput OmnilogTM-based indirect phage assay has also been reported for the real time detection of B. anthracis [].
Phage specific PCR and immunoassay methods: In an attempt to speed up and reduce the time ‘from sample to result’ of the assay, a real-time PCR assay was developed to detect the amplification of phage DNA []. This reduced the detection time to 5 h and had the added benefit of determining the viability of the bacteria in test sample since only metabolically active B anthracis cells would allow for phage DNA amplification. Alternatively, a method that detects the initial burst of γ phage RNA upon infection by RT-PCR allows for shortened incubation times and the ability to determine antibiotic sensitivity due to the suppressed metabolic state in the presence of antibiotics []. Another approach to using phage amplification as a means to detect vegetative B. anthracis cells is to use phage specific antibodies in an immunoassay format. Using the lateral flow immunoassay (LFI) format, Cox et. al. was able to detect 8 × 105 cells in as little as 2 h and down to 1.5 × 105 cells in 4 h [].
Phage reporter assays: An indirect measurement of phage growth to detect viable B. anthracis cells is the use of reporter phages. One of the first attempts on this idea was to engineer the parental phage of γ, phage Wβ []. Two phage genes considered non-essential were replaced with the bacterial luciferase genes luxAB from Vibrio harveyi. As expected, the reporter phage is inert and unable to express the luxAB genes until the phage is able to attach and inject its genome into the host cell. At this point, the bacterial host transcriptional and translational machinery is taken over by the phage and upon expression of phage genes will also express the luciferase enzyme which produces bioluminescence upon the addition of a substrate. With this reporter phage, vegetative B. anthracis cells could be detected down to about a thousand cells/mL, 60 min post infection and as fast as 16 min at higher cell densities. By using the temperate phage Wβ, the luciferase enzyme can accumulate within the bacterial host to increase the amount of bioluminescence produced, thus increasing the sensitivity of the assay. The drawback of using the phages γ and Wβ is that they can infect only vegetative cells and not spores; in order to detect encapsulated B. anthracis spores they need first to be cultured in germinating conditions to allow for infection, thus adding additional step and time to the assay. The Wβ reporter phage displayed 100% inclusivity (38 of 38 strains) for B. anthracis []. Six out of the 126 non-anthracis Bacillus near neighbors such as B. cereus, B. mycoides, and B. weihenstephanensis, (95% specificity) elicited bioluminescent signals, albeit at reduced intensities compared to B. anthracis. Species outside the Bacillus genus tested negative for bioluminescence. After protocol optimization, this phage was able to detect germinated B. anthracis spores in a range of spiked matrices including blood, ground beef, milk, and other dairy products [,]. After optimization of the reporter, increased sensitivity allowed for detection of spores in environmental soil samples and water samples from pond, lake or brackish waters, demonstrating a wide range of application for spore detection [,].
Receptor binding proteins (RBPs) as tools for detection: The first step in phage life cycle is the attachment of the phage to its cognate bacterial host through interaction between a phage component known as the receptor binding protein or RBP and the bacterial receptor on the cell surface. RBPs provide the same specificity as the whole phage in targeting the host and RBPs have been successfully used for detection of B. anthracis. In tailed phages, it is the tail fiber protein that is most often responsible for determining the specificity of the phage. RBPs can be used as a detection reagent independent of the whole phage. One approach is to fuse RBPs to fluorescent reporters. For phage γ, fusing the RBP (gp14) to green fluorescent protein (GFP) allows for fluorescent labeling of B. anthracis []. Using a bioinformatics approach, Braun, P. et. al. were able to predict a putative RBP (P28 + P29) encoded by phage AP50c based on genomic synteny and small identical stretches of protein sequence with the RBP (P24) polypeptide sequences from phage Wip1 []. Phages AP50c and Wip1 are similar phages belonging to the Tectiviridae family. Using the RBP (gp14) sequence from phage γ, they were also able to identify another putative RBP of the prophage λBa03 present on the B. anthracis genome. These RBPs fused to mCherry fluorescent proteins were evaluated for their ability to bind to and detect cultured B. anthracis cells by fluorescence microscopy. Full length as well as truncated versions of the RBPs were evaluated and RPB::mCherry reporters from Wip1, AP50c, and truncated λBa03 were found to work best in terms of producing fluorescence signals. Bacterial detection was time and growth phase dependent after germination from spores and only vegetative cells could be detected. Inactivation of B. anthracis cells by heat, formaldehyde or peracetic acid did not prevent the detection of several virulent strains using the RBP reporters but sensitivity varied depending on inactivation method. Presence of the poly-γ-D-glutamic acid (PGA) capsule found on cell surface did not prevent detection of virulent strains. Specificity of detection of B. anthracis was good; of the 56 non-anthracis Bacillus spp. strains tested, most were not bound by any of the three RBP:: mCherry proteins at all, and a small number showed only marginal binding.
Phage components as tools for detection: Another approach for detection of B. anthracis involved using components of the phages known to specifically target their bacterial host. The first example of this was the use of the γ phage protein PlyG lysin []. Gamma phage uses the PlyG to hydrolyze the peptidoglycan components of the cell wall and lyse the cell to release progeny phage. Addition of purified PlyG externally to a B. anthracis containing sample, rapidly lysed the bacteria, releasing the cellular ATP. Upon further addition of the luciferase enzyme and the substrate luciferin, as with the reporter phage example described above, light is emitted, allowing for detection of as few as 100 cells [].
The PlyG protein has also been used for its binding properties and a non-catalytic truncated version was demonstrated to be a capture agent for the detection of B. anthracis, both encapsulated and non-encapsulated cells []. The C-terminal region of the protein is responsible for specific binding to the cell wall of B. anthracis but do not bind to B. subtilis, B. cereus, B. thuringiensis, and three other Bacillus species when used in a dot blot format []. In another study, short 10–20 amino acid peptides from the C-terminal end of PlyG were shown to bind to B. anthracis using the same dot blot assay as used with the truncated protein []. By coupling one of the peptides with Quantum dots (Qdots), B. anthracis cells could be detected in spiked plasma by fluorescence microscopy or by plate reader with the same specificity as the phage γ assay [].
Engineered phages as tools for detection: Additional peptides that can be used in the Qdot assay for B. anthracis detection were discovered by employing a commercial M13 phage display peptide library against B. cereus 4342, which is a rare strain also susceptible to phage γ []. These peptides BBP-1 and BBP-2 were specific to B. anthracis and B. cereus 4342 and did not bind to other B. cereus or B. thuringiensis strains. Based on 2D gel electrophoresis, immunoblotting and peptide extraction and sequencing the authors hypothesize that the BBP-1 and BBP-2 peptides bind to the S-layer protein EA1 from these specific Bacillus strains. EA1 gene diversity is quite extensive and it is possible that the EA1 in strains such as B. cereus 11778 is diverged enough to preclude binding by these peptides. Comparison of the EA1 gene sequences of B. cereus 11778 to B. anthracis, B. cereus 4342, and other strains can potentially reveal motifs critical for binding of the peptide.
One of the common criticisms about phage-based detection is that it is useful only for bacterial detection and cannot be used for detection of other agents such as spores, viruses, and toxins. However, it is possible to engineer specificities for these non-bacterial agents in a phage-based platform. For example, bioengineering of phages is a powerful tool as demonstrated by the use of the filamentous phage display system of M13 phage for antibody discovery. Many recent reviews describe the application of the M13 phage display system [,,,,,]. One variation of the phage display approach is the landscape phages which are also filamentous phages but instead of fusing scFv or peptide libraries to the pIII minor coat protein, the peptide libraries are fused to the pVIII major coat protein. In phage fd, there are thousands of copies of pVIII repeated along the phage surface [,,]. This allows creating libraries that express the peptides on thousands of copies of major coat protein in a fixed conformation and can be screened for specific binding peptides to a targeted structure. These phages have been used as substitutes for antibodies [,]. This approach was used to identify peptide sequences that could bind to B. anthracis spores in an ELISA format. Three of these peptides were evaluated for their specificity using a co-precipitation assay and while distant Bacillus spores weakly bound to these phages, there was stronger cross-reactivity to closer Bacillus relatives of B. cereus and B. thuringiensis []. One of these phages was used as the capture reagent for magnetoelastic resonating particles. Detection of B. anthracis spores was concentration dependent down to 103 spores/mL. Scanning electron micrographs showed spores bound to the phage coated particles but not the uncoated particles. When other Bacillus spores B. cereus and B. megaterium were mixed in, there was some cross-reactivity consistent with the earlier co-precipitation studies []. Similar approaches can be taken to develop phages that are specific for viruses and toxin detection.
2.2. Plague
Yersinia pestis is the etiological agent of the plague. It is a zoonotic disease that affects rodents and can be transmitted from animal to animal by fleas, which happens to be the most common mode of transmission to humans []. When bitten by a Y. pestis infected flea, humans develop swollen lymph nodes, or buboes, that can develop into septicemia or occasionally into pneumonia. Pneumonic plague infections are transmissible between humans through respiratory droplets and is highly virulent. The three great plagues have been well described elsewhere but today plague is still endemic in certain parts of the globe including the Democratic Republic of the Congo, Madagascar, and Peru. Y. pestis is also considered a Category A pathogen with great potential for use in a bioterrorist attack due to its rapid disease progression and high mortality rate. Y. pestis grows slowly and diagnosis by culturing methods can take 2–5 days. Alternatively, a positive diagnosis can be determined by observing a 4-fold difference in antibody titers to F1 capsular antigen []. Rapid test using ELISA, LFI, PCR, or real-time PCR have been developed to speed up the detection or diagnosis of plague [,,,,,,,].
Culture-based detection: Similar to γ phage, the CDC recommended Y. pestis specific phage for identification and plague diagnosis is φA1122 []. As with γ phage, φA1122 is used in the plaque assay format by the CDC, World Health Organization and U.S. Army Research Institute of Infectious Diseases []. φA1122 is able to infect most Y. pestis strains tested and will only infect the near neighbor Y. pseudotuberculosis under certain temperature constraints [,,]. Two other phages are used for plague diagnosis, the Pokrovskaya phage and the L-413C phage [,].
Phage molecular assay: Sergueev et. al. developed a quantitative real-time PCR (real time-qPCR) assay for φA1122 and L-413C that could be used to detect replicating phage DNA in live Y. pestis with a sensitivity of 103 cfu/mL to 105 cfu/mL or one bacterium to 100 cfu per qPCR sample, respectively []. This assay proved to be suitable for both lab-based (LightCycler 2.0) and field-deployable (JBAIDS) real time-qPCR systems []. The assay was sensitive in simulated blood specimens, serum, and organ samples. Bacteriophage φA1122 showed some degree of amplification on 4 of the 20 Y. pseudotuberculosis strains tested at 28 °C. The specificity of this assay was increased to almost 100% by incubating the bacteria at 24 °C. φA1122 did not lyse any of the 17 Y. enterocolitica strains tested. L-413C demonstrated greater specificity, regardless of temperature, but was less sensitive for Y. pestis detection.
Reporter phage assay: The φA1122 phage has also been used to develop a reporter phage by inserting the luxAB genes into a non-coding region downstream of the A1, A2, A3 promoters, which are highly active early during infection []. In log phase cultures grown at either 28 °C or 37 °C the production of bioluminescence signal is rapid (<15 min) and has a sensitivity of ~800 cells on a plate reader. To demonstrate its clinical relevance, serum samples spiked with Y. pestis were evaluated for production of signal upon addition of the reporter phage. Serum did increase the time to detect and damped the strength of signal but detection in less than an hour was possible. Furthermore, blood samples spiked with Y. pestis and diluted with LB or transferred to commercial blood culture bottles were compatible for detection with the reporter phage despite a reduction in overall bioluminescence. Cultured samples were detected in as little as 20 min by the reporter phage. Antibiotic susceptibility can also be determined with the application of the reporter phage as the strength of signal diminishes in the presence of antibiotics in a dose dependent manner []. After optimizing the reporter phage by integrating the luxAB genes under control of the major capsid gene promoter gp10, this new reporter phage allowed for a more rapid and greater dynamic range of detection. After adding phage 1-2 h after adding either chloramphenicol, tetracycline, or streptomycin, bioluminescence was measured. As bioluminescence production is dependent on cell growth, there was an inverse correlation of signal to antibiotic susceptibility [].
RBP protein fusions as tools for detection: As described before with Bacillus phages, fluorescent protein fusions to the RBP of the Y. pestis phages φA1122 (gp17) and L-413C (gpH) have been used to detect the attenuated EV76 strain of Y. pestis species by fluorescent microscopy []. This detection capability was dependent on the temperature and time of the culture conditions. Both RBP fusions were able to differentiate Y. pestis from other related risk group 2 pathogens Y. pseudotuberculosis, Y. enterocolitica subsp. enterocolitica and Y. enterocolitica subsp. palearctica, Y. wautersii, and Y. similis when grown at 28 °C. However, when grown at 37 °C, the φA1122 RBP fusion could detect Y. pseudotuberculosis, consistent with the ability of φA1122 to form plaques on Y. pseudotuberculosis when grown at this temperature. Therefore, the authors suggest that the L-413C RBP fusion would be better suited for clinical diagnosis. Finally, the process of inactivation by heat, peracetic acid, ethanol, and paraformaldehyde (PFA) was evaluated in this study. It was determined that 4% PFA was ideal for inactivation and used to demonstrate detection of six different risk group 3 strains of Y. pestis [].
Phage protein-based assay: Another application of using φA1122 as a surrogate agent for rapid Y. pestis identification is the use of phage proteins detected by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) []. MALDI-TOF MS has been a valuable tool for rapid bacterial detection but often requires pure cultures grown overnight or more under specific conditions. Infection and detection by MS of an increase in phage proteins during the phage life cycle serves as a surrogate for bacterial detection and eliminates the need for overnight culturing thereby reducing time to result. A drawback to this approach is the need for repetitive, time-consuming sample preparations and analysis over the course of phage infection to determine the detection limit of the phage proteins. However, using a modified version of the Payne and Jansen phage therapy model, specific equations and information such as burst size and time can be used to predict when the phage will be detectable by MALDI-TOF MS in the presence of various concentrations of bacteria and multiplicities of infection (MOI) []. The starting concentration of phage used was based on the experimental limit of detection in order to differentiate between the inoculating phage and their progeny. Modeling φA1122 replication in Y. pestis demonstrated how these models can be used to predict when the phage 15.8 kDa capsid assembly protein can be detected and effectively reducing the time to detection compared to detecting the bacteria directly. However, time to detection is dependent on the MOI (concentration of both phage and bacteria) with the most rapid detection of an hour in the presence of 107 Y. pestis cells []. It is noteworthy that the limit of detection of purified phage needs to be determined for each phage and host model for a particular MALDI-TOF MS system.
2.3. Brucella Species
Brucellosis is a debilitating and incapacitating bacterial zoonotic disease that infects hundreds of thousands of people all across the globe. In addition, it causes reproductive losses in livestock each year [,,]. The four main species of Brucella that cause disease in humans and livestock are, B. melitensis, B. abortus, B. suis, and B. canis. Brucella spp. and were weaponized during the Cold War and today the CDC still classifies B. melitensis, B. abortus, and B. suis as Category B Select Agents. Diagnosis of brucellosis is challenging as symptoms are not specific and the infection can affect many organs and systems. Laboratory diagnosis usually consist of three modalities: culture, serology, and PCR [,,,,,,,,]. All of these tests have their own set of challenges from long culture times, delayed and false positive serology results, and false negative PCR results due to low bacteremia or post antibiotic treatment and symptomatic false positives, as well as PCR inhibitors in blood and tissues [,]. Phage-based testing using lytic phages specific for Brucella has been around for decades [,].
Culture-based phage assay: Brucellaphages Tbilisi (Tb), Firenze (Fz), Weybridge (Wb), S708, Berkeley (Bk), R/C, and Izatnagar (Iz) are used at two different concentrations for identification of Brucella at both genus and species level, including differentiation between smooth and rough strains of B. abortus, B. suis, B. melitensis, B. neotomae, B. canis, and B. ovis [,,]. As with other phage lysis assays, this is a multi-day assay requiring pure cultures of Brucella isolates from samples
Phage molecular assay: Similar to Y. pestis phages, Brucella phages can be used for indirect detection of Brucella spp. Although not as quick as the Y. pestis phage PCR assays, the qPCR assay using phage S708 could detect live B. abortus S19 cells at concentration from 106–108 CFU/mL in 24 h after the addition of phage to the culture. Extending this incubation time to 48 h increased detection sensitivity to 103–105 CFU/mL range and 72 h 1–100 cells/mL range []. A detection sensitivity of one cell per mL of test culture is a remarkable sensitivity considering the low concentration of bacteria in early Brucellosis. Four other phages, Bk, Fz, Tb, and Wb, also showed robust sensitivity as a surrogate qPCR marker for live Brucella. Phage Bk has the ability lyse many smooth Brucella spp. and therefore would be suited for diagnosis of human brucellosis. Testing on samples of near neighbor bacterial species O. anthropi and A. felis as well as the serological relevant cross reactive species Y. enterocolitica serovar O:9, demonstrated the specificity of Bk phage for Brucella spp. Mixing in the other species dampened the qPCR assay for Bk but did not inhibit it adversely for good detection. The assay did not detect phage amplification in several Brucella spiked commercial sheep blood products; so, the assay method had to be modified to remove suspected inhibitors of Brucella growth before qPCR amplification of phages could be achieved [].
Phage biosensor-based detection: The detection of Brucella using phages has also been demonstrated using biosensors that employ surface-enhanced Raman scattering (SERS) technology combined with multilayer octupolar nanostructures []. By functionalizing these nanostructures with the Brucella phage Tb using 4-mercaptobenzoic acid, the authors were able to specifically detect inactivated or live B. abortus at concentrations of 104 CFU/mL and the ability to take measurements at the single cell level in less than an hour. It is unclear how this biosensor would perform with clinical or environmental samples since the samples of Brucella were from cultures re-suspended in water. Another SERS substrate made of plasmonic nanocavities in a photonic quasicrystal arrangement allowed for the detection of femtomolar amounts of Tb but the authors did not test the ability of this biosensor to detect bacteria [].
2.4. Burkholderia
The gram-negative bacterium Burkholderia pseudomallei is an opportunistic pathogen that causes the life-threatening infection melioidosis. It is endemic to tropical and subtropical regions of the world and recent studies suggest that it is more widespread than previously thought [,,]. Melioidosis is known to have mortality rates ranging from 30–60% and in the U.S. it is considered to be a Category A select agent due to its aerosol transmissibility and infectivity, severity, and world-wide distribution. Current treatment for the disease involves a strict 20 week regimen of antibiotics, but even with this, relapse and mortality rates remain high []. More troubling, B. pseudomallei is intrinsically resistant to many antibiotic treatments, particularly aminoglycosides, due to an efflux pump mechanism that works in concert with other resistance mechanisms present in this organism to further elevate multidrug resistance [].
Protein-based phage assay: As described for Y. pestis, in silico prediction of phage amplification combined with MALDI-TOF MS has been used to detect the presence of live B. pseudomallei. Using the lytic phage φX216, three major proteins are detectable corresponding to phage tail protein, major capsid protein and its doubly charged ion []. After determining the limit of detection of the phage proteins in purified phage samples, this model accurately predicted the detection of phage proteins in 2 h at bacteria concentrations of 3.2 × 105 CFU/mL using phage inoculum concentrations below the pre-determined limit of detection. As described above, the need for metabolically active cells for phages to replicate can be leveraged to determine antibiotic sensitivity. The authors were able to demonstrate phage proteins could not be detected in a sensitive B. pseudomallei strains in the presence of ceftazidime but when an isogenic antibiotic resistant strain was used, phage proteins, but not bacterial proteins, were detectable in the 2 h time frame predicted in silico, despite the presence of antibiotics.
2.5. Future Directions
The current Gold Standard for pathogen detection continues to be PCR. The analytical sensitivity of PCR is extremely high thanks to the huge amplification of the target nucleic acid segment; theoretically a single target copy can be amplified to billion copies in 40 cycles. The analytical specificity of PCR is largely dependent on target selection, i.e., present in target pathogen and not present in other organisms. Among the immunoassay category, LFI is the predominant method for pathogen detection. They are faster, cheaper and easy to use. The analytical sensitivity of LFI is relatively low compared to PCR due to the fact that there is no amplification of the target antigen and is determined by the kinetics of antigen-antibody interactions used in the assay. Similar to PCR, the specificity of the assay is largely determined the presence of the antigen only in the target pathogen and absence in other organisms. Both PCR and LFI do not discriminate live or dead bacterial pathogen. The sensitivity of phage-based assay is largely determined by the detection modality of the assay, i.e., methods such as fluorescence microscopy can detect single target pathogen. If the assay is designed to detect the first step in phage’s life cycle which is adsorption of the phage particle to the cell surface, it can be faster than other methods. The kinetics of interaction of phage-bacteria has not been compared to antigen-antibody interactions. However, both involve protein-protein interactions and are expected to be quite comparable. The specificity of phage-based assay is quite exquisite due to coevolution of the pair. Hence, the phages are extremely specific to the target host species and sometimes even to the strain level. There are exceptions to this rule where a phage specific for a particular species can infect strains of near neighbor species most likely due to the presence of the phage receptor gene or its functional variant.
There are many more examples of how phages are used for pathogen detection for non-biothreat organisms that are well reviewed [,,,,,]. This is in part due to the greater interest in bacteria that cause food borne illness and other common bacterial pathogens that are easier to work with in the laboratory and do not have biosecurity concerns. If we are to take advantage of these phage-based approaches for biodefense, there needs to be more investment into the discovery of phages for biothreat agents. For example, there are no known virulent or temperate phages for the causative agents for Q fever, Coxiella burnetii. The chromosome does not contain any prophage sequences nor are there any CRISPR arrays in all except potentially one strain, Coxiella burnetii str. Namibia, suggesting most Coxiella spp. may not have needed to evolve a defense against phages [,,]. There are also no known natural phages for Francisella tularensis. However, the F. tularensis. subsp. novicida strain U112 genome has both a putative phage and CRISPR/Cas system []. Other Ft. subsp. lack a CRISPR array do not appear to have a functional CRISRP/Cas system according to the CRISPR-Cas++ database.
Since the dawn of next generation sequencing about 15 years ago, there has been an explosion of whole genome sequences of many biodefense pathogen and near neighbors. Systematically analyzing whole genome sequences for the presence of prophages can expand the repertoire of phages that can be potentially used for detection and diagnostics [].
Bioprospecting for new phages is a robust method for discovery of new phages. There are dozens of phages for the four biothreat agents described in this review (Table S1) but other biothreats are lacking in numbers and diversity [,]. Expanding the host range of known phages is another strategy that could increase the number of phages available for biothreats. Various approaches have been used to expand the host range of phages [,,,,,,,,,]. This was demonstrated in T4 phage using an error prone PCR strategy [] and in T3 phage via site-directed mutagenesis libraries [] of tail fiber genes, specifically in the host-range-determining regions (HRDRs). Both approaches resulted in mutant phages that were able to infect Y. ruckeri and Y. pseudotuberculosis strains, respectfully, suggesting these approaches could be used to expand host range to Y. pestis as well. By replacing portions of the tail fiber gene gp17 of T3 phage with sequence from Yersinia phage R, Ando et al. was able to change the E. coli strain host range of these phages to infect and replicate in two Yersinia pseudotuberculosis stains []. Although not demonstrated, it is likely that this engineered phage host range would also be expanded to Y. pestis and could be used for detection purposes. The host range of phages can also be altered by continuous propagation of the phage in the presence of multiple host strains with varying degrees of adsorption when one or more of the strains is restrictive for phage propagation. Using an innovative chemostat system, Holtzman et al. demonstrated that the host range of T7 can evolve to avoid restrictive strains by altering its RBP, even when those restrictive strains have better adsorption properties of the LPS receptor []. Host range expansion or changes is also possible for gram positive host specific phages. After identifying and confirming the RBP of the Listeria monocytogenes phage PSA, a library of modified PSA phages with mutations in the RBP were generated by error prone PCR, followed by whole genome reconstruction by Gibson assembly []. RBP mutants able to shift host serotype specificity were identified and found to have single point mutations in the C-terminal end of the RBP. By creating a chimeric PSA phage encoding both wild-type and mutant RBPs, it was possible to generate phages that had a host range of both serotypes []. Furthermore, by mining published sequences of PSA-like prophages, combined with structural knowledge of the RBP, it was possible to redirect phage specificity by exchanging the RBP head domains from these pro-phages with the wild-type domain []. These approaches open up the possibility of engineering phages of near neighbor hosts to infect biothreat pathogens that currently have few or no known phages due to their added security and laboratory restrictions constraining phage discovery against these bacteria.
Some phages have the ability to modify their tropism through diversity-generating retroelements (DGRs) encoded by them. This was first demonstrated with the Bordetella bacteriophage, BPP-1, which can switch tropism between the infectious and environmental phases of Bordetella subspecies [,]. By modifying its own RBPs, BPP-1 can produce variants of a small defined region in the RBP that recognize distinct surface receptors of its host []. This is accomplished through the use of a reverse transcriptase (RT) encoded in a variability generating cassette. This cassette functions to introduce nucleotide substitutions at 23 sites in a 134-base-pair (bp) variability region (VR) present at the 3′ end of the mtd locus, which encodes the RBP. Sites of variability in VR correspond to adenine residues in a nearby homologous template repeat (TR), which is invariant and essential for RT mediated mutagenesis. The process involves transcription of TR to produce TR-RNA, which is then reverse transcribed by the RT to produce TR-cDNA. The TR-cDNA, which has specific errors due to the unfaithful copying of adenines, homes to and replaces VR through an undefined mechanism. This process can theoretically produce 1013–1014 variable nucleotide sequences at the C-terminal end of the RBP [,]. DGRs are found widely among prokaryotes and are not limited to phages [,,,]. However, DGRs could provide powerful tools for biothreat detection by directed RBP evolution via in vivo DNA diversification [].
3. Conclusions
Biodefense related bacterial pathogens form a unique group due to their notoriety of being highly virulent, easily weaponized, and high consequence nature. Biosurveillance and clinical diagnosis of these pathogens traditionally relied on microbiological, clinical, molecular, and immunological approaches using a variety of platforms and assays. Phage-based detection of these pathogens is still not commonplace and has relied on conventional culture-based plaque assays. Recent more modern rapid approaches include detection of genomic or protein components that exploit the amplification of these components during the active growth of phages inside a given bacterium. Other approaches use reporter phages or very rarely sensor-based approaches. A more comprehensive phage-based rapid and plug and play platform technology for the entire biodefense panel including viruses, spores and toxins is desirable and needed for a unified biodefense posture. Tapping into ongoing revolution in next generation sequencing technologies and other nanosensor platforms will provide a sensitive detection platform for biodefense pathogens. There is an untapped, easy to achieve phage potential that has not been fully exploited despite the fact there is renewed interest in phage therapy around the globe.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4915/12/12/1393/s1, Table S1: General genomic features of the bacteriophages of biodefense pathogens.
Author Contributions
S.S. and E.R.H. contributed equally to this review. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Anany, H.; Chou, Y.; Cucic, S.; Derda, R.; Evoy, S.; Griffiths, M.W. From Bits and Pieces to Whole Phage to Nanomachines: Pathogen Detection Using Bacteriophages. Annu. Rev. Food Sci. Technol. 2017, 8, 305–329. [Google Scholar] [CrossRef] [PubMed]
- Richter, L.; Janczuk-Richter, M.; Niedziolka-Jonsson, J.; Paczesny, J.; Holyst, R. Recent advances in bacteriophage-based methods for bacteria detection. Drug Discov. Today 2018, 23, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Chelliah, R.; Rubab, M.; Oh, D.H.; Uddin, M.J.; Ahn, J. Bacteriophages as Potential Tools for Detection and Control of Salmonella spp. in Food Systems. Microorganisms 2019, 7, 570. [Google Scholar] [CrossRef] [PubMed]
- Kilcher, S.; Loessner, M.J. Engineering Bacteriophages as Versatile Biologics. Trends Microbiol. 2019, 27, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Dams, D.; Brondsted, L.; Drulis-Kawa, Z.; Briers, Y. Engineering of receptor-binding proteins in bacteriophages and phage tail-like bacteriocins. Biochem. Soc. Trans. 2019, 47, 449–460. [Google Scholar] [CrossRef]
- Moon, J.S.; Choi, E.J.; Jeong, N.N.; Sohn, J.R.; Han, D.W.; Oh, J.W. Research Progress of M13 Bacteriophage-Based Biosensors. Nanomaterials 2019, 9, 1448. [Google Scholar] [CrossRef] [PubMed]
- Meile, S.; Kilcher, S.; Loessner, M.J.; Dunne, M. Reporter Phage-Based Detection of Bacterial Pathogens: Design Guidelines and Recent Developments. Viruses 2020, 12, 944. [Google Scholar] [CrossRef]
- Paczesny, J.; Richter, L.; Holyst, R. Recent Progress in the Detection of Bacteria Using Bacteriophages: A Review. Viruses 2020, 12, 845. [Google Scholar] [CrossRef]
- Aliakbar Ahovan, Z.; Hashemi, A.; De Plano, L.M.; Gholipourmalekabadi, M.; Seifalian, A. Bacteriophage Based Biosensors: Trends, Outcomes and Challenges. Nanomaterials 2020, 10, 501. [Google Scholar] [CrossRef]
- Schmidt, W.C.; Jeffries, C.D. Bacteriophage typing of gram-negative rod-shaped bacteria. CRC Crit. Rev. Clin. Lab. Sci. 1975, 6, 201–246. [Google Scholar] [CrossRef]
- Brown, E.R.; Cherry, W.B. Specific Identification of Bacillus anthracis by Means of a Variant Bacteriophage. J. Infect. Dis. 1955, 96, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Henry, M.; Biswas, B.; Vincent, L.; Mokashi, V.; Schuch, R.; Bishop-Lilly, K.A.; Sozhamannan, S. Development of a high throughput assay for indirectly measuring phage growth using the OmniLog(TM) system. Bacteriophage 2012, 2, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Reiman, R.W.; Atchley, D.H.; Voorhees, K.J. Indirect detection of Bacillus anthracis using real-time PCR to detect amplified gamma phage DNA. J. Microbiol. Methods 2007, 68, 651–653. [Google Scholar] [CrossRef] [PubMed]
- Malagon, F.; Estrella, L.A.; Stockelman, M.G.; Hamilton, T.; Teneza-Mora, N.; Biswas, B. Phage-Mediated Molecular Detection (PMMD): A Novel Rapid Method for Phage-Specific Bacterial Detection. Viruses 2020, 12, 435. [Google Scholar] [CrossRef] [PubMed]
- Cox, C.R.; Jensen, K.R.; Mondesire, R.R.; Voorhees, K.J. Rapid detection of Bacillus anthracis by gamma phage amplification and lateral flow immunochromatography. J. Microbiol. Methods 2015, 118, 51–56. [Google Scholar] [CrossRef]
- Schofield, D.A.; Westwater, C. Phage-mediated bioluminescent detection of Bacillus anthracis. J. Appl. Microbiol. 2009, 107, 1468–1478. [Google Scholar] [CrossRef]
- Schuch, R.; Fischetti, V.A. Detailed genomic analysis of the Wbeta and gamma phages infecting Bacillus anthracis: Implications for evolution of environmental fitness and antibiotic resistance. J. Bacteriol. 2006, 188, 3037–3051. [Google Scholar] [CrossRef]
- Braun, P.; Wolfschlager, I.; Reetz, L.; Bachstein, L.; Jacinto, A.C.; Tocantins, C.; Poppe, J.; Grass, G. Rapid Microscopic Detection of Bacillus anthracis by Fluorescent Receptor Binding Proteins of Bacteriophages. Microorganisms 2020, 8, 934. [Google Scholar] [CrossRef]
- Schuch, R.; Nelson, D.; Fischetti, V.A. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 2002, 418, 884–889. [Google Scholar] [CrossRef]
- Fujinami, Y.; Hirai, Y.; Sakai, I.; Yoshino, M.; Yasuda, J. Sensitive detection of Bacillus anthracis using a binding protein originating from gamma-phage. Microbiol. Immunol. 2007, 51, 163–169. [Google Scholar] [CrossRef]
- Kikkawa, H.; Fujinami, Y.; Suzuki, S.; Yasuda, J. Identification of the amino acid residues critical for specific binding of the bacteriolytic enzyme of gamma-phage, PlyG, to Bacillus anthracis. Biochem. Biophys. Res. Commun. 2007, 363, 531–535. [Google Scholar] [CrossRef] [PubMed]
- Sainathrao, S.; Mohan, K.V.; Atreya, C. Gamma-phage lysin PlyG sequence-based synthetic peptides coupled with Qdot-nanocrystals are useful for developing detection methods for Bacillus anthracis by using its surrogates, B. anthracis-Sterne and B. cereus-4342. BMC Biotechnol. 2009, 9, 67. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Johnson, M.L.; Guntupalli, R.; Petrenko, V.A.; Chin, B.A. Detection of Bacillus anthracis spores in liquid using phage-based magnetoelastic micro-resonators. Sens. Actuators B Chem. 2007, 127, 559–566. [Google Scholar] [CrossRef]
- Garcia, E.; Elliott, J.M.; Ramanculov, E.; Chain, P.S.; Chu, M.C.; Molineux, I.J. The genome sequence of Yersinia pestis bacteriophage phiA1122 reveals an intimate history with the coliphage T3 and T7 genomes. J. Bacteriol. 2003, 185, 5248–5262. [Google Scholar] [CrossRef]
- Sergueev, K.V.; He, Y.; Borschel, R.H.; Nikolich, M.P.; Filippov, A.A. Rapid and sensitive detection of Yersinia pestis using amplification of plague diagnostic bacteriophages monitored by real-time PCR. PLoS ONE 2010, 5, e11337. [Google Scholar] [CrossRef] [PubMed]
- Sergueev, K.V.; Nikolich, M.P.; Filippov, A.A. Field and clinical applications of advanced bacteriophage-based detection of Yersinia pestis. Adv. Exp. Med. Biol. 2012, 954, 135–141. [Google Scholar] [CrossRef]
- Schofield, D.A.; Molineux, I.J.; Westwater, C. Diagnostic bioluminescent phage for detection of Yersinia pestis. J. Clin. Microbiol. 2009, 47, 3887–3894. [Google Scholar] [CrossRef]
- Schofield, D.A.; Molineux, I.J.; Westwater, C. Rapid identification and antibiotic susceptibility testing of Yersinia pestis using bioluminescent reporter phage. J. Microbiol. Methods 2012, 90, 80–82. [Google Scholar] [CrossRef]
- Born, F.; Braun, P.; Scholz, H.C.; Grass, G. Specific Detection of Yersinia pestis Based on Receptor Binding Proteins of Phages. Pathogens 2020, 9, 611. [Google Scholar] [CrossRef]
- Cox, C.R.; Rees, J.C.; Voorhees, K.J. Modeling bacteriophage amplification as a predictive tool for optimized MALDI-TOF MS-based bacterial detection. J. Mass Spectrom. 2012, 47, 1435–1441. [Google Scholar] [CrossRef]
- Corbel, M.J.; Thomas, E. The Brucella-Phages: Their Properties, Characterisation and Applications; Ministry of Agriculture, Fisheries and Food: Pinner, UK, 1983; Volume 2266. [Google Scholar]
- Drozevkina, M.S. The Present Position in Brucella Phage Research. A Review of the Literature. Bull. World Health Organ. 1963, 29, 43–57. [Google Scholar] [PubMed]
- Kersters, K.; De Vos, P.; Gillis, M.; Swings, J.; Vandamme, P.; Stackebrandt, E. The Prokaryotes: A Handbook on the Biology of Bacteria; Springer: New York, NY, USA, 2006. [Google Scholar]
- Lipsitz, R.; Garges, S.; Aurigemma, R.; Baccam, P.; Blaney, D.D.; Cheng, A.C.; Currie, B.J.; Dance, D.; Gee, J.E.; Larsen, J.; et al. Workshop on treatment of and postexposure prophylaxis for Burkholderia pseudomallei and B. mallei Infection, 2010. Emerg. Infect. Dis. 2012, 18, e2. [Google Scholar] [CrossRef] [PubMed]
- Abshire, T.G.; Brown, J.E.; Ezzell, J.W. Production and validation of the use of gamma phage for identification of Bacillus anthracis. J. Clin. Microbiol. 2005, 43, 4780–4788. [Google Scholar] [CrossRef]
- Popovic, T.; Hoffmaster, A.; Ezzell, J.W.; Abshire, T.G.; Brown, J.E.; Aoac. Validation of methods for confirmatory identification of presumptive isolates of Bacillus anthracis. J. AOAC Int. 2005, 88, 175–177. [Google Scholar] [CrossRef] [PubMed]
- Schofield, D.A.; Sharp, N.J.; Vandamm, J.; Molineux, I.J.; Spreng, K.A.; Rajanna, C.; Westwater, C.; Stewart, G.C. Bacillus anthracis diagnostic detection and rapid antibiotic susceptibility determination using ‘bioluminescent’ reporter phage. J. Microbiol. Methods 2013, 95, 156–161. [Google Scholar] [CrossRef]
- Sharp, N.J.; Vandamm, J.P.; Molineux, I.J.; Schofield, D.A. Rapid Detection of Bacillus anthracis in Complex Food Matrices Using Phage-Mediated Bioluminescence. J. Food Prot. 2015, 78, 963–968. [Google Scholar] [CrossRef]
- Sharp, N.J.; Molineux, I.J.; Page, M.A.; Schofield, D.A. Rapid Detection of Viable Bacillus anthracis Spores in Environmental Samples by Using Engineered Reporter Phages. Appl. Environ. Microbiol. 2016, 82, 2380–2387. [Google Scholar] [CrossRef]
- Nguyen, C.; Makkar, R.; Sharp, N.J.; Page, M.A.; Molineux, I.J.; Schofield, D.A. Detection of Bacillus anthracis spores from environmental water using bioluminescent reporter phage. J. Appl. Microbiol. 2017, 123, 1184–1193. [Google Scholar] [CrossRef]
- Sainath Rao, S.; Mohan, K.V.; Nguyen, N.; Abraham, B.; Abdouleva, G.; Zhang, P.; Atreya, C.D. Peptides panned from a phage-displayed random peptide library are useful for the detection of Bacillus anthracis surrogates B. cereus 4342 and B. anthracis Sterne. Biochem. Biophys. Res. Commun. 2010, 395, 93–98. [Google Scholar] [CrossRef]
- Ledsgaard, L.; Kilstrup, M.; Karatt-Vellatt, A.; McCafferty, J.; Laustsen, A.H. Basics of Antibody Phage Display Technology. Toxins 2018, 10, 236. [Google Scholar] [CrossRef]
- Shim, H. Antibody Phage Display. Adv. Exp. Med. Biol. 2017, 1053, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Kugler, J.; Zantow, J.; Meyer, T.; Hust, M. Oligopeptide m13 phage display in pathogen research. Viruses 2013, 5, 2531. [Google Scholar] [CrossRef] [PubMed]
- Georgieva, Y.; Konthur, Z. Design and screening of M13 phage display cDNA libraries. Molecules 2011, 16, 1667. [Google Scholar] [CrossRef] [PubMed]
- Pande, J.; Szewczyk, M.M.; Grover, A.K. Phage display: Concept, innovations, applications and future. Biotechnol. Adv. 2010, 28, 849–858. [Google Scholar] [CrossRef]
- Sidhu, S.S. Engineering M13 for phage display. Biomol. Eng. 2001, 18, 57–63. [Google Scholar] [CrossRef]
- Petrenko, V.A.; Smith, G.P.; Gong, X.; Quinn, T. A library of organic landscapes on filamentous phage. Protein Eng. 1996, 9, 797–801. [Google Scholar] [CrossRef]
- Malik, P.; Terry, T.D.; Gowda, L.R.; Langara, A.; Petukhov, S.A.; Symmons, M.F.; Welsh, L.C.; Marvin, D.A.; Perham, R.N. Role of capsid structure and membrane protein processing in determining the size and copy number of peptides displayed on the major coat protein of filamentous bacteriophage. J. Mol. Biol. 1996, 260, 9–21. [Google Scholar] [CrossRef]
- Marvin, D.A.; Hale, R.D.; Nave, C.; Helmer-Citterich, M. Molecular models and structural comparisons of native and mutant class I filamentous bacteriophages Ff (fd, f1, M13), If1 and IKe. J. Mol. Biol. 1994, 235, 260–286. [Google Scholar] [CrossRef]
- Petrenko, V.A.; Smith, G.P. Phages from landscape libraries as substitute antibodies. Protein Eng. 2000, 13, 589–592. [Google Scholar] [CrossRef]
- Brigati, J.; Williams, D.D.; Sorokulova, I.B.; Nanduri, V.; Chen, I.H.; Turnbough, C.L., Jr.; Petrenko, V.A. Diagnostic probes for Bacillus anthracis spores selected from a landscape phage library. Clin. Chem. 2004, 50, 1899–1906. [Google Scholar] [CrossRef]
- Valles, X.; Stenseth, N.C.; Demeure, C.; Horby, P.; Mead, P.S.; Cabanillas, O.; Ratsitorahina, M.; Rajerison, M.; Andrianaivoarimanana, V.; Ramasindrazana, B.; et al. Human plague: An old scourge that needs new answers. PLoS Negl. Trop. Dis. 2020, 14, e0008251. [Google Scholar] [CrossRef] [PubMed]
- Butler, T. Plague and Other Yersinia Infections; Springer Science & Business Media: Berlin, Germany, 2012. [Google Scholar]
- Chu, M. Laboratory Manual of Plague Diagnostic Tests; Centers for Disease Control and Prevention, World Health Organization: Geneva, Switzerland, 2000. [Google Scholar]
- Chanteau, S.; Rahalison, L.; Ratsitorahina, M.; Mahafaly; Rasolomaharo, M.; Boisier, P.; O’Brien, T.; Aldrich, J.; Keleher, A.; Morgan, C.; et al. Early diagnosis of bubonic plague using F1 antigen capture ELISA assay and rapid immunogold dipstick. Int. J. Med. Microbiol. 2000, 290, 279–283. [Google Scholar] [CrossRef]
- Chanteau, S.; Rahalison, L.; Ralafiarisoa, L.; Foulon, J.; Ratsitorahina, M.; Ratsifasoamanana, L.; Carniel, E.; Nato, F. Development and testing of a rapid diagnostic test for bubonic and pneumonic plague. Lancet 2003, 361, 211–216. [Google Scholar] [CrossRef]
- Splettstoesser, W.D.; Rahalison, L.; Grunow, R.; Neubauer, H.; Chanteau, S. Evaluation of a standardized F1 capsular antigen capture ELISA test kit for the rapid diagnosis of plague. FEMS Immunol. Med. Microbiol. 2004, 41, 149–155. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Matero, P.; Pasanen, T.; Laukkanen, R.; Tissari, P.; Tarkka, E.; Vaara, M.; Skurnik, M. Real-time multiplex PCR assay for detection of Yersinia pestis and Yersinia pseudotuberculosis. APMIS 2009, 117, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Rajerison, M.; Melocco, M.; Andrianaivoarimanana, V.; Rahajandraibe, S.; Rakotoarimanana, F.; Spiegel, A.; Ratsitorahina, M.; Baril, L. Performance of plague rapid diagnostic test compared to bacteriology: A retrospective analysis of the data collected in Madagascar. BMC Infect. Dis. 2020, 20, 90. [Google Scholar] [CrossRef]
- Jullien, S.; Dissanayake, H.A.; Chaplin, M. Rapid diagnostic tests for plague. Cochrane Database Syst. Rev. 2020, 6, CD013459. [Google Scholar] [CrossRef]
- Prentice, K.W.; DePalma, L.; Ramage, J.G.; Sarwar, J.; Parameswaran, N.; Petersen, J.; Yockey, B.; Young, J.; Joshi, M.; Thirunavvukarasu, N.; et al. Comprehensive Laboratory Evaluation of a Lateral Flow Assay for the Detection of Yersinia pestis. Health Secur. 2019, 17, 439–453. [Google Scholar] [CrossRef]
- Harrison, D.N.; Cavanaugh, D.C.; Rust, J.H., Jr.; Marshall, J.D., Jr. Characteristics of a bacteriophage-infected strain of Pasteurella pestis isolated from a human case of plague. Infect. Immun. 1971, 4, 85–87. [Google Scholar] [CrossRef]
- Mingle, C.K. Brucellosis, in livestock: Control and eradication. Ann. N.Y. Acad. Sci. 1958, 70, 557–563. [Google Scholar] [CrossRef]
- Goodwin, Z.I.; Pascual, D.W. Brucellosis vaccines for livestock. Vet. Immunol. Immunopathol. 2016, 181, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Poester, F.P.; Samartino, L.E.; Santos, R.L. Pathogenesis and pathobiology of brucellosis in livestock. Rev. Sci. Tech. 2013, 32, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Al-Attas, R.A.; Al-Khalifa, M.; Al-Qurashi, A.R.; Badawy, M.; Al-Gualy, N. Evaluation of PCR, culture and serology for the diagnosis of acute human brucellosis. Ann. Saudi Med. 2000, 20, 224–228. [Google Scholar] [CrossRef] [PubMed]
- Bricker, B.J. PCR as a diagnostic tool for brucellosis. Vet. Microbiol. 2002, 90, 435–446. [Google Scholar] [CrossRef]
- Navarro, E.; Casao, M.A.; Solera, J. Diagnosis of human brucellosis using PCR. Expert Rev. Mol. Diagn. 2004, 4, 115–123. [Google Scholar] [CrossRef]
- Sanjuan-Jimenez, R.; Morata, P.; Bermudez, P.; Bravo, M.J.; Colmenero, J.D. Comparative clinical study of different multiplex real time PCR strategies for the simultaneous differential diagnosis between extrapulmonary tuberculosis and focal complications of brucellosis. PLoS Negl. Trop. Dis. 2013, 7, e2593. [Google Scholar] [CrossRef]
- Faddoul, L.; Azar, R.; Haddad, A.; Hanna, A. The importance of blood culture and serology for Brucellosis diagnosis and treatment. J. Infect. Dev. Ctries 2018, 12, 19S. [Google Scholar] [CrossRef][Green Version]
- Damon, S.R.; Donham, C.R.; Hutchings, L.M.; Simms, B.T.; Steele, J.H. Serology of brucellosis in rural Indiana. Public Health Rep. 1953, 68, 563–571. [Google Scholar] [CrossRef]
- Nielsen, K. Diagnosis of brucellosis by serology. Vet. Microbiol. 2002, 90, 447–459. [Google Scholar] [CrossRef]
- Nimri, L.F. Diagnosis of recent and relapsed cases of human brucellosis by PCR assay. BMC Infect. Dis. 2003, 3, 5. [Google Scholar] [CrossRef]
- Navarro, E.; Fernandez, J.A.; Escribano, J.; Solera, J. PCR assay for diagnosis of human brucellosis. J. Clin. Microbiol. 1999, 37, 1654–1655. [Google Scholar] [PubMed]
- Lopez-Goni, I.; Garcia-Yoldi, D.; Marin, C.M.; de Miguel, M.J.; Munoz, P.M.; Blasco, J.M.; Jacques, I.; Grayon, M.; Cloeckaert, A.; Ferreira, A.C.; et al. Evaluation of a multiplex PCR assay (Bruce-ladder) for molecular typing of all Brucella species, including the vaccine strains. J. Clin. Microbiol. 2008, 46, 3484–3487. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Z.; Zhang, Y.; Bai, L.; Zhao, Y.; Liu, C.; Ma, A.; Yu, H. Polymerase chain reaction-based assays for the diagnosis of human brucellosis. Ann. Clin. Microbiol. Antimicrob. 2014, 13, 31. [Google Scholar] [CrossRef] [PubMed]
- Sergueev, K.V.; Filippov, A.A.; Nikolich, M.P. Highly Sensitive Bacteriophage-Based Detection of Brucella abortus in Mixed Culture and Spiked Blood. Viruses 2017, 9, 144. [Google Scholar] [CrossRef]
- Rippa, M.; Castagna, R.; Pannico, M.; Musto, P.; Borriello, G.; Paradiso, R.; Galiero, G.; Bolletti Censi, S.; Zhou, J.; Zyss, J.; et al. Octupolar Metastructures for a Highly Sensitive, Rapid, and Reproducible Phage-Based Detection of Bacterial Pathogens by Surface-Enhanced Raman Scattering. ACS Sens. 2017, 2, 947–954. [Google Scholar] [CrossRef]
- Rippa, M.; Castagna, R.; Zhou, J.; Paradiso, R.; Borriello, G.; Bobeico, E.; Petti, L. Dodecagonal plasmonic quasicrystals for phage-based biosensing. Nanotechnology 2018, 29, 405501. [Google Scholar] [CrossRef]
- Wiersinga, W.J.; Birnie, E.; Weehuizen, T.A.; Alabi, A.S.; Huson, M.A.; Huis in ‘t Veld, R.A.; Mabala, H.K.; Adzoda, G.K.; Raczynski-Henk, Y.; Esen, M.; et al. Clinical, environmental, and serologic surveillance studies of melioidosis in Gabon, 2012–2013. Emerg. Infect. Dis. 2015, 21, 40–47. [Google Scholar] [CrossRef]
- Wiersinga, W.J.; Currie, B.J.; Peacock, S.J. Melioidosis. N. Engl. J. Med. 2012, 367, 1035–1044. [Google Scholar] [CrossRef]
- Wiersinga, W.J.; van der Poll, T.; White, N.J.; Day, N.P.; Peacock, S.J. Melioidosis: Insights into the pathogenicity of Burkholderia pseudomallei. Nat. Rev. Microbiol. 2006, 4, 272–282. [Google Scholar] [CrossRef]
- Rhodes, K.A.; Schweizer, H.P. Antibiotic resistance in Burkholderia species. Drug Resist. Updates 2016, 28, 82–90. [Google Scholar] [CrossRef]
- Cox, C.R.; Saichek, N.R.; Schweizer, H.P.; Voorhees, K.J. Rapid Burkholderia pseudomallei identification and antibiotic resistance determination by bacteriophage amplification and MALDI-TOF MS. Bacteriophage 2014, 4, e29011. [Google Scholar] [CrossRef] [PubMed]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007, 35, W52–W57. [Google Scholar] [CrossRef] [PubMed]
- Abby, S.S.; Neron, B.; Menager, H.; Touchon, M.; Rocha, E.P. MacSyFinder: A program to mine genomes for molecular systems with an application to CRISPR-Cas systems. PLoS ONE 2014, 9, e110726. [Google Scholar] [CrossRef] [PubMed]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Neron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef] [PubMed]
- Schunder, E.; Rydzewski, K.; Grunow, R.; Heuner, K. First indication for a functional CRISPR/Cas system in Francisella tularensis. Int. J. Med. Microbiol. 2013, 303, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Shin, H.; Ryu, S. Characterization and comparative genomic analysis of bacteriophages infecting members of the Bacillus cereus group. Arch. Virol. 2014, 159, 871–884. [Google Scholar] [CrossRef]
- Jonczyk-Matysiak, E.; Klak, M.; Weber-Dabrowska, B.; Borysowski, J.; Gorski, A. Possible use of bacteriophages active against Bacillus anthracis and other B. cereus group members in the face of a bioterrorism threat. Biomed. Res. Int. 2014, 2014, 735413. [Google Scholar] [CrossRef]
- Zhao, X.; Skurnik, M. Bacteriophages of Yersinia pestis. Adv. Exp. Med. Biol. 2016, 918, 361–375. [Google Scholar] [CrossRef]
- Ando, H.; Lemire, S.; Pires, D.P.; Lu, T.K. Engineering Modular Viral Scaffolds for Targeted Bacterial Population Editing. Cell Syst. 2015, 1, 187–196. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, L.; Abdelgader, S.A.; Yu, L.; Xu, J.; Yao, H.; Lu, C.; Zhang, W. Alterations in gp37 Expand the Host Range of a T4-Like Phage. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, L.; Xin, S.; Yao, H.; Lu, C.; Zhang, W. Inducible Prophage Mutant of Escherichia coli Can Lyse New Host and the Key Sites of Receptor Recognition Identification. Front. Microbiol. 2017, 8, 147. [Google Scholar] [CrossRef] [PubMed]
- Gebhart, D.; Williams, S.R.; Scholl, D. Bacteriophage SP6 encodes a second tailspike protein that recognizes Salmonella enterica serogroups C2 and C3. Virology 2017, 507, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Heilpern, A.J.; Waldor, M.K. pIIICTX, a predicted CTXphi minor coat protein, can expand the host range of coliphage fd to include Vibrio cholerae. J. Bacteriol. 2003, 185, 1037–1044. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.Y.; Lo, Y.H.; Tseng, P.W.; Chang, S.F.; Lin, Y.T.; Chen, T.S. A T3 and T7 recombinant phage acquires efficient adsorption and a broader host range. PLoS ONE 2012, 7, e30954. [Google Scholar] [CrossRef]
- Nguyen, A.H.; Molineux, I.J.; Springman, R.; Bull, J.J. Multiple genetic pathways to similar fitness limits during viral adaptation to a new host. Evolution 2012, 66, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Pouillot, F.; Blois, H.; Iris, F. Genetically engineered virulent phage banks in the detection and control of emergent pathogenic bacteria. Biosecur. Bioterrorism 2010, 8, 155–169. [Google Scholar] [CrossRef]
- Yoichi, M.; Abe, M.; Miyanaga, K.; Unno, H.; Tanji, Y. Alteration of tail fiber protein gp38 enables T2 phage to infect Escherichia coli O157:H7. J. Biotechnol. 2005, 115, 101–107. [Google Scholar] [CrossRef]
- Yosef, I.; Goren, M.G.; Globus, R.; Molshanski-Mor, S.; Qimron, U. Extending the Host Range of Bacteriophage Particles for DNA Transduction. Mol. Cell 2017, 66, 721–728.e723. [Google Scholar] [CrossRef]
- Yehl, K.; Lemire, S.; Yang, A.C.; Ando, H.; Mimee, M.; Torres, M.T.; de la Fuente-Nunez, C.; Lu, T.K. Engineering Phage Host-Range and Suppressing Bacterial Resistance through Phage Tail Fiber Mutagenesis. Cell 2019, 179, 459–469.e9. [Google Scholar] [CrossRef]
- Holtzman, T.; Globus, R.; Molshanski-Mor, S.; Ben-Shem, A.; Yosef, I.; Qimron, U. A continuous evolution system for contracting the host range of bacteriophage T7. Sci. Rep. 2020, 10, 307. [Google Scholar] [CrossRef]
- Dunne, M.; Rupf, B.; Tala, M.; Qabrati, X.; Ernst, P.; Shen, Y.; Sumrall, E.; Heeb, L.; Pluckthun, A.; Loessner, M.J.; et al. Reprogramming Bacteriophage Host Range through Structure-Guided Design of Chimeric Receptor Binding Proteins. Cell Rep. 2019, 29, 1336–1350.e4. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Deora, R.; Doulatov, S.R.; Gingery, M.; Eiserling, F.A.; Preston, A.; Maskell, D.J.; Simons, R.W.; Cotter, P.A.; Parkhill, J.; et al. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science 2002, 295, 2091–2094. [Google Scholar] [CrossRef] [PubMed]
- Doulatov, S.; Hodes, A.; Dai, L.; Mandhana, N.; Liu, M.; Deora, R.; Simons, R.W.; Zimmerly, S.; Miller, J.F. Tropism switching in Bordetella bacteriophage defines a family of diversity-generating retroelements. Nature 2004, 431, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Hodes, A.; Hui, W.H.; Gingery, M.; Miller, J.F.; Zhou, Z.H. Three-dimensional structure of tropism-switching Bordetella bacteriophage. Proc. Natl. Acad. Sci. USA 2010, 107, 4347–4352. [Google Scholar] [CrossRef] [PubMed]
- Medhekar, B.; Miller, J.F. Diversity-generating retroelements. Curr. Opin. Microbiol. 2007, 10, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Handa, S.; Jiang, Y.; Tao, S.; Foreman, R.; Schinazi, R.F.; Miller, J.F.; Ghosh, P. Template-assisted synthesis of adenine-mutagenized cDNA by a retroelement protein complex. Nucleic Acids Res. 2018, 46, 9711–9725. [Google Scholar] [CrossRef]
- Schillinger, T.; Zingler, N. The low incidence of diversity-generating retroelements in sequenced genomes. Mob. Genet. Elem. 2012, 2, 287–291. [Google Scholar] [CrossRef]
- Schillinger, T.; Lisfi, M.; Chi, J.; Cullum, J.; Zingler, N. Analysis of a comprehensive dataset of diversity generating retroelements generated by the program DiGReF. BMC Genom. 2012, 13, 430. [Google Scholar] [CrossRef]
- Ye, Y. Identification of diversity-generating retroelements in human microbiomes. Int. J. Mol. Sci. 2014, 15, 4234. [Google Scholar] [CrossRef]
- Paul, B.G.; Bagby, S.C.; Czornyj, E.; Arambula, D.; Handa, S.; Sczyrba, A.; Ghosh, P.; Miller, J.F.; Valentine, D.L. Targeted diversity generation by intraterrestrial archaea and archaeal viruses. Nat. Commun. 2015, 6, 6585. [Google Scholar] [CrossRef]
- Guo, H.; Tse, L.V.; Nieh, A.W.; Czornyj, E.; Williams, S.; Oukil, S.; Liu, V.B.; Miller, J.F. Target site recognition by a diversity-generating retroelement. PLoS Genet. 2011, 7, e1002414. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).