Recent Progress in the Detection of Bacteria Using Bacteriophages: A Review
Abstract
1. Introduction
2. Whole Phage-Based Bacteria Sensing
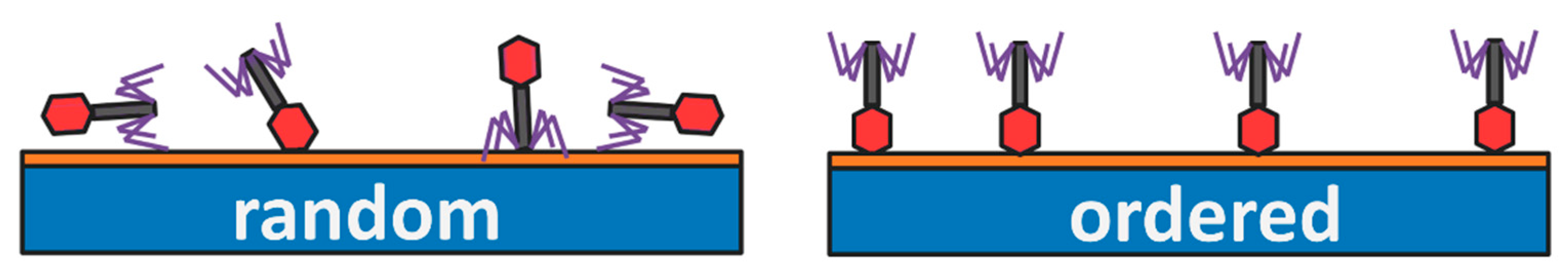
2.1. Bacteriophages Deposited on Solid Substrates
Oriented Layers of Bacteriophages
2.2. Bacteriophage Based Bioconjugates
2.3. Genetically Modified Phages
2.4. Phage Amplification
2.5. Detection of Bacterial Metabolites
3. Parts of Phages Used for Detection
4. Concluding Remarks
5. Future of Phage-Based Biosensing
Author Contributions
Funding
Conflicts of Interest
References
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2015. [Google Scholar] [CrossRef]
- Wang, R.; Van Dorp, L.; Shaw, L.P.; Bradley, P.; Wang, Q.; Wang, X.; Jin, L.; Zhang, Q.; Liu, Y.; Rieux, A.; et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 2018, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lambert, G.; Liao, D.; Kim, H.; Robin, K.; Tung, C.K.; Pourmand, N.; Austin, R.H. Acceleration of emergence of bacterial antibiotic resistance in connected microenvironments. Science 2011, 333, 1764–1767. [Google Scholar] [CrossRef]
- Matuła, K.; Richter, Ł.; Janczuk-Richter, M.; Nogala, W.; Grzeszkowiak, M.; Peplińska, B.; Jurga, S.; Wyroba, E.; Suski, S.; Bilski, H.; et al. Phenotypic plasticity of Escherichia coli upon exposure to physical stress induced by ZnO nanorods. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Ramdas, K.; Darzi, A.; Jain, S. “Test, re-test, re-test”: Using inaccurate tests to greatly increase the accuracy of COVID-19 testing. Nat. Med. 2020. [Google Scholar] [CrossRef]
- Yang, Y.; Rajendran, V.; Jayaraman, V.; Wang, T.; Bei, K.; Krishna, K.; Rajasekaran, K.; Rajasekaran, J.J.; Krishnamurthy, H. Evaluation of the Vibrant DNA microarray for the high-throughput multiplex detection of enteric pathogens in clinical samples. Gut Pathog. 2019, 11, 1–12. [Google Scholar] [CrossRef]
- KrishnanNair Geetha, D.; Sivaraman, B.; Rammohan, R.; Venkatapathy, N.; Solai Ramatchandirane, P. A SYBR Green based multiplex Real-Time PCR assay for rapid detection and differentiation of ocular bacterial pathogens. J. Microbiol. Methods 2020, 171, 105875. [Google Scholar] [CrossRef]
- Belgrader, P.; Benett, W.; Hadley, D.; Richards, J.; Stratton, P.; Mariella, R.; Milanovich, F.; Niemeyer, D.M.; Tang, Y.W.; Procop, G.W.; et al. PCR detection of bacteria in seven minutes. Science 1999, 284, 449–450. [Google Scholar] [CrossRef]
- Jackson, N.; Wu, T.Z.; Adams-Sapper, S.; Satoorian, T.; Geisberg, M.; Murthy, N.; Lee, L.; Riley, L.W. A multiplexed, indirect enzyme-linked immunoassay for the detection and differentiation of E. coli from other Enterobacteriaceae and P. aeruginosa from other glucose non-fermenters. J. Microbiol. Methods 2019, 158, 52–58. [Google Scholar] [CrossRef]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 1–16. [Google Scholar] [CrossRef]
- Chen, J.; Andler, S.M.; Goddard, J.M.; Nugen, S.R.; Rotello, V.M. Integrating recognition elements with nanomaterials for bacteria sensing. Chem. Soc. Rev. 2017, 46, 1272–1283. [Google Scholar] [CrossRef]
- Farooq, U.; Yang, Q.; Ullah, M.W.; Wang, S. Bacterial biosensing: Recent advances in phage-based bioassays and biosensors. Biosens. Bioelectron. 2018, 118, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Richter, Ł.; Janczuk-Richter, M.; Niedziółka-Jönsson, J.; Paczesny, J.; Hołyst, R. Recent advances in bacteriophage-based methods for bacteria detection. Drug Discov. Today 2018, 23, 448–455. [Google Scholar] [CrossRef]
- Sharma, S.; Chatterjee, S.; Datta, S.; Prasad, R.R.K.R.; Dubey, D.; Prasad, R.R.K.R.; Vairale, M.G. Bacteriophages and its applications: An overview. Folia Microbiol. 2017, 62, 17–55. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, H.-W.W. 5500 Phages examined in the electron microscope. Arch. Virol. 2007, 152, 227–243. [Google Scholar] [CrossRef] [PubMed]
- Brigati, J.R.; Petrenko, V.A. Thermostability of landscape phage probes. Anal. Bioanal. Chem. 2005, 382, 1346–1350. [Google Scholar] [CrossRef] [PubMed]
- Nobrega, F.L.; Costa, A.R.; Santos, J.F.; Siliakus, M.F.; Van Lent, J.W.M.; Kengen, S.W.M.; Azeredo, J.; Kluskens, L.D. Genetically manipulated phages with improved pH resistance for oral administration in veterinary medicine. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Olofsson, L.; Ankarloo, J.; Nicholls, I.A. Phage viability in organic media: Insights into phage stability. J. Mol. Recognit. 1998, 11, 91–93. [Google Scholar] [CrossRef]
- Olofsson, L.; Ankarloo, J.; Andersson, P.O.; Nicholls, I.A. Filamentous bacteriophage stability in non-aqueous media. Chem. Biol. 2001, 8, 661–671. [Google Scholar] [CrossRef]
- Stone, E.; Campbell, K.; Grant, I.; McAuliffe, O. Understanding and exploiting phage–host interactions. Viruses 2019, 11, 567. [Google Scholar] [CrossRef]
- Nobrega, F.L.; Vlot, M.; de Jonge, P.A.; Dreesens, L.L.; Beaumont, H.J.E.; Lavigne, R.; Dutilh, B.E.; Brouns, S.J.J. Targeting mechanisms of tailed bacteriophages. Nat. Rev. Microbiol. 2018, 16, 760–773. [Google Scholar] [CrossRef] [PubMed]
- Krueger, A.P. The sorption of bacteriophage by living and dead susceptible bacteria. J. Gen. Physiol. 1931, 14, 493–516. [Google Scholar] [CrossRef] [PubMed]
- De Jonge, P.A.; Nobrega, F.L.; Brouns, S.J.J.; Dutilh, B.E. Molecular and Evolutionary Determinants of Bacteriophage Host Range. Trends Microbiol. 2019, 27, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Faure, G.; Shmakov, S.A.; Yan, W.X.; Cheng, D.R.; Scott, D.A.; Peters, J.E.; Makarova, K.S.; Koonin, E.V. CRISPR–Cas in mobile genetic elements: Counter-defence and beyond. Nat. Rev. Microbiol. 2019, 17, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Trasanidou, D.; Gerós, A.S.; Mohanraju, P.; Nieuwenweg, A.C.; Nobrega, F.L.; Staals, R.H.J. Keeping crispr in check: Diverse mechanisms of phage-encoded anti-crisprs. FEMS Microbiol. Lett. 2019, 366, 1–14. [Google Scholar] [CrossRef]
- Farooq, U.; Ullah, M.W.; Yang, Q.; Aziz, A.; Xu, J.; Zhou, L.; Wang, S. High-density phage particles immobilization in surface-modified bacterial cellulose for ultra-sensitive and selective electrochemical detection of Staphylococcus aureus. Biosens. Bioelectron. 2020, 157, 112163. [Google Scholar] [CrossRef]
- Yan, C.; Zhang, Y.; Yang, H.; Yu, J.; Wei, H. Combining phagomagnetic separation with immunoassay for specific, fast and sensitive detection of Staphylococcus aureus. Talanta 2017, 170, 291–297. [Google Scholar] [CrossRef]
- Bhardwaj, N.; Bhardwaj, S.K.; Mehta, J.; Nayak, M.K.; Deep, A. Bacteriophage conjugated IRMOF-3 as a novel opto-sensor for: S. arlettae. New J. Chem. 2016, 40, 8068–8073. [Google Scholar] [CrossRef]
- Bhardwaj, N.; Bhardwaj, S.K.; Mehta, J.; Kim, K.H.; Deep, A. MOF-bacteriophage biosensor for highly sensitive and specific detection of staphylococcus aureus. ACS Appl. Mater. Interfaces 2017, 9, 33589–33598. [Google Scholar] [CrossRef]
- Neufeld, T.; Schwartz-Mittelmann, A.; Biran, D.; Ron, E.Z.; Rishpon, J. Combined phage typing and amperometric detection of released enzymatic activity for the specific identification and quantification of bacteria. Anal. Chem. 2003, 75, 580–585. [Google Scholar] [CrossRef]
- Neufeld, T.; Mittelman, A.S.; Buchner, V.; Rishpon, J. Electrochemical phagemid assay for the specific detection of bacteria using Escherichia coli TG-1 and the M13KO7 phagemid in a model system. Anal. Chem. 2005, 77, 652–657. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Marar, A.; Kner, P.; Ramasamy, R.P. Charge-Directed Immobilization of Bacteriophage on Nanostructured Electrode for Whole-Cell Electrochemical Biosensors. Anal. Chem. 2017, 89, 5734–5741. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Chen, J.; Nugen, S.R. Electrochemical Detection of Escherichia coli from Aqueous Samples Using Engineered Phages. Anal. Chem. 2017, 89, 1650–1657. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Xie, G.; Qiu, J.; Zhou, D.; Gou, D.; Tao, Y.; Li, Y.Y.; Chen, H. A new biosensor based on the recognition of phages and the signal amplification of organic-inorganic hybrid nanoflowers for discriminating and quantitating live pathogenic bacteria in urine. Sens. Actuators B Chem. 2018, 258, 803–812. [Google Scholar] [CrossRef]
- Sedki, M.; Chen, X.; Chen, C.; Ge, X.; Mulchandani, A. Non-lytic M13 phage-based highly sensitive impedimetric cytosensor for detection of coliforms. Biosens. Bioelectron. 2020, 148, 111794. [Google Scholar] [CrossRef]
- Xu, J.; Zhao, C.; Chau, Y.; Lee, Y.K. The synergy of chemical immobilization and electrical orientation of T4 bacteriophage on a micro electrochemical sensor for low-level viable bacteria detection via Differential Pulse Voltammetry. Biosens. Bioelectron. 2020, 151, 111914. [Google Scholar] [CrossRef]
- Ferapontova, E.E. Electrochemical assays for microbial analysis: How far they are from solving microbiota and microbiome challenges. Curr. Opin. Electrochem. 2020, 19, 153–161. [Google Scholar] [CrossRef]
- Janczuk-Richter, M.; Marinović, I.; Niedziółka-Jönsson, J.; Szot-Karpińska, K. Recent applications of bacteriophage-based electrodes: A mini-review. Electrochem. Commun. 2019, 99, 11–15. [Google Scholar] [CrossRef]
- Xu, J.; Chau, Y.; Lee, Y. kuen Phage-based Electrochemical Sensors: A Review. Micromachines 2019, 10, 855. [Google Scholar] [CrossRef]
- Moon, J.S.; Choi, E.J.; Jeong, N.N.; Sohn, J.R.; Han, D.W.; Oh, J.W. Research progress of M13 bacteriophage-based biosensors. Nanomaterials 2019, 9, 1448. [Google Scholar] [CrossRef]
- Yue, H.; He, Y.; Fan, E.; Wang, L.; Lu, S.; Fu, Z. Label-free electrochemiluminescent biosensor for rapid and sensitive detection of pseudomonas aeruginosa using phage as highly specific recognition agent. Biosens. Bioelectron. 2017, 94, 429–432. [Google Scholar] [CrossRef]
- Hiremath, N.; Guntupalli, R.; Vodyanoy, V.; Chin, B.A.; Park, M.K. Detection of methicillin-resistant Staphylococcus aureus using novel lytic phage-based magnetoelastic biosensors. Sens. Actuators B Chem. 2015, 210, 129–136. [Google Scholar] [CrossRef]
- Hiremath, N.; Chin, B.A.; Park, M.K. Effect of Competing Foodborne Pathogens on the Selectivity and Binding Kinetics of a Lytic Phage for Methicillin-Resistant Staphylococcus aureus Detection. J. Electrochem. Soc. 2017, 164, B142–B146. [Google Scholar] [CrossRef]
- Chen, I.-H.; Horikawa, S.; Bryant, K.; Riggs, R.; Chin, B.A.; Barbaree, J.M. Bacterial assessment of phage magnetoelastic sensors for Salmonella enterica Typhimurium detection in chicken meat. Food Control 2017, 71, 273–278. [Google Scholar] [CrossRef]
- Mack, J.D.; Yehualaeshet, T.; Park, M.K.; Tameru, B.; Samuel, T.; Chin, B.A. Phage-Based Biosensor and Optimization of Surface Blocking Agents to Detect Salmonella Typhimurium on Romaine Lettuce. J. Food Saf. 2017, 37. [Google Scholar] [CrossRef]
- Moram, S.S.B.; Shaik, A.K.; Byram, C.; Hamad, S.; Soma, V.R. Instantaneous trace detection of nitro-explosives and mixtures with nanotextured silicon decorated with Ag–Au alloy nanoparticles using the SERS technique. Anal. Chim. Acta 2020, 1101, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Xiao, T.-H.; Wu, Y.; Li, W.; Zeng, Q.-G.; Long, L.; Li, Z.-Y. Roadmap for single-molecule surface-enhanced Raman spectroscopy. Adv. Photonics 2020, 2, 1. [Google Scholar] [CrossRef]
- Fan, M.; Andrade, G.F.S.; Brolo, A.G. A review on recent advances in the applications of surface-enhanced Raman scattering in analytical chemistry. Anal. Chim. Acta 2020, 1097, 1–29. [Google Scholar] [CrossRef]
- Srivastava, S.K.; Hamo, H.B.; Kushmaro, A.; Marks, R.S.; Grüner, C.; Rauschenbach, B.; Abdulhalim, I. Highly sensitive and specific detection of E. coli by a SERS nanobiosensor chip utilizing metallic nanosculptured thin films. Analyst 2015, 140, 3201–3209. [Google Scholar] [CrossRef]
- Rippa, M.; Castagna, R.; Zhou, J.; Paradiso, R.; Borriello, G.; Bobeico, E.; Petti, L. Dodecagonal plasmonic quasicrystals for phage-based biosensing. Nanotechnology 2018, 29. [Google Scholar] [CrossRef]
- Rippa, M.; Castagna, R.; Pannico, M.; Musto, P.; Borriello, G.; Paradiso, R.; Galiero, G.; Censi, S.B.; Zhou, J.; Zyss, J.; et al. Octupolar metastructures for a highly sensitive, rapid, and reproducible phage-based detection of bacterial pathogens by surface-enhanced Raman scattering. ACS Sens. 2017, 2, 947–954. [Google Scholar] [CrossRef] [PubMed]
- Lai, A.; Almaviva, S.; Spizzichino, V.; Luciani, D.; Palucci, A.; Mengali, S.; Marquette, C.; Berthuy, O.; Jankiewicz, B.; Pierno, L. Bacillus spp. Cells Captured Selectively by Phages and Identified by Surface Enhanced Raman Spectroscopy Technique. Proceedings 2017, 1, 519. [Google Scholar] [CrossRef]
- Lakshmanan, R.S.; Guntupalli, R.; Hu, J.; Kim, D.J.; Petrenko, V.A.; Barbaree, J.M.; Chin, B.A. Phage immobilized magnetoelastic sensor for the detection of Salmonella typhimurium. J. Microbiol. Methods 2007, 71, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Handa, H.; Gurczynski, S.; Jackson, M.P.; Auner, G.; Walker, J.; Mao, G. Recognition of Salmonella typhimurium by immobilized phage P22 monolayers. Surf. Sci. 2008, 602, 1392–1400. [Google Scholar] [CrossRef] [PubMed]
- Tolba, M.; Minikh, O.; Brovko, L.Y.; Evoy, S.; Griffiths, M.W. Oriented immobilization of bacteriophages for biosensor applications. Appl. Environ. Microbiol. 2010, 76, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Anany, H.; Chen, W.; Pelton, R.; Griffiths, M.W. Biocontrol of Listeria monocytogenes and Escherichia coli O157:H7 in meat by using phages immobilized on modified cellulose membranes. Appl. Environ. Microbiol. 2011, 77, 6379–6387. [Google Scholar] [CrossRef]
- Richter, Ł.; Matuła, K.; Leśniewski, A.; Kwaśnicka, K.; Łoś, J.; Łoś, M.; Paczesny, J.; Hołyst, R. Ordering of bacteriophages in the electric field: Application for bacteria detection. Sens. Actuators B Chem. 2016, 224, 233–240. [Google Scholar] [CrossRef]
- Liana, A.E.; Chia, E.W.; Marquis, C.P.; Gunawan, C.; Gooding, J.J.; Amal, R. Adsorption of T4 bacteriophages on planar indium tin oxide surface via controlled surface tailoring. J. Colloid Interface Sci. 2016, 468, 192–199. [Google Scholar] [CrossRef]
- Bone, S.; Alum, A.; Markovski, J.; Hristovski, K.; Bar-Zeev, E.; Kaufman, Y.; Abbaszadegan, M.; Perreault, F. Physisorption and chemisorption of T4 bacteriophages on amino functionalized silica particles. J. Colloid Interface Sci. 2018, 532, 68–76. [Google Scholar] [CrossRef]
- Richter, Ł.; Bielec, K.; Leśniewski, A.; Łoś, M.; Paczesny, J.; Hołyst, R. Dense Layer of Bacteriophages Ordered in Alternating Electric Field and Immobilized by Surface Chemical Modification as Sensing Element for Bacteria Detection. ACS Appl. Mater. Interfaces 2017, 9, 19622–19629. [Google Scholar] [CrossRef]
- Wang, L.; Gong, C.; Yuan, X.; Wei, G. Controlling the self-assembly of biomolecules into functional nanomaterials through internal interactions and external stimulations: A review. Nanomaterials 2019, 9, 285. [Google Scholar] [CrossRef] [PubMed]
- Tronolone, J.J.; Orrill, M.; Song, W.; Kim, H.S.; Lee, B.Y.; Leblanc, S. Electric field assisted self-assembly of viruses into colored thin films. Nanomaterials 2019, 9, 1310. [Google Scholar] [CrossRef] [PubMed]
- Richter, Ł.; Albrycht, P.; Księżopolska-Gocalska, M.; Poboży, E.; Bachliński, R.; Sashuk, V.; Paczesny, J.; Hołyst, R. Fast and efficient deposition of broad range of analytes on substrates for surface enhanced Raman spectroscopy. Biosens. Bioelectron. 2020, 156. [Google Scholar] [CrossRef]
- Franco, D.; De Plano, L.M.; Rizzo, M.G.; Scibilia, S.; Lentini, G.; Fazio, E.; Neri, F.; Guglielmino, S.P.P.; Mezzasalma, A.M. Bio-hybrid gold nanoparticles as SERS probe for rapid bacteria cell identification. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 224, 117394. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Chen, I.A. Rapid Colorimetric Detection of Bacterial Species through the Capture of Gold Nanoparticles by Chimeric Phages. ACS Nano 2019, 13, 1244–1252. [Google Scholar] [CrossRef] [PubMed]
- Imai, M.; Mine, K.; Tomonari, H.; Uchiyama, J.; Matuzaki, S.; Niko, Y.; Hadano, S.; Watanabe, S. Dark-Field Microscopic Detection of Bacteria using Bacteriophage-Immobilized SiO2@AuNP Core-Shell Nanoparticles. Anal. Chem. 2019, 91, 12352–12357. [Google Scholar] [CrossRef] [PubMed]
- Janczuk, M.; Richter, Ł.; Hoser, G.; Kawiak, J.; Łoś, M.; Niedziółka-Jönsson, J.; Paczesny, J.; Hołyst, R. Bacteriophage-Based Bioconjugates as a Flow Cytometry Probe for Fast Bacteria Detection. Bioconjug. Chem. 2017, 28, 419–425. [Google Scholar] [CrossRef]
- Zhou, Y.; Ramasamy, R.P. Isolation and separation of Listeria monocytogenes using bacteriophage P100-modified magnetic particles. Colloids Surfaces B Biointerfaces 2019, 175, 421–427. [Google Scholar] [CrossRef]
- Liana, A.E.; Marquis, C.P.; Gunawan, C.; Gooding, J.J.; Amal, R. T4 bacteriophage conjugated magnetic particles for E. coli capturing: Influence of bacteriophage loading, temperature and tryptone. Colloids Surfaces B Biointerfaces 2017, 151, 47–57. [Google Scholar] [CrossRef]
- Pires, D.P.; Cleto, S.; Sillankorva, S.; Azeredo, J.; Lu, T.K. Genetically Engineered Phages: A Review of Advances over the Last Decade. Microbiol. Mol. Biol. Rev. 2016, 80, 523–543. [Google Scholar] [CrossRef]
- Bárdy, P.; Pantůček, R.; Benešík, M.; Doškař, J. Genetically modified bacteriophages in applied microbiology. J. Appl. Microbiol. 2016, 121, 618–633. [Google Scholar] [CrossRef]
- Pizarro-Bauerle, J.; Ando, H. Engineered Bacteriophages for Practical Applications. Biol. Pharm. Bull. 2020, 43, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Wisuthiphaet, N.; Yang, X.; Young, G.M.; Nitin, N. Rapid detection of Escherichia coli in beverages using genetically engineered bacteriophage T7. AMB Express 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Kretzer, J.W.; Schmelcher, M.; Loessner, M.J. Ultrasensitive and Fast Diagnostics of Viable Listeria Cells by CBD Magnetic Separation Combined with A511::luxAB Detection. Viruses 2018, 10, 626. [Google Scholar] [CrossRef] [PubMed]
- Pulkkinen, E.M.; Hinkley, T.C.; Nugen, S.R. Utilizing in vitro DNA assembly to engineer a synthetic T7 Nanoluc reporter phage for Escherichia coli detection. Integr. Biol. 2019, 11, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Hinkley, T.C.; Singh, S.; Garing, S.; Le Ny, A.L.M.; Nichols, K.P.; Peters, J.E.; Talbert, J.N.; Nugen, S.R. A phage-based assay for the rapid, quantitative, and single CFU visualization of E. coli (ECOR #13) in drinking water. Sci. Rep. 2018, 8, 1–8. [Google Scholar] [CrossRef]
- Hinkley, T.C.; Garing, S.; Singh, S.; Le Ny, A.L.M.; Nichols, K.P.; Peters, J.E.; Talbert, J.N.; Nugen, S.R. Reporter bacteriophage T7NLC utilizes a novel NanoLuc::CBM fusion for the ultrasensitive detection of: Escherichia coli in water. Analyst 2018, 143, 4074–4082. [Google Scholar] [CrossRef]
- Hinkley, T.C.; Garing, S.; Jain, P.; Williford, J.; Le Ny, A.-L.M.; Nichols, K.P.; Peters, J.E.; Talbert, J.N.; Nugen, S.R. A Syringe-Based Biosensor to Rapidly Detect Low Levels of Escherichia Coli (ECOR13) in Drinking Water Using Engineered Bacteriophages. Sensors 2020, 20, 1953. [Google Scholar] [CrossRef]
- Vinay, M.; Franche, N.; Grégori, G.; Fantino, J.R.; Pouillot, F.; Ansaldi, M. Phage-based fluorescent biosensor prototypes to specifically detect enteric bacteria such as E. coli and Salmonella enterica Typhimurium. PLoS ONE 2015, 10, 1–17. [Google Scholar] [CrossRef]
- Franche, N.; Vinay, M.; Ansaldi, M. Substrate-independent luminescent phage-based biosensor to specifically detect enteric bacteria such as E. coli. Environ. Sci. Pollut. Res. 2017, 24, 42–51. [Google Scholar] [CrossRef]
- Kim, J.; Kim, M.; Kim, S.; Ryu, S. Sensitive detection of viable Escherichia coli O157:H7 from foods using a luciferase-reporter phage phiV10lux. Int. J. Food Microbiol. 2017, 254, 11–17. [Google Scholar] [CrossRef]
- Rondón, L.; Urdániz, E.; Latini, C.; Payaslian, F.; Matteo, M.; Sosa, E.J.; Do Porto, D.F.; Turjanski, A.G.; Nemirovsky, S.; Hatfull, G.F.; et al. Fluoromycobacteriophages can detect viable Mycobacterium tuberculosis and determine phenotypic rifampicin resistance in 3-5 days from sputum collection. Front. Microbiol. 2018, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Jiang, M.; Xiong, J.; Li, J.; Zhang, X.; Wei, H.; Yu, J. Exploring a phage-based real-time PCR assay for diagnosing Acinetobacter baumannii bloodstream infections with high sensitivity. Anal. Chim. Acta 2018, 1044, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Jiang, M.; Hiong, J.; Li, J.; Wei, H.; Yu, J. Rapid Ultrasensitive Diagnosis of Pneumonia Caused by Acinetobacter Baumannii Using a Combination of Enrichment and Phage-Based qPCR Assay; Research Square: Durham, NC, USA, 2020; pp. 1–25. [Google Scholar] [CrossRef]
- Garrido-Maestu, A.; Fuciños, P.; Azinheiro, S.; Carvalho, C.; Carvalho, J.; Prado, M. Specific detection of viable Salmonella Enteritidis by phage amplification combined with qPCR (PAA-qPCR) in spiked chicken meat samples. Food Control. 2019, 99, 79–83. [Google Scholar] [CrossRef]
- Sergueev, K.V.; Filippov, A.A.; Nikolich, M.P. Highly sensitive bacteriophage-based detection of Brucella abortus in mixed culture and spiked blood. Viruses 2017, 9, 144. [Google Scholar] [CrossRef] [PubMed]
- Anany, H.; Brovko, L.; El Dougdoug, N.K.; Sohar, J.; Fenn, H.; Alasiri, N.; Jabrane, T.; Mangin, P.; Monsur Ali, M.; Kannan, B.; et al. Print to detect: A rapid and ultrasensitive phage-based dipstick assay for foodborne pathogens. Anal. Bioanal. Chem. 2018, 410, 1217–1230. [Google Scholar] [CrossRef] [PubMed]
- Mido, T.; Schaffer, E.M.; Dorsey, R.W.; Sozhamannan, S.; Hofmann, E.R. Sensitive detection of live Escherichia coli by bacteriophage amplification-coupled immunoassay on the Luminex® MAGPIX instrument. J. Microbiol. Methods 2018, 152, 143–147. [Google Scholar] [CrossRef]
- Said, M.B.; Saad, M.B.; Achouri, F.; Bousselmi, L.; Ghrabi, A. Detection of active pathogenic bacteria under stress conditions using lytic and specific phage. Water Sci. Technol. 2019, 80, 282–289. [Google Scholar] [CrossRef]
- Tilton, L.; Das, G.; Yang, X.; Wisuthiphaet, N.; Kennedy, I.M.; Nitin, N. Nanophotonic Device in Combination with Bacteriophages for Enhancing Detection Sensitivity of Escherichia coli in Simulated Wash Water. Anal. Lett. 2019, 52, 2203–2213. [Google Scholar] [CrossRef]
- He, Y.; Wang, M.; Fan, E.; Ouyang, H.; Yue, H.; Su, X.; Liao, G.; Wang, L.; Lu, S.; Fu, Z. Highly Specific Bacteriophage-Affinity Strategy for Rapid Separation and Sensitive Detection of Viable Pseudomonas aeruginosa. Anal. Chem. 2017, 89, 1916–1921. [Google Scholar] [CrossRef]
- He, Y.; Shi, Y.; Liu, M.; Wang, Y.; Wang, L.; Lu, S.; Fu, Z. Nonlytic Recombinant Phage Tail Fiber Protein for Specific Recognition of Pseudomonas aeruginosa. Anal. Chem. 2018, 90, 14462–14468. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; He, Y.; Bhattacharyya, S.; Lu, S.; Fu, Z. Recombinant Bacteriophage Cell-Binding Domain Proteins for Broad-Spectrum Recognition of Methicillin-Resistant Staphylococcus aureus Strains. Anal. Chem. 2020, 92, 3340–3345. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Torres, N.; Dunne, M.; Garde, S.; Meijers, R.; Narbad, A.; Ávila, M.; Mayer, M.J. Development of a specific fluorescent phage endolysin for in situ detection of Clostridium species associated with cheese spoilage. Microb. Biotechnol. 2018, 11, 332–345. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Wang, Y.; Han, L.; Cai, Y.; Ren, H.; Ma, T.; Li, X.; Petrenko, V.A.; Liu, A. Colorimetric Assay of Bacterial Pathogens Based on Co 3 O 4 Magnetic Nanozymes Conjugated with Specific Fusion Phage Proteins and Magnetophoretic Chromatography. ACS Appl. Mater. Interfaces 2020, 12, 9090–9097. [Google Scholar] [CrossRef] [PubMed]
- Niyomdecha, S.; Limbut, W.; Numnuam, A.; Kanatharana, P.; Charlermroj, R.; Karoonuthaisiri, N.; Thavarungkul, P. Phage-based capacitive biosensor for Salmonella detection. Talanta 2018, 188, 658–664. [Google Scholar] [CrossRef]
- Laube, T.; Cortés, P.; Llagostera, M.; Alegret, S.; Pividori, M.I. Phagomagnetic immunoassay for the rapid detection of Salmonella. Appl. Microbiol. Biotechnol. 2014, 98, 1795–1805. [Google Scholar] [CrossRef]
- Ulitzur, N.; Ulitzur, S. New rapid and simple methods for detection of bacteria and determination of their antibiotic susceptibility by using phage mutants. Appl. Environ. Microbiol. 2006, 72, 7455–7459. [Google Scholar] [CrossRef][Green Version]
- Wu, L.; Song, Y.; Luan, T.; Ma, L.; Su, L.; Wang, S.; Yan, X. Specific detection of live Escherichia coli O157: H7 using tetracysteine-tagged PP01 bacteriophage. Biosens. Bioelectron. 2016, 86, 102–108. [Google Scholar] [CrossRef]
- Edgar, R.; McKinstry, M.; Hwang, J.; Oppenheim, A.B.; Fekete, R.A.; Giulian, G.; Merril, C.; Nagashima, K.; Adhya, S. High-sensitivity bacterial detection using biotin-tagged phage and quantum-dot nanocomplexes. Proc. Natl. Acad. Sci. USA 2006, 103, 4841–4845. [Google Scholar] [CrossRef]
- Alam, N.; Oskam, E.; Stassen, P.M.; van Exter, P.; van de Ven, P.M.; Haak, H.R.; Holleman, F.; van Zanten, A.; van Leeuwen-Nguyen, H.; Bon, V.; et al. Prehospital antibiotics in the ambulance for sepsis: A multicentre, open label, randomised trial. Lancet Respir. Med. 2018, 6, 40–50. [Google Scholar] [CrossRef]
- Brun-Buison, C.; Abrouk, F.; Legrand, P.; Huet, Y.; Larabi, S.; Rapin, M. Diagnosis of Central Venous Catheter-Related Sepsis. Surv. Anesthesiol. 1987, 31, 369. [Google Scholar] [CrossRef]
- Akrong, M.O.; Amu-Mensah, F.K.; Amu-Mensah, M.A.; Darko, H.; Addico, G.N.D.; Ampofo, J.A. Seasonal analysis of bacteriological quality of drinking water sources in communities surrounding Lake Bosomtwe in the Ashanti Region of Ghana. Appl. Water Sci. 2019, 9, 82. [Google Scholar] [CrossRef]

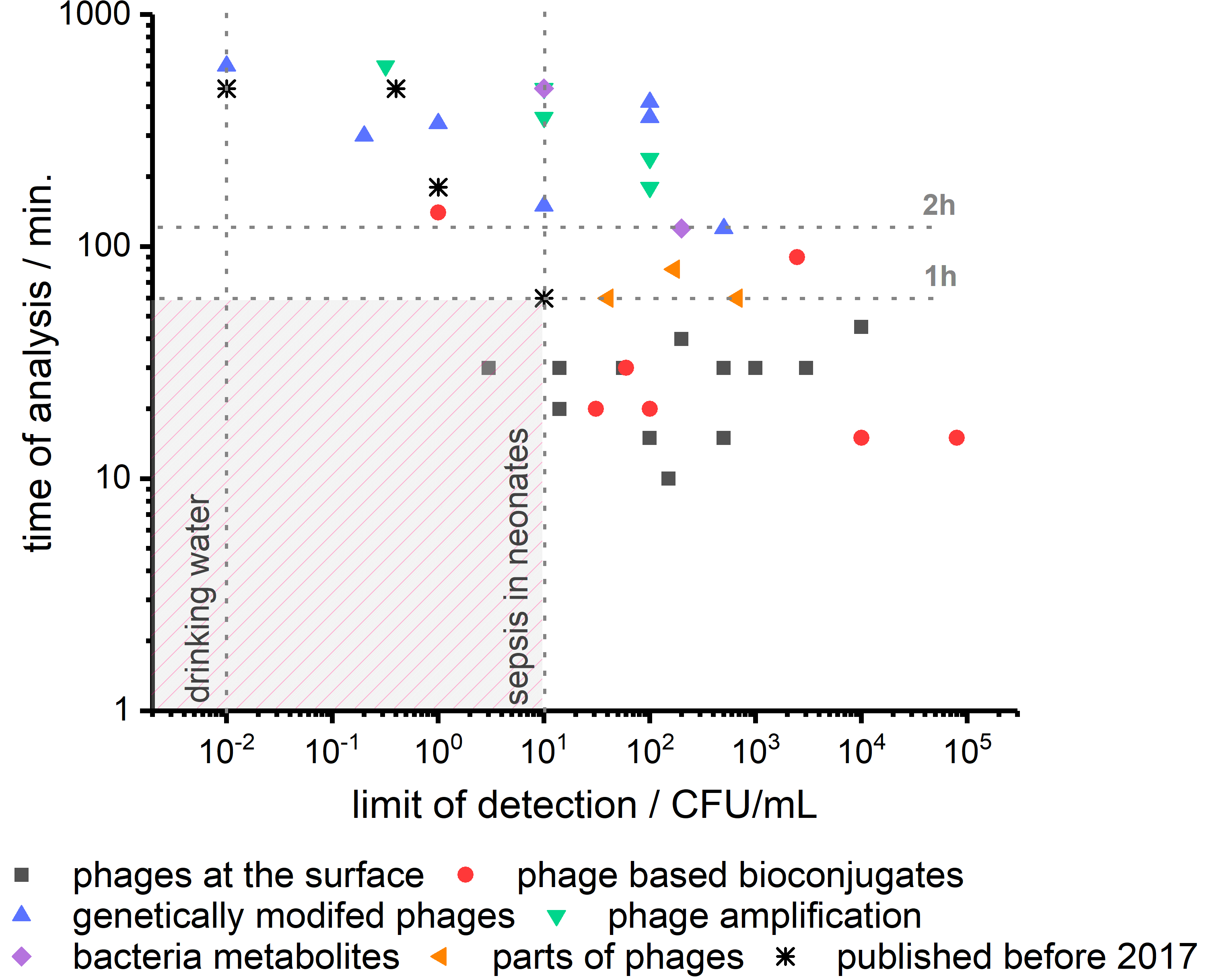
| Bioreceptor | Bacteria | Method | LOD | Time | Comments | Reference |
|---|---|---|---|---|---|---|
| Phages at the surface | ||||||
| T4 phage | Escherichia coli BL21 DE3 | microscopic | 102–103 CFU/mL | 15 min of incubation | virions properly oriented in the constant electric field | [57] |
| T2 phage | Escherichia coli B ATCC 11303 | electrochemical impedance spectroscopy | 103 CFU/mL | 30 min | virions correctly oriented according to charge driven assembly on carbon nanotube-based impedimetric biosensor | [32] |
| T4 phage | Escherichia coli BL21 DE3 | microscopic | 102 CFU/mL | 15 min of incubation | virions oriented correctly in the alternating electric field | [60] |
| T4 phage | Escherichia coli B | differential pulse voltammetry | 14 ± 5 CFU/mL | 20 min | virions properly oriented in the alternating electric field on the micro-electrochemical sensor | [36] |
| lytic phage isolated from the hospital sewage water | Staphylococcus aureus CCTCC AB2013186 | differential pulse voltammetry | 3 CFU/mL in PBS | 30 min | the best balance between LOD and time of analysis reported to date | [26] |
| 5 CFU/mL in milk | ||||||
| T4 phage | Escherichia coli B, ATCC 11303, Escherichia coli XLMRF | SERS | 1.5 × 102 CFU/mL | 10 min of incubation | reusable biosensor | [49] |
| Tbilisi bacteriophage | Brucella abortus | SERS | 104 CFU/mL | 45 min | [51] | |
| Gamma-phages | Bacillus thuringiensis | SERS | 104 CFU/mL | 45 min | the principal component analysis was used for data processing | [52] |
| M13 phage | Escherichia coli XL1-Blue and K12 strains | electrochemical impedance spectroscopy | 14 CFU/mL | 30 min of incubation | virions chemisorbed on glassy carbon electrode decorated with gold nanoparticles | [35] |
| M13 phage displaying specific peptide NRPDSAQFWLHHGGGSC (MSal020417) | Salmonella spp. | the capacitive flow injection system | 2 × 102 CFU/mL | 40 min | reusable (up to 40 times) biosensors; virions immobilized on a polytyramine/gold surface | [96] |
| PaP1 phage | Pseudomonas aeruginosa | electrochemiluminescence | 56 CFU/mL | 30 min | carboxyl graphene-PaP1 composite was dropped onto the glassy carbon electrode | [41] |
| C4-22 phage | Salmonella enterica | magnetoelastic | 7.86 × 103 CFU/mm2 | 2 min of incubation | bacteria were captured from the surface of raw chicken breast filet | [44] |
| phage 12600 | methicillin-resistant Staphylococcus aureus | magnetoelastic | 3 × 103 CFU/mL | 30 min | the method is based on sensors previously reported by the same group [42] | [43] |
| E2 phage | Salmonella Typhimurium ATCC 1331 | magnetoelastic | 5 × 102 CFU/mL | 30 min | bacteria were captured from the surface of romaine lettuce | [45] |
| Bacteriophage based bioconjugates | ||||||
| S13 phage | Staphylococcus aureus SA27 | dark field microscopy | 8 × 104 CFU/mL | 15–20 min | virions were oriented according to charge driven assembly on the surface of core−shell nanoparticles | [66] |
| P9b phage displaying the specific peptide (QRKLAAKLT) | Pseudomonas aeruginosa ATCC 27853 | SERS | NA | around 2h | gold nanoparticles were used | [64] |
| chemically modified and genetically engineered M13 phages | Escherichia coli (2 strains), Pseudomonas aeruginosa, Vibrio cholerae, Xanthomonas campestris (2 strains) | colorimetric sensor | 60 to 102 cell/mL | 30 min | gold nanoparticles were used | [65] |
| T4 phage | Escherichia coli BL21 | flow cytometry | 104 CFU/mL | 15 min of incubation | magnetic and fluorescent particles were used | [67] |
| P-S. aureus-9, isolated from an environmental water sample | Staphylococcus aureus (18 clinical strains) | isolation and separation by magnetic bioconjugates + immunoassay | 2.47 × 103 CFU/mL in PBS | 90 min | no pre-enrichment | [27] |
| 8.9 × 103 CFU/mL in juice | ||||||
| temperate phages isolated from environment samples | Staphylococcus arlettae | fluorescence quenching | 102 CFU/mL | 20 min of incubation | IRMOF-3 was used | [28] |
| Staphylococcus aureus | fluorescence quenching | 31 CFU/mL | 20 min of incubation | NH2-MIL-53(Fe) was used | [29] | |
| T4 phage | Escherichia coli ATCC 11303 | differential pulse voltammetry | 1 CFU/mL | 140 min | Cu3(PO4)2 nanoflowers loaded with glucose oxidase, horseradish peroxidase, thionine, and gold nanoparticles to which virions attached were used as the electrochemical signal amplification system | [34] |
| Genetically modified phages | ||||||
| T7-ALP phage | Escherichia coli BL21 | fluorescence imaging and image analysis | around 102 bacteria per gram of sample | 6 h | fluorescent substrate for alkaline phosphatase activity was added; detection in model beverage samples | [73] |
| A511::luxAB | Listeria monocytogenes WSLC 1001, ScottA, EGDe, Listeria innocua WSLC 2012, Listeria ivanovii WSLC 3009 | magnetic separation combined with fluorescence | around 102 cells/mL | 6 h | magnetic beads with cell wall-binding domains from Listeria phage endolysins were used for magnetic separation | [74] |
| NRGp6 phage (T7 with NanoLuc luciferase expression cassette | Escherichia coli BL21 | spectroscopic detection | 5 × 102 CFU/mL | 2 h of incubation | NanoGlo substrate was added | [75] |
| T7 phage with luciferase or an alkaline phosphatase fused with CBM | Escherichia coli | visualization of colonies | 1 CFU/100 mL | 10 h | filtration based method; enzymatic substrate was added | [76] |
| T7 phage with NanoLuc-CBM | Escherichia coli | luminescence of cellulose bound fused proteins | <10 CFU/mL | 2.5 h | NanoGlo substrate was added | [77] |
| T7 phage with NanoLuc-CBM | Escherichia coli ECOR13 | luminescence | 20 CFU/100 mL | 5 h | NanoGlo substrate was added | [78] |
| phiV10lux phage | Escherichia coli O157:H7 | bioluminescent intensity | around 1 CFU/mL | 40 min after 5 h of incubation | LOD in artificially contaminated romaine lettuce 10 CFU/cm2, apple juice 13 CFU/mL, ground beef 17 CFU/g | [81] |
| T7lacZ | Escherichia coli | differential pulse voltammetry | 102 CFU/mL | 7 h | 4-aminophenyl-β-galactopyranoside was added as a substrate for β-galactosidase | [33] |
| mCherrybombφ | Mycobacterium tuberculosis | fluorescence microscopy | NA | at least 48 h to 96 h | the method allowed for a phenotypic determination of rifampicin resistance; sputum samples were collected from patients | [82] |
| Phage amplification | ||||||
| p53 phage | Acinetobacter baumannii (15 various clinical isolates) | qPCR | 102 CFU/mL in serum | 4 h | [83] | |
| p53 phage | Acinetobacter baumannii | qPCR | 10 CFU/mL sputum samples | 6 h | [84] | |
| vB_SenS_PVP-SE2 phage | Salmonella Enteritidis | qPCR | 8 CFU/25 g in chicken samples | 10 h | [85] | |
| brucellaphage | Brucella abortus | qPCR | 1 CFU/mL | 72 h | in mixed cultures and blood samples | [86] |
| rV5 phage | Escherichia coli O157:H7 | qPCR, phages printed onto paper strips using modified inkjet | 10–50 CFU/mL | 8 h | in spinach and broth | [87] |
| AG2A phage | Escherichia coli O45:H2 | in ground beef | ||||
| CGG4-1 phage | Salmonella Newport | in chicken samples | ||||
| MS2 phage | Escherichia coli C-3000 | bead-based sandwich-type immunoassay | 102 cells/mL | 3 h | [88] | |
| Detection of bacterial metabolites | ||||||
| T7 phage | Escherichia coli BL21 | fluorescence | 10 CFU/mL in simulated spinach wash | 8 h | resorufin β -D-galactopyranoside was added after lysis | [90] |
| PAP1 phage | Pseudomonas aeruginosa | luminescence | 2 × 102 CFU/mL | 2 h | firefly luciferase-adenosine triphosphate bioluminescence system was used | [91] |
| Phage fragments | ||||||
| pVIII protein | Staphylococcus aureus | magnetophoretic chromatography in the external magnetic field combined with colorimetric readout due to enzymatic activity of nanozyme | 8 CFU/mL | NA | magnetic nanozyme Co3O4 MNE@fusion-pVIII was used | [95] |
| cell-binding domain (CBD) | methicillin-resistant Staphylococcus aureus (6 strains) | flow cytometry | 40 CFU/mL | Around 1 h (2x 30 min incubation + washing steps) | The CBD-GFP fusion protein was used, broad host recognition due to CBD; no lysis | [93] |
| bacteriophage endolysin CTP1L | Clostridium tyrobutyricum (17 strains) | fluorescence microscopy | 3 spores per g of cheese | around 35 min + washing steps | GFP-CTP1L and GFP-CBD were used; also bind to clostridial spores | [94] |
| fiber protein (P069) | Pseudomonas aeruginosa (4 strains) | bioluminescence | 6.7 × 102 CFU/mL | around 60 min | two very different detection approaches. BL based on magnetic beads, FL on the interactions with modified surface | [92] |
| fluorescence | 1.7 × 102 CFU/mL | around 80 min | ||||
| Best performing phage-based methods reported before 2017 [13] | ||||||
| Lambda phage | Escherichia coli | amperometric | 1 CFU/100 mL | 6–8 h | detection of metabolites | [30] |
| P22 phage | Salmonella | colorimetric | 1 CFU/24 mL | 6 h | phagomagnetic separation of bacteria labeled with antibodies conjugated with horseradish peroxide | [97] |
| AR1 phage | Escherichia coli | plaque count method | 1 CFU/mL | 3 h | phage amplification | [98] |
| PP01 phage | Escherichia coli | fluorescence | 1 CFU/mL | 3 h | genetically modified phages | [99] |
| M13 phage | Escherichia coli | amperometric | 1 CFU/mL | 3 h | detection of metabolites | [31] |
| HK620 phage | Escherichia coli | flow cytometry | 10 CFU/mL | 1 h | genetically modified phages | [79] |
| P22 phage | Salmonella | |||||
| T7 phage | Escherichia coli | flow cytometry | 10 CFU/mL | 1 h | conjugates of biotinylated phages and streptavidin bound quantum dots | [100] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paczesny, J.; Richter, Ł.; Hołyst, R. Recent Progress in the Detection of Bacteria Using Bacteriophages: A Review. Viruses 2020, 12, 845. https://doi.org/10.3390/v12080845
Paczesny J, Richter Ł, Hołyst R. Recent Progress in the Detection of Bacteria Using Bacteriophages: A Review. Viruses. 2020; 12(8):845. https://doi.org/10.3390/v12080845
Chicago/Turabian StylePaczesny, Jan, Łukasz Richter, and Robert Hołyst. 2020. "Recent Progress in the Detection of Bacteria Using Bacteriophages: A Review" Viruses 12, no. 8: 845. https://doi.org/10.3390/v12080845
APA StylePaczesny, J., Richter, Ł., & Hołyst, R. (2020). Recent Progress in the Detection of Bacteria Using Bacteriophages: A Review. Viruses, 12(8), 845. https://doi.org/10.3390/v12080845





