Purification Methods and the Presence of RNA in Virus Particles and Extracellular Vesicles
Abstract
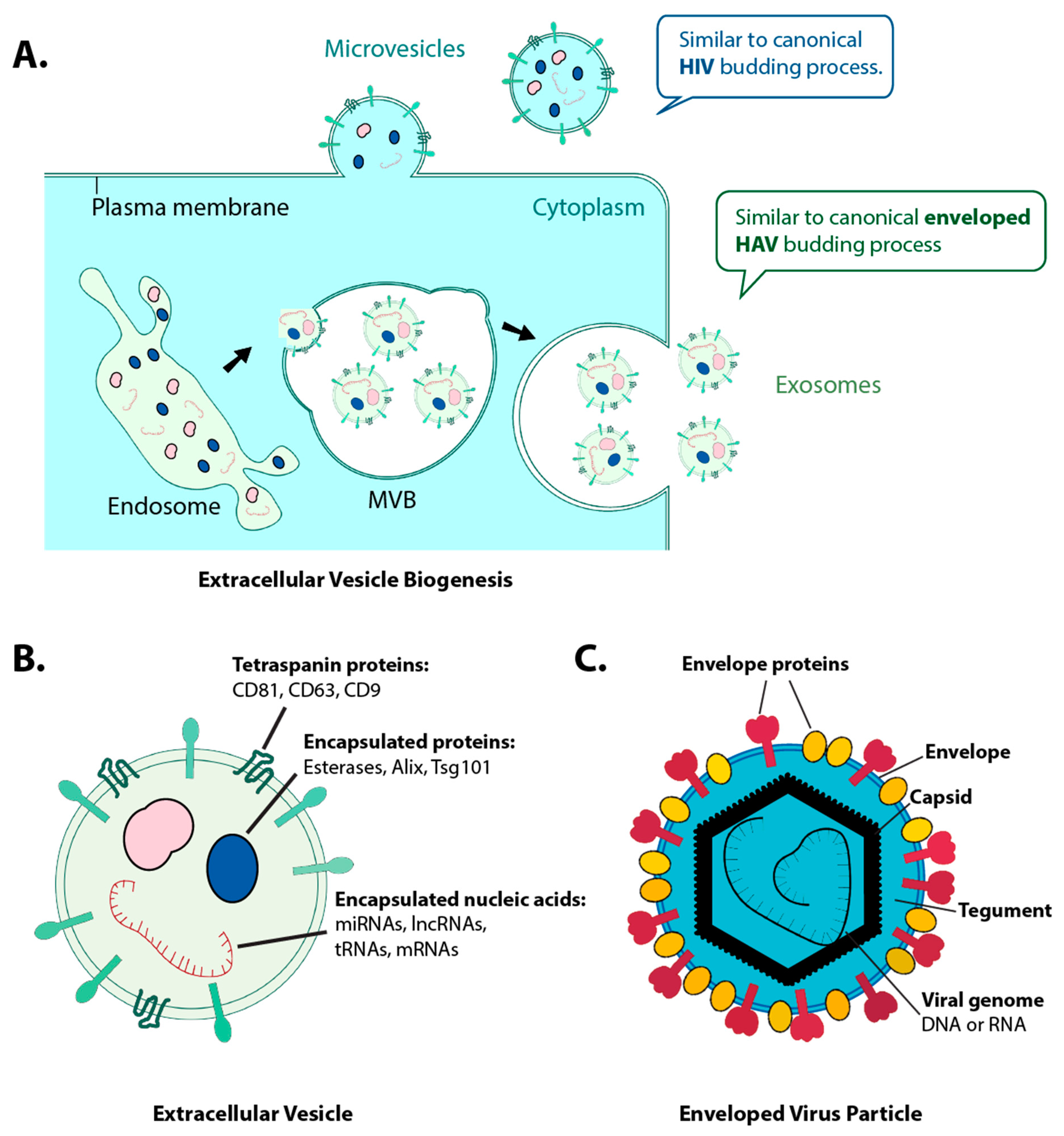
1. Introduction
2. How do RNAs Co-Purify with Viruses and EV by Different Purification Methods?
3. What Have We Learned from Different Purification Methods?
4. Do DNA Viruses Package RNAs in Virus Particles?
5. How Many miRNAs are in an Exosome?
6. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Ulmer, J.B.; Valley, U.; Rappuoli, R. Vaccine manufacturing: Challenges and solutions. Nat. Biotechnol. 2006, 24, 1377–1383. [Google Scholar] [CrossRef] [PubMed]
- Roizman, B. Redefining virology. Science 2000, 288, 2327–2328. [Google Scholar] [CrossRef] [PubMed]
- Sciortino, M.-T.; Suzuki, M.; Taddeo, B.; Roizman, B. RNAs Extracted from Herpes Simplex Virus 1 Virions: Apparent Selectivity of Viral but Not Cellular RNAs Packaged in Virions. J. Virol. 2001, 75, 8105–8116. [Google Scholar] [CrossRef]
- Sciortino, M.T.; Taddeo, B.; Poon, A.P.W.; Mastino, A.; Roizman, B. Of the three tegument proteins that package mRNA in herpes simplex virions, one (VP22) transports the mRNA to uninfected cells for expression prior to viral infection. Proc. Natl. Acad. Sci. USA 2002, 99, 8318–8323. [Google Scholar] [CrossRef]
- Prichard, M.N.; Jairath, S.; Penfold, M.E.T.; Jeor, S.S.; Bohlman, M.C.; Pari, G.S. Identification of Persistent RNA-DNA Hybrid Structures within the Origin of Replication of Human Cytomegalovirus. J. Virol. 1998, 72, 6997–7004. [Google Scholar] [CrossRef]
- Greijer, A.E.; Dekkers, C.A.J.; Middeldorp, J.M. Human Cytomegalovirus Virions Differentially Incorporate Viral and Host Cell RNA during the Assembly Process. J. Virol. 2000, 74, 9078–9082. [Google Scholar] [CrossRef]
- Cliffe, A.R.; Nash, A.A.; Dutia, B.M. Selective Uptake of Small RNA Molecules in the Virion of Murine Gammaherpesvirus 68. J. Virol. 2009, 83, 2321–2326. [Google Scholar] [CrossRef]
- Lin, X.; Li, X.; Liang, D.; Lan, K. MicroRNAs and Unusual Small RNAs Discovered in Kaposi’s Sarcoma-Associated Herpesvirus Virions. J. Virol. 2012, 86, 12717–12730. [Google Scholar] [CrossRef]
- Bechtel, J.; Grundhoff, A.; Ganem, D. RNAs in the Virion of Kaposi’s Sarcoma-Associated Herpesvirus. J. Virol. 2005, 79, 10138–10146. [Google Scholar] [CrossRef]
- Amen, M.A.; Griffiths, A. Identification and Expression Analysis of Herpes B Virus-Encoded Small RNAs. J. Virol. 2011, 85, 7296–7311. [Google Scholar] [CrossRef]
- Chung, S.W.; Arnott, J.A.; Yang, Y.; Wong, P.M. Presence of prepackaged mRNA in virions of DNA adenovirus. J. Biol. Chem. 2003, 278, 50635–50640. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Tikoo, S.K. Viral RNAs detected in virions of porcine adenovirus type 3. Virology 2004, 321, 372–382. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bresnahan, W.A.; Shenk, T. A subset of viral transcripts packaged within human cytomegalovirus particles. Science 2000, 288, 2373–2376. [Google Scholar] [CrossRef] [PubMed]
- Jochum, S.; Ruiss, R.; Moosmann, A.; Hammerschmidt, W.; Zeidler, R. RNAs in Epstein–Barr virions control early steps of infection. Proc. Natl. Acad. Sci. USA 2012, 109, E1396–E1404. [Google Scholar] [CrossRef]
- Dalgleish, A.G.; Beverley, P.C.L.; Clapham, P.R.; Crawford, D.H.; Greaves, M.F.; Weiss, R.A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 1984, 312, 763–767. [Google Scholar] [CrossRef]
- Wu, L.; LaRosa, G.; Kassam, N.; Gordon, C.J.; Heath, H.; Ruffing, N.; Chen, H.; Humblias, J.; Samson, M.; Parmentier, M.; et al. Interaction of Chemokine Receptor CCR5 with its Ligands: Multiple Domains for HIV-1 gp120 Binding and a Single Domain for Chemokine Binding. J. Exp. Med. 1997, 186, 1373–1381. [Google Scholar] [CrossRef]
- McNamara, R.P.; Dittmer, D.P. Modern Techniques for the Isolation of Extracellular Vesicles and Viruses. J. Neuroimmune Pharm. 2019, 1–14. [Google Scholar] [CrossRef]
- Hurley, J.H. ESCRT complexes and the biogenesis of multivesicular bodies. Curr. Opin. Cell Biol. 2008, 20, 4–11. [Google Scholar] [CrossRef]
- Van Niel, G.; Charrin, S.; Simoes, S.; Romao, M.; Rochin, L.; Saftig, P.; Marks, M.S.; Rubinstein, E.; Raposo, G. The Tetraspanin CD63 Regulates ESCRT-Independent and-Dependent Endosomal Sorting during Melanogenesis. Dev. Cell 2011, 21, 708–721. [Google Scholar] [CrossRef]
- Chevillet, J.R.; Kang, Q.; Ruf, I.K.; Briggs, H.A.; Vojtech, L.N.; Hughes, S.M.; Cheng, H.H.; Arroyo, J.D.; Meredith, E.K.; Gallichotte, E.N.; et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. USA 2014, 111, 14888–14893. [Google Scholar] [CrossRef] [PubMed]
- Caby, M.-P.; Lankar, D.; Vincendeau-Scherrer, C.; Raposo, G.; Bonnerot, C. Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 2005, 17, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef] [PubMed]
- Rein, A. RNA Packaging in HIV. Trends Microbiol. 2019, 27, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Van der Grein, S.G.; Defourny, K.A.Y.; Rabouw, H.H.; Galiveti, C.R.; Langereis, M.A.; Wauben, M.H.M.; Arkesteijn, G.J.A.; van Kuppeveld, F.J.M.; Nolte-‘t Hoen, E.N.M. Picornavirus infection induces temporal release of multiple extracellular vesicle subsets that differ in molecular composition and infectious potential. PLoS Pathog. 2019, 15, e1007594. [Google Scholar] [CrossRef] [PubMed]
- Gould, S.J.; Booth, A.M.; Hildreth, J.E. The Trojan exosome hypothesis. Proc. Natl. Acad. Sci. USA 2003, 100, 10592–10597. [Google Scholar] [CrossRef] [PubMed]
- Meckes, D.G.; Raab-Traub, N. Microvesicles and Viral Infection. J. Virol. 2011, 85, 12844–12854. [Google Scholar] [CrossRef]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Price, C.A. Centrifugation in Density Gradients; Academic Press: New York, NY, USA, 1982. [Google Scholar]
- Mateescu, B.; Kowal, E.J.K.; van Balkom, B.W.M.; Bartel, S.; Bhattacharyya, S.N.; Buzás, E.I.; Buck, A.H.; de Candia, P.; Chow, F.W.N.; Das, S.; et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA—An ISEV position paper. J. Extracell. Vesicles 2017, 6, 1286095. [Google Scholar] [CrossRef]
- Raab-Traub, N.; Dittmer, D.P. Viral effects on the content and function of extracellular vesicles. Nat. Rev. Microbiol. 2017, 15, 559–572. [Google Scholar] [CrossRef]
- Kalamvoki, M.; Du, T.; Roizman, B. Cells infected with herpes simplex virus 1 export to uninfected cells exosomes containing STING, viral mRNAs, and microRNAs. Proc. Natl. Acad. Sci. USA 2014, 111, E4991–E4996. [Google Scholar] [CrossRef] [PubMed]
- Kalamvoki, M.; Deschamps, T. Extracellular vesicles during Herpes Simplex Virus type 1 infection: An inquire. Virol. J. 2016, 13, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.R.; Alberts, B.M.; Benzinger, R.; Lawhorne, L.; Treiber, G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology 1970, 40, 734–744. [Google Scholar] [CrossRef]
- Adams, A. Concentration of Epstein-Barr Virus from Cell Culture Fluids with Polyethylene Glycol. J. Gen. Virol. 1973, 20, 391–394. [Google Scholar] [CrossRef]
- Chugh, P.E.; Sin, S.H.; Ozgur, S.; Henry, D.H.; Menezes, P.; Griffith, J.; Eron, J.J.; Damania, B.; Dittmer, D.P. Systemically circulating viral and tumor-derived microRNAs in KSHV-associated malignancies. PLoS Pathog. 2013, 9, e1003484. [Google Scholar] [CrossRef]
- Atha, D.H.; Ingham, K.C. Mechanism of precipitation of proteins by polyethylene glycols. Analysis in terms of excluded volume. J. Biol. Chem. 1981, 256, 12108–12117. [Google Scholar]
- Lobb, R.J.; Becker, M.; Wen Wen, S.; Wong, C.S.F.; Wiegmans, A.P.; Leimgruber, A.; Möller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4, 27031. [Google Scholar] [CrossRef]
- Besnard, L.; Fabre, V.; Fettig, M.; Gousseinov, E.; Kawakami, Y.; Laroudie, N.; Scanlan, C.; Pattnaik, P. Clarification of vaccines: An overview of filter based technology trends and best practices. Biotechnol. Adv. 2016, 34, 1–13. [Google Scholar] [CrossRef]
- McNamara, R.P.; Caro-Vegas, C.P.; Costantini, L.M.; Landis, J.T.; Griffith, J.D.; Damania, B.A.; Dittmer, D.P. Large-scale, cross-flow based isolation of highly pure and endocytosis-competent extracellular vesicles. J. Extracell. Vesicles 2018, 7, 1541396. [Google Scholar] [CrossRef]
- Kallel, H.; Kamen, A.A. Large-scale adenovirus and poxvirus-vectored vaccine manufacturing to enable clinical trials. Biotechnol. J. 2015, 10, 741–747. [Google Scholar] [CrossRef]
- Tomono, T.; Hirai, Y.; Okada, H.; Adachi, K.; Ishii, A.; Shimada, T.; Onodera, M.; Tamaoka, A.; Okada, T. Ultracentrifugation-free chromatography-mediated large-scale purification of recombinant adeno-associated virus serotype 1 (rAAV1). Mol. Ther. Methods Clin. Dev. 2016, 3, 15058. [Google Scholar] [CrossRef] [PubMed]
- Hagen, A.; Aunins, J.; DePhillips, P.; Oswald, C.B.; Hennessey Jr, J.P.; Lewis, J.; Armstrong, M.; Oliver, C.; Orella, C.; Buckland, B.; et al. Development, preparation, and testing of VAQTA®, a highly purified hepatitis A vaccine. Bioprocess Eng. 2000, 23, 439–449. [Google Scholar] [CrossRef]
- Kalbfuss, B.; Wolff, M.; Morenweiser, R.; Reichl, U. Purification of cell culture-derived human influenza A virus by size-exclusion and anion-exchange chromatography. Biotechnol. Bioeng. 2007, 96, 932–944. [Google Scholar] [CrossRef] [PubMed]
- Reiter, K.; Aguilar, P.P.; Wetter, V.; Steppert, P.; Tover, A.; Jungbauer, A. Separation of virus-like particles and extracellular vesicles by flow-through and heparin affinity chromatography. J. Chromatogr. A 2019, 1588, 77–84. [Google Scholar] [CrossRef]
- McNamara, R.P.; Chugh, P.E.; Bailey, A.; Costantini, L.M.; Ma, Z.; Bigi, R.; Cheves, A.; Eason, A.B.; Landis, J.T.; Host, K.M.; et al. Extracellular vesicles from Kaposi Sarcoma-associated herpesvirus lymphoma induce long-term endothelial cell reprogramming. PLoS Pathog. 2019, 15, e1007536. [Google Scholar] [CrossRef]
- McNamara, R.P.; Costantini, L.M.; Myers, T.A.; Schouest, B.; Maness, N.J.; Griffith, J.D.; Damania, B.A.; MacLean, A.G.; Dittmer, D.P. Nef Secretion into Extracellular Vesicles or Exosomes Is Conserved across Human and Simian Immunodeficiency Viruses. MBio 2018, 9. [Google Scholar] [CrossRef]
- Cantin, R.; Diou, J.; Bélanger, D.; Tremblay, A.M.; Gilbert, C. Discrimination between exosomes and HIV-1: Purification of both vesicles from cell-free supernatants. J. Immunol. Methods 2008, 338, 21–30. [Google Scholar] [CrossRef]
- Deschamps, T.; Kalamvoki, M. Extracellular Vesicles Released by Herpes Simplex Virus 1-Infected Cells Block Virus Replication in Recipient Cells in a STING-Dependent Manner. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Temoche-Diaz, M.M.; Shurtleff, M.J.; Nottingham, R.M.; Yao, J.; Fadadu, R.P.; Lambowitz, A.M.; Schekman, R. Distinct mechanisms of microRNA sorting into cancer cell-derived extracellular vesicle subtypes. eLife 2019, 8. [Google Scholar] [CrossRef]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef]
- Brandariz-Nuñez, A.; Robinson, S.J.; Evilevitch, A. Pressurized DNA state inside herpes capsids—A novel antiviral target. PLoS Pathog. 2020, 16, e1008604. [Google Scholar] [CrossRef] [PubMed]
- Cvjetkovic, A.; Jang, S.C.; Konečná, B.; Höög, J.L.; Sihlbom, C.; Lässer, C.; Lötvall, J. Detailed Analysis of Protein Topology of Extracellular Vesicles–Evidence of Unconventional Membrane Protein Orientation. Sci. Rep. 2016, 6, 36338. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Serrano, E.E.; González-López, O.; Das, A.; Lemon, S.M. Cellular entry and uncoating of naked and quasi-enveloped human hepatoviruses. Elife 2019, 8, e43983. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef]
- Skalsky, R.L.; Cullen, B.R. Viruses, microRNAs, and Host Interactions. Annu. Rev. Microbiol. 2010, 64, 123–141. [Google Scholar] [CrossRef]
- Weigel, T.; Solomaier, T.; Peuker, A.; Pathapati, T.; Wolff, M.W.; Reichl, U. A flow-through chromatography process for influenza A and B virus purification. J. Virol Methods 2014, 207, 45–53. [Google Scholar] [CrossRef]
- Tseng, Y.-F.; Weng, T.-C.; Lai, C.-C.; Chen, P.-L.; Lee, M.-S.; Hu, A.Y.-C. A fast and efficient purification platform for cell-based influenza viruses by flow-through chromatography. Vaccine 2018, 36, 3146–3152. [Google Scholar] [CrossRef]
- Mundle, S.T.; Kishko, M.; Groppo, R.; DiNapoli, J.; Hamberger, J.; McNeil, B.; Kleanthous, H.; Parrington, M.; Zhang, L.; Anderson, S.F. Core bead chromatography for preparation of highly pure, infectious respiratory syncytial virus in the negative purification mode. Vaccine 2016, 34, 3690–3696. [Google Scholar] [CrossRef]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Martin, A.B.; Bojmar, L.; et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef]
- Guo, B.B.; Bellingham, S.A.; Hill, A.F. The Neutral Sphingomyelinase Pathway Regulates Packaging of the Prion Protein into Exosomes. J. Biol. Chem. 2015, 290, 3455–3467. [Google Scholar] [CrossRef]
- Naik, J.; Hau, C.M.; ten Bloemendaal, L.; Mok, K.S.; Hajji, N.; Wehman, A.M.; Meisner, S.; Muncan, V.; Paauw, N.J.; de Vries, H.E.; et al. The P4-ATPase ATP9A is a novel determinant of exosome release. PLoS ONE 2019, 14, e0213069. [Google Scholar] [CrossRef] [PubMed]
- Beer, K.B.; Rivas-Castillo, J.; Kuhn, K.; Fazeli, G.; Karmann, B.; Nance, J.F.; Stigloher, C.; Wehman, A.M. Extracellular vesicle budding is inhibited by redundant regulators of TAT-5 flippase localization and phospholipid asymmetry. Proc. Natl. Acad. Sci. USA 2018, 115, E1127–E1136. [Google Scholar] [CrossRef] [PubMed]
- Durcin, M.; Fleury, A.; Taillebois, E.; Hilairet, G.; Krupova, Z.; Henry, C.; Truchet, S.; Trötzmüller, M.; Köfeler, H.; Mabilleau, G.; et al. Characterisation of adipocyte-derived extracellular vesicle subtypes identifies distinct protein and lipid signatures for large and small extracellular vesicles. J. Extracell. Vesicles 2017, 6, 1305677. [Google Scholar] [CrossRef] [PubMed]
- Atkin-Smith, G.K.; Tixeira, R.; Paone, S.; Mathivanan, S.; Collins, C.; Liem, M.; Goodall, K.J.; Ravichandran, K.S.; Hulett, M.D.; Poon, I.K.H. A novel mechanism of generating extracellular vesicles during apoptosis via a beads-on-a-string membrane structure. Nat. Commun. 2015, 6, 1–10. [Google Scholar] [CrossRef]
- Ratner, L.; Haseltine, W.; Patarca, R.; Livak, K.J.; Starcich, B.; Josephs, S.F.; Doran, E.R.; Rafalski, J.A.; Whitehorn, E.A.; Baumeister, K.; et al. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature 1985, 313, 277–284. [Google Scholar] [CrossRef]
- Cen, S.; Khorchid, A.; Javanbakht, H.; Gabor, J.; Stello, T.; Shiba, K.; Musier-Forsyth, K.; Kleiman, L. Incorporation of Lysyl-tRNA Synthetase into Human Immunodeficiency Virus Type 1. J. Virol. 2001, 75, 5043–5048. [Google Scholar] [CrossRef]
- Rulli Jr, S.J.; Hibbert, C.S.; Mirro, J.; Pederson, T.; Biswal, S.; Rein, A. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J. Virol. 2007, 81, 6623–6631. [Google Scholar] [CrossRef]
- Comas-Garcia, M.; Datta, S.A.K.; Baker, L.; Varma, R.; Gudla, P.R.; Rein, A. Dissection of specific binding of HIV-1 Gag to the ‘packaging signal’ in viral RNA. eLife 2017, 6, e27055. [Google Scholar] [CrossRef]
- Rein, A.; Henderson, L.E.; Levin, J.G. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: Significance for viral replication. Trends Biochem. Sci. 1998, 23, 297–301. [Google Scholar] [CrossRef]
- Bogerd, H.P.; Kennedy, E.M.; Whisnant, A.W.; Cullen, B.R. Induced Packaging of Cellular MicroRNAs into HIV-1 Virions Can Inhibit Infectivity. MBio 2017, 8. [Google Scholar] [CrossRef]
- Eckwahl, M.J.; Arnion, H.; Kharytonchyk, S.; Zang, T.; Bieniasz, P.D.; Telesnitsky, A.; Wolin, S.L. Analysis of the human immunodeficiency virus-1 RNA packageome. RNA 2016, 22, 1228–1238. [Google Scholar] [CrossRef] [PubMed]
- Šimonová, A.; Svojanovská, B.; Trylčová, J.; Hubálek, M.; Moravčík, O.; Zavřel, M.; Pávová, M.; Hodek, J.; Weber, J.; Cvačka, J.; et al. LC/MS analysis and deep sequencing reveal the accurate RNA composition in the HIV-1 virion. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Meckes, D.G.; Shair, K.H.Y.; Marquitz, A.R.; Kung, C.-P.; Edwards, R.H.; Raab-Traub, N. Human tumor virus utilizes exosomes for intercellular communication. Proc. Natl. Acad. Sci. USA 2010, 107, 20370–20375. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M.; Cosmopoulos, K.; Thorley-Lawson, D.A.; van Eijndhoven, M.A.J.; Hopmans, E.S.; Lindenberg, J.L.; de Gruijl, T.D.; Würdinger, T.; Middeldorp, J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 6328–6333. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M.; van de Garde, M.D.B.; Middeldorp, J.M. Viral miRNAs exploiting the endosomal–exosomal pathway for intercellular cross-talk and immune evasion. Biochim. Biophys. Acta Gene Regul. Mech. 2011, 1809, 715–721. [Google Scholar] [CrossRef]
- Bess, J.W.; Gorelick, R.J.; Bosche, W.J.; Henderson, L.E.; Arthur, L.O. Microvesicles Are a Source of Contaminating Cellular Proteins Found in Purified HIV-1 Preparations. Virology 1997, 230, 134–144. [Google Scholar] [CrossRef]
- Renne, R.; Zhong, W.; Herndier, B.; McGrath, M.; Abbey, N.; Kedes, D.; Ganem, D. Lytic growth of Kaposi’s sarcoma–associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 1996, 2, 342–346. [Google Scholar] [CrossRef]
- Gottwein, E.; Corcoran, D.L.; Mukherjee, N.; Skalsky, R.L.; Hafner, M.; Nusbaum, J.D.; Shamulailatpam, P.; Love, C.L.; Dave, S.S.; Tuschl, T.; et al. Viral MicroRNA Targetome of KSHV-Infected Primary Effusion Lymphoma Cell Lines. Cell Host Microbe 2011, 10, 515–526. [Google Scholar] [CrossRef]
- Jenner, R.G.; Albà, M.M.; Boshoff, C.; Kellam, P. Kaposi’s Sarcoma-Associated Herpesvirus Latent and Lytic Gene Expression as Revealed by DNA Arrays. J. Virol. 2001, 75, 891–902. [Google Scholar] [CrossRef]
- Cai, X.; Schäfer, A.; Lu, S.; Bilello, J.P.; Desrosiers, R.C.; Edwards, R.; Raab-Traub, N.; Cullen, B.R. Epstein–Barr Virus MicroRNAs Are Evolutionarily Conserved and Differentially Expressed. PLoS Pathog. 2006, 2, e23. [Google Scholar] [CrossRef]
- Mori, M.A.; Ludwig, R.G.; Garcia-Martin, R.; Brandao, B.B.; Kahn, C.R. Extracellular miRNAs: From Biomarkers to Mediators of Physiology and Disease. Cell Metab. 2019, 30, 656–673. [Google Scholar] [CrossRef] [PubMed]
- Hoshina, S.; Sekizuka, T.; Kataoka, M.; Hasegawa, H.; Hamada, H.; Kuroda, M.; Katano, H. Profile of Exosomal and Intracellular microRNA in Gamma-Herpesvirus-Infected Lymphoma Cell Lines. PLoS ONE 2016, 11, e0162574. [Google Scholar] [CrossRef] [PubMed]
- Devhare, P.B.; Sasaki, R.; Shrivastava, S.; Di Bisceglie, A.M.; Ray, R.; Ray, R.B. Exosome-Mediated Intercellular Communication between Hepatitis C Virus-Infected Hepatocytes and Hepatic Stellate Cells. J. Virol 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Lee, C.H.; Lee, S.W. Exosomal Transmission of MicroRNA from HCV Replicating Cells Stimulates Transdifferentiation in Hepatic Stellate Cells. Mol. Ther. Nucleic Acids 2019, 14, 483–497. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Salvi, V.; Gianello, V.; Busatto, S.; Bergese, P.; Andreoli, L.; D’Oro, U.; Zingoni, A.; Tincani, A.; Sozzani, S.; Bosisio, D. Exosome-delivered microRNAs promote IFN-alpha secretion by human plasmacytoid DCs via TLR7. JCI Insight 2018, 3, e98204. [Google Scholar] [CrossRef]
- Li, J.; Liu, K.; Liu, Y.; Xu, Y.; Zhang, F.; Yang, H.; Liu, J.; Pan, T.; Chen, J.; Wu, M.; et al. Exosomes mediate the cell-to-cell transmission of IFN-alpha-induced antiviral activity. Nat. Immunol. 2013, 14, 793–803. [Google Scholar] [CrossRef]
- Wei, Z.; Batagov, A.O.; Schinelli, S.; Wang, J.; Wang, Y.; El Fatimy, R.; Rabinovsky, R.; Balaj, L.; Chen, C.C.; Hochberg, F.; et al. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat. Commun. 2017, 8, 1–15. [Google Scholar] [CrossRef]
- Yogev, O.; Henderson, S.; Hayes, M.J.; Marelli, S.S.; Ofir-Birin, Y.; Regev-Rudzki, N.; Herrero, J.; Enver, T. Herpesviruses shape tumour microenvironment through exosomal transfer of viral microRNAs. PLoS Pathog. 2017, 13, e1006524. [Google Scholar] [CrossRef]
- McCann, J.V.; Liu, A.; Musante, L.; Erdbrugger, U.; Lannigan, J.; Dudley, A.C. A miRNA signature in endothelial cell-derived extracellular vesicles in tumor-bearing mice. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef]
- Shurtleff, M.J.; Temoche-Diaz, M.M.; Schekman, R. Extracellular Vesicles and Cancer: Caveat Lector. Annu. Rev. Cancer Biol. 2018, 2, 395–411. [Google Scholar] [CrossRef]
- de Jong, O.G.; Murphy, D.E.; Mager, I.; Willms, E.; Garcia-Guerra, A.; Gitz-Francois, J.J.; Lefferts, J.; Gupta, D.; Steenbeek, S.C.; van Rheenen, J.; et al. A CRISPR-Cas9-based reporter system for single-cell detection of extracellular vesicle-mediated functional transfer of RNA. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- O’Hara, A.J.; Wang, L.; Dezube, B.J.; Harrington, W.J.J.; Damania, B.; Dittmer, D.P. Tumor suppressor microRNAs are underrepresented in primary effusion lymphoma and Kaposi sarcoma. Blood 2009, 113, 5938–5941. [Google Scholar] [CrossRef] [PubMed]
- Gottwein, E.; Cullen, B.R. Viral and Cellular MicroRNAs as Determinants of Viral Pathogenesis and Immunity. Cell Host Mircobe 2008, 3, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Lu, S.; Zhang, Z.; Gonzalez, C.M.; Damania, B.; Cullen, B.R. Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. USA 2005, 102, 5570–5575. [Google Scholar] [CrossRef]
- Samols, M.A.; Hu, J.; Skalsky, R.L.; Renne, R. Cloning and Identification of a MicroRNA Cluster within the Latency-Associated Region of Kaposi’s Sarcoma-Associated Herpesvirus. J. Virol. 2005, 79, 9301–9305. [Google Scholar] [CrossRef]
- O’Hara, A.J.; Vahrson, W.; Dittmer, D.P. Gene alteration and precursor and mature microRNA transcription changes contribute to the miRNA signature of primary effusion lymphoma. Blood 2008, 111, 2347–2353. [Google Scholar] [CrossRef]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Narayanan, A.; Iordanskiy, S.; Das, R.; Van Duyne, R.; Santos, S.; Jaworski, E.; Guendel, I.; Sampey, G.; Dalby, E.; Iglesias-Ussel, M.; et al. Exosomes derived from HIV-1-infected cells contain trans-activation response element RNA. J. Biol. Chem. 2013, 288, 20014–20033. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and Exosomal MicroRNA: Trafficking, Sorting, and Function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Ono, M.; Kosaka, N.; Tominaga, N.; Yoshioka, Y.; Takeshita, F.; Takahashi, R.U.; Yoshida, M.; Tsuda, H.; Tamura, K.; Ochiya, T. Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Sci. Signal. 2014, 7, ra63. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.; Chin, A.R.; et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, J.; Chen, J.; Wang, H.; Yang, L.; Chen, F.; Fan, S.; Wang, J.; Shao, B.; Yin, D.; et al. A serum microRNA signature predicts trastuzumab benefit in HER2-positive metastatic breast cancer patients. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]

| Methods | Separation | Concentration | Scale Range |
|---|---|---|---|
| Ultracentrifugation | +++ | +++ | 5 to 250 mL |
| Normal flow filtration | + | ++ | 0.5 to 1000 mL |
| Tangential flow filtration | + | ++ | 100 to 5000 mL |
| Precipitation | - | +++ | 0.2 mL to >3 L |
| Size exclusion chromatography | + | - | 0.5 mL to >3 L |
| Ion exchange chromatography | ++ | + | 0.5 mL to >3 L |
| Affinity purification | ++++ | ++ | 0.5 mL to >3 L |
| Purification Method | Gradient or Size Limit | Reference | Virion/EV Separation * | Virus | Detected RNA |
|---|---|---|---|---|---|
| Dextran gradient centrifugation | 1.04–1.09 g/cm3 | [4,5] | - | HSV-1 | mRNA |
| Sucrose gradient centrifugation | 35%, 30%–60% | [6] | - | HCMV | vRNA |
| 20%–40% | [7] | Detergent treatment, Infectivity | HCMV | mRNA | |
| 20%, 10%–55% | [8] | - | MHV-68 | vtRNA | |
| 30%–60% | [9] | Banding, WB | KSHV | miRNA, usRNA | |
| Histodenz gradient centrifugation | 20%–35% | [10] | - | KSHV | mRNA |
| CsCl gradient centrifugation | n.a. | [12] | - | Adenovirus | mRNA |
| [13] | - | Adenovirus | mRNA | ||
| Sorbitol cushion centrifugation | 20% | [11] | - | HBV | miRNA |
| Sorbitol cushion, Glycerol-tartrate gradient, CsCl gradient centrifugation | n.a. | [14] | Banding | HCMV | mRNA |
| Filtration | 0.8 µm | [15] | - | EBV | mRNA, non-coding RNA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; McNamara, R.P.; Dittmer, D.P. Purification Methods and the Presence of RNA in Virus Particles and Extracellular Vesicles. Viruses 2020, 12, 917. https://doi.org/10.3390/v12090917
Zhou Y, McNamara RP, Dittmer DP. Purification Methods and the Presence of RNA in Virus Particles and Extracellular Vesicles. Viruses. 2020; 12(9):917. https://doi.org/10.3390/v12090917
Chicago/Turabian StyleZhou, Yijun, Ryan P. McNamara, and Dirk P. Dittmer. 2020. "Purification Methods and the Presence of RNA in Virus Particles and Extracellular Vesicles" Viruses 12, no. 9: 917. https://doi.org/10.3390/v12090917
APA StyleZhou, Y., McNamara, R. P., & Dittmer, D. P. (2020). Purification Methods and the Presence of RNA in Virus Particles and Extracellular Vesicles. Viruses, 12(9), 917. https://doi.org/10.3390/v12090917





