Characterisation of SARS-CoV-2 Lentiviral Pseudotypes and Correlation between Pseudotype-Based Neutralisation Assays and Live Virus-Based Micro Neutralisation Assays
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Line Cultures
2.2. Serum Samples
2.3. Production, Quantification and Characterisation of Lentiviral Pseudotypes with S Protein from SARS-CoV-2
2.3.1. Plasmids
2.3.2. SARS-CoV-2 Pseudotypes Titration
2.4. p24 Capsid ELISA
2.5. Western Blotting
2.6. SARS-CoV-2 Pseudotypes Tropism Study
2.7. Pseudotype-Based Neutralisation Assays
2.8. Live Virus, Titration and Microneutralisation Assay
2.9. Statistical Analyses
2.9.1. Calculation of SARS-CoV-2 Pseudotype Titres
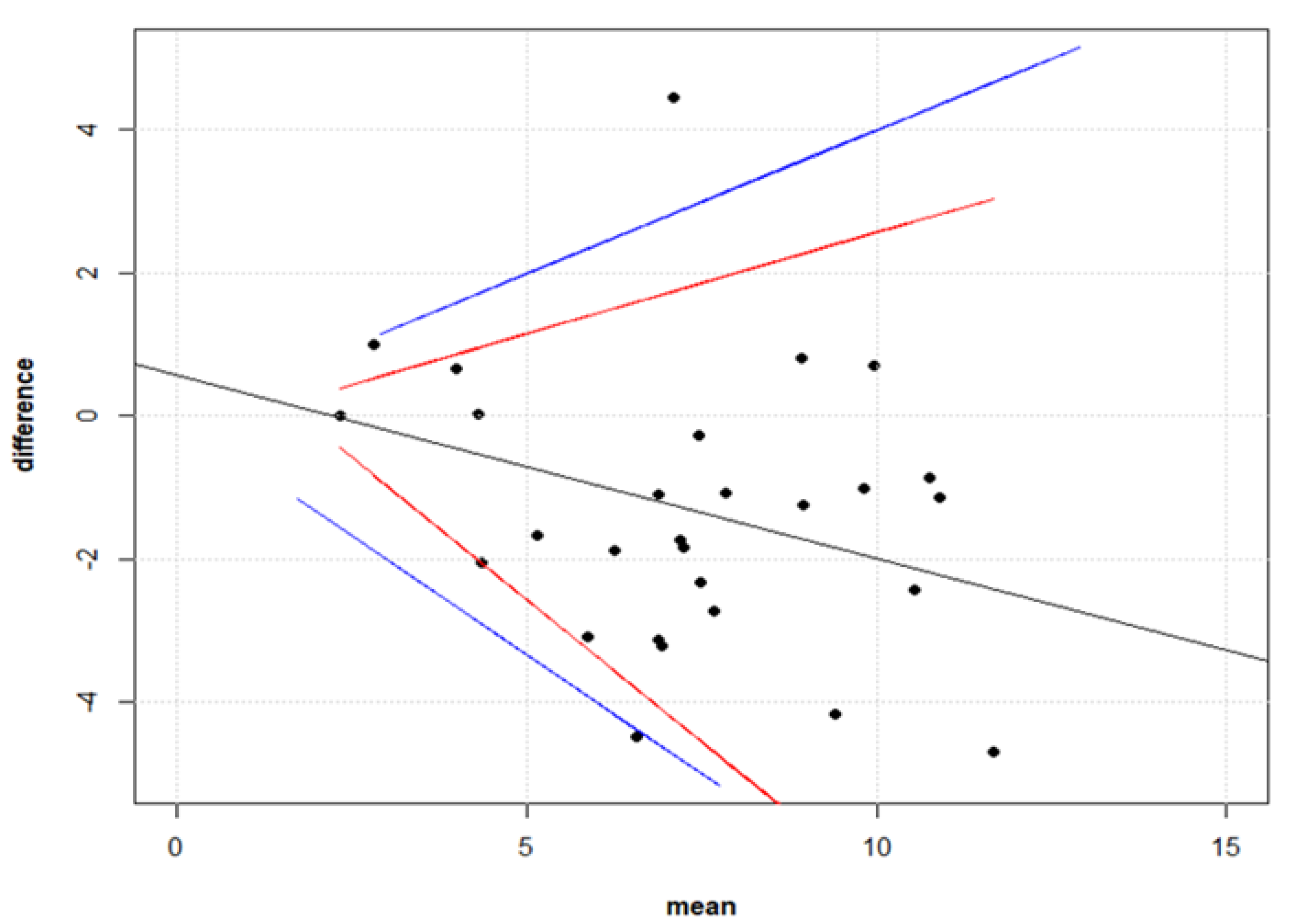
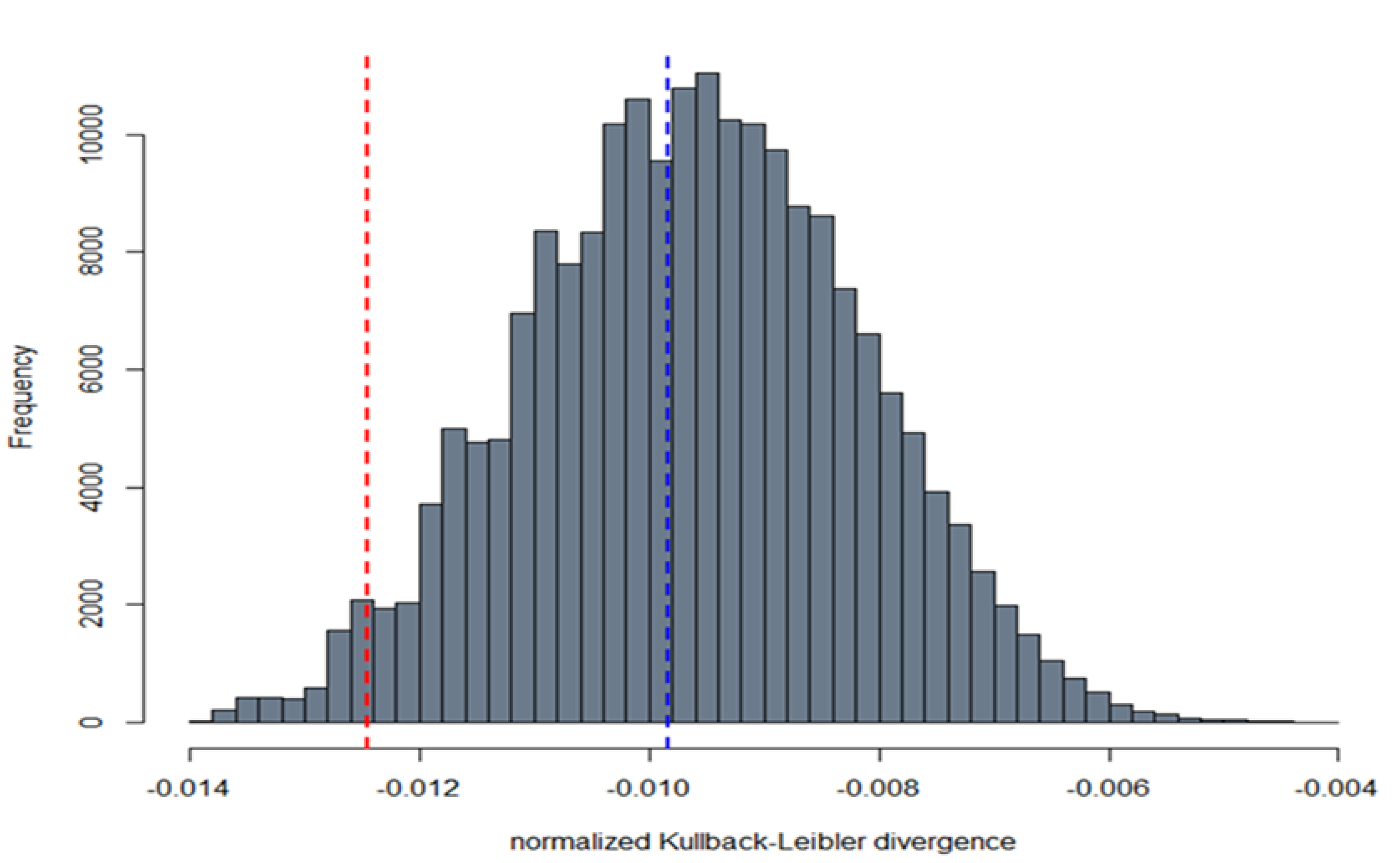
2.9.2. Comparison between Live Virus Microneutralisation Titres (MNT) and Pseudotype Neutralising Titres (PNT)
3. Results
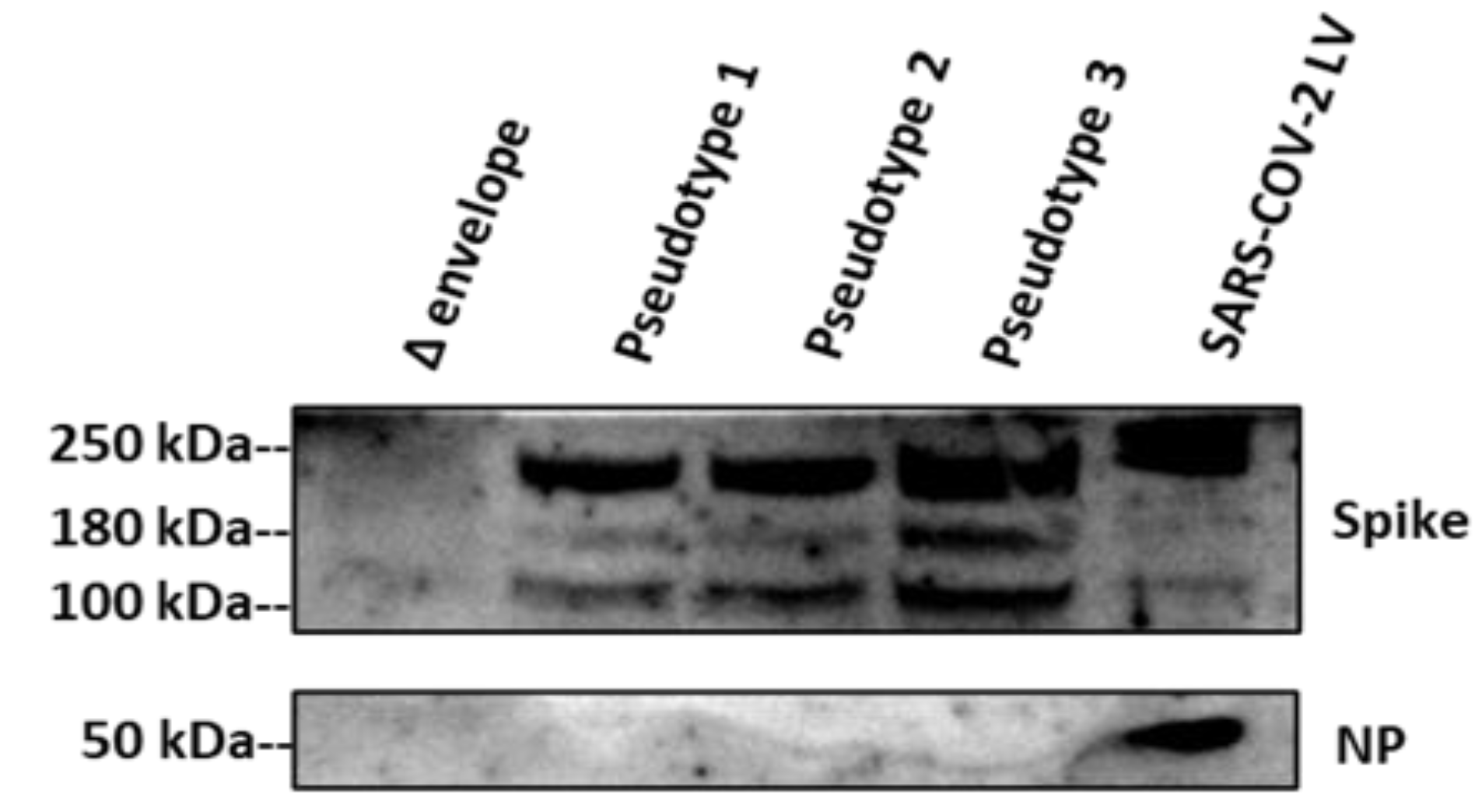
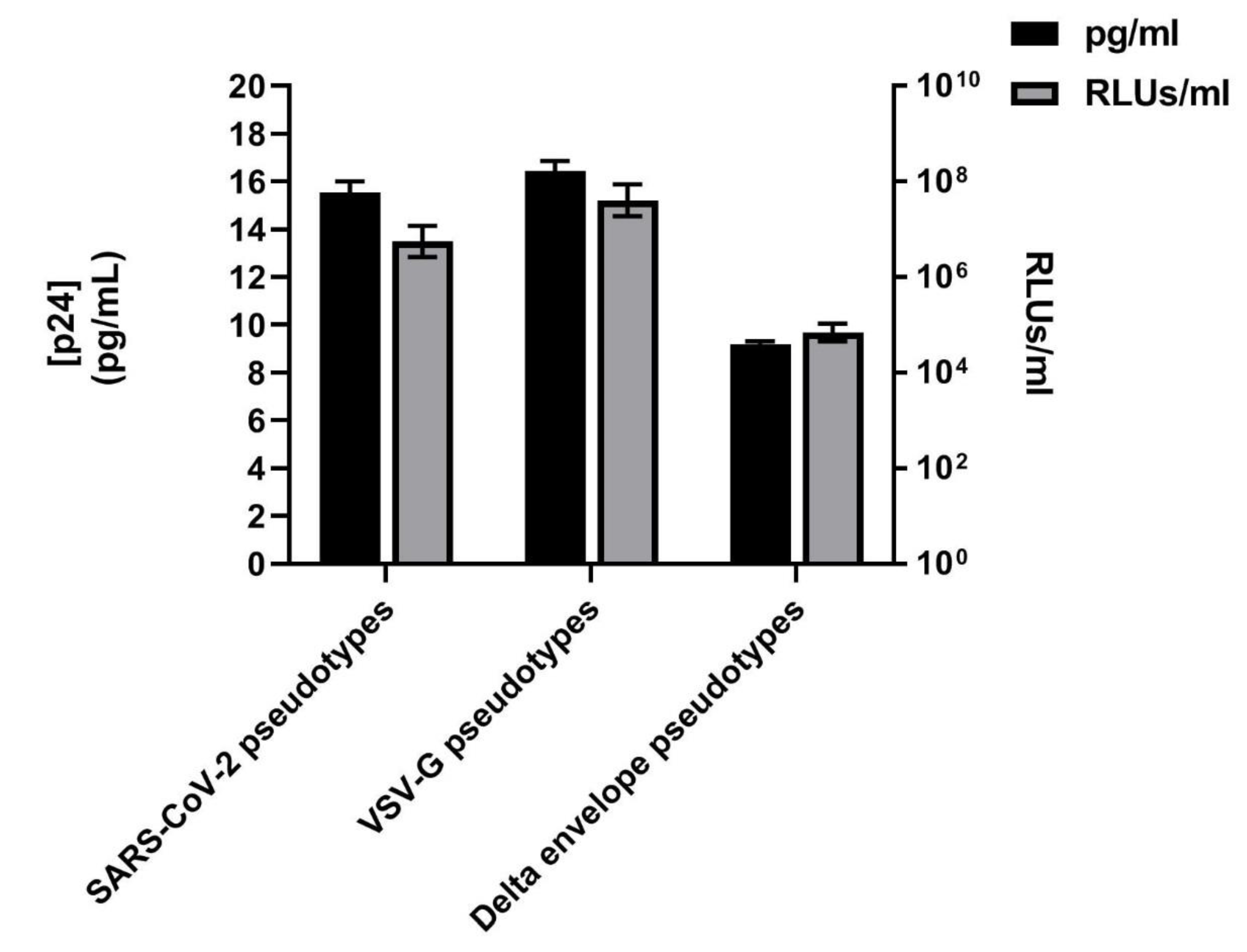
3.1. SARS-CoV-2 Spike Protein Expression Evaluated by Western Blotting and p24 Quantification
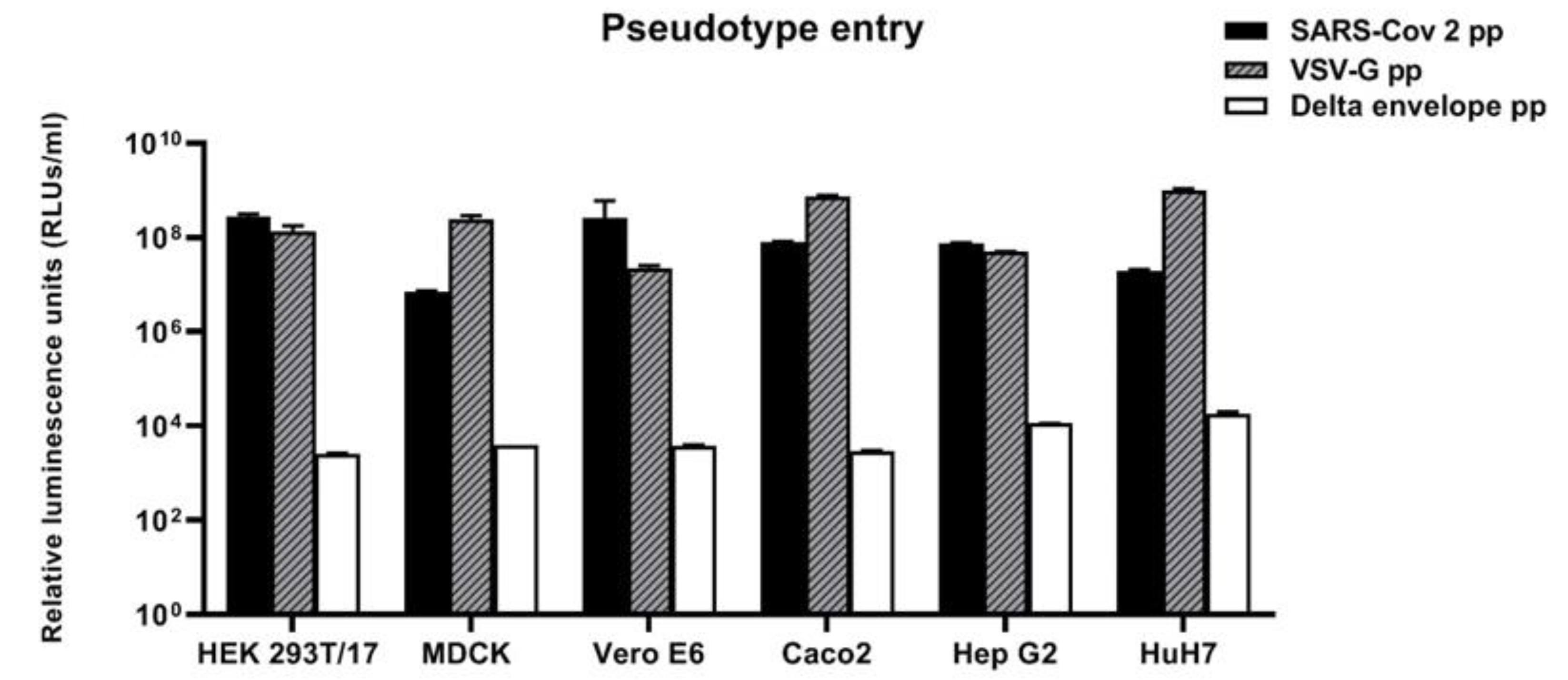
3.2. Susceptibility of Cell Lines Panel to SARS-CoV-2 for Pseudotype-Based Neutralisation Assays
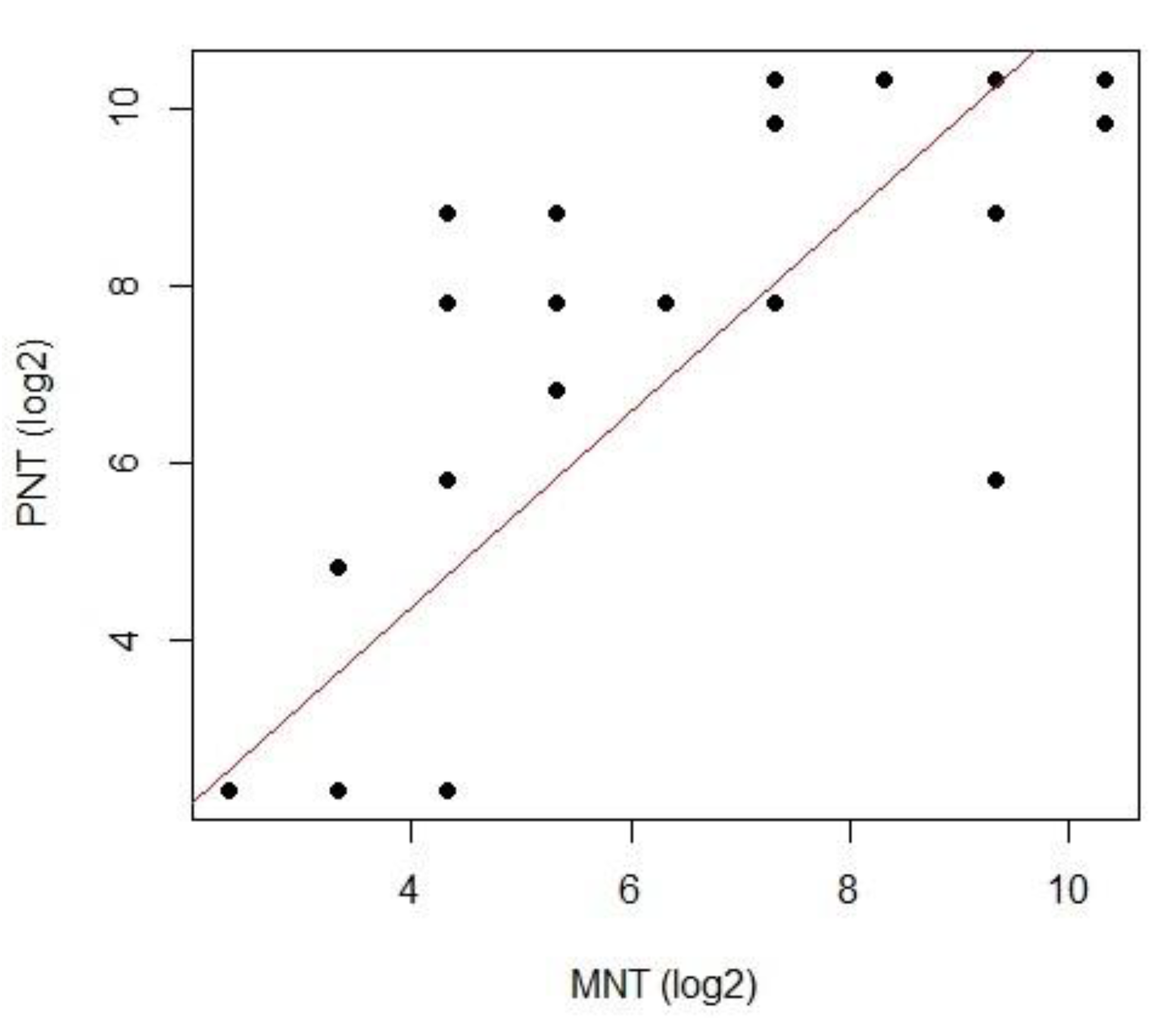
3.3. Correlation between Live Virus MN and Pseudotype-Based Neutralisation Assays
4. Discussions
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chan, J.F.-W.; Yuan, S.; Kok, K.-H.; To, K.K.-W.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.-Y.; Poon, R.W.-S.; et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef] [Green Version]
- WHO 2020, Coronavirus Disease 2019 (COVID19) Situation Report 23. Coronavirus Disease 2019 (COVID-19) Situation Report—23. Available online: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200212-sitrep-23-ncov.pdf?sfvrsn=41e9fb78_4 (accessed on 10 July 2020).
- Fehr, A.R.; Perlman, S. Coronaviruses: An Overview of Their Replication and Pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wormser, G.P.; Aitken, C. Clinical Virology, 3rd ed.; Richman, D.D., Whitley, R.J., Hayden, F.G., Eds.; ASM Press: Washington, DC, USA, 2009; 1408p. [Google Scholar]
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Meng, J.; Zhu, Z.; Zhang, Z.; Wang, J.; et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe 2020, 27, 325–328. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Zhang, C.; Sui, J.; Kuhn, J.H.; Moore, M.J.; Luo, S.; Wong, S.K.; Huang, I.C.; Xu, K.; Vasilieva, N.; et al. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005, 24, 1634–1643. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Li, W.; Farzan, M.; Harrison, S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 2005, 309, 1864–1868. [Google Scholar] [CrossRef]
- Zhang, H.; Penninger, J.M.; Li, Y.; Zhong, N.; Slutsky, A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: Molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020, 46, 586–590. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Glowacka, I.; Bertram, S.; Müller, M.A.; Allen, P.; Soilleux, E.; Pfefferle, S.; Steffen, I.; Tsegaye, T.S.; He, Y.; Gnirss, K.; et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011, 85, 4122–4134. [Google Scholar] [CrossRef] [Green Version]
- Wong, S.K.; Li, W.; Moore, M.J.; Choe, H.; Farzan, M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004, 279, 3197–3201. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Xiao, G.; Chen, Y.; He, Y.; Niu, J.; Escalante, C.R.; Xiong, H.; Farmar, J.; Debnath, A.K.; Tien, P.; et al. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: Implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet 2004, 363, 938–947. [Google Scholar] [CrossRef] [Green Version]
- Simmons, G.; Gosalia, D.N.; Rennekamp, A.J.; Reeves, J.D.; Diamond, S.L.; Bates, P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA 2005, 102, 11876–11881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosch, B.J.; Martina, B.E.E.; Van Der Zee, R.; Lepault, J.; Haijema, B.J.; Versluis, C.; Heck, A.J.R.; De Groot, R.; Osterhaus, A.D.M.E.; Rottier, P.J.M. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. USA 2004, 101, 8455–8460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, F.; Wang, A.; Liu, M.; Wang, Q.; Chen, J.; Xia, S.; Ling, Y.; Zhang, Y.; Xun, J.; Lu, L.; et al. Neutralizing Antibody Responses to SARS-CoV-2 in a COVID-19 Recovered Patient Cohort and Their Implications. medRxiv 2020. [Google Scholar] [CrossRef]
- Cheng, H.Y.; Jian, S.W.; Liu, D.P.; Ng, T.C.; Huang, W.T.; Lin, H.H. High transmissibility of COVID-19 near symptom onset. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Crawford, K.H.D.; Eguia, R.; Dingens, A.S.; Loes, A.N.; Malone, K.D.; Wolf, C.R.; Chu, H.Y.; Tortorici, M.A.; Veesler, D.; Murphy, M.; et al. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses 2020, 12, 513. [Google Scholar] [CrossRef]
- Millet, J.K.; Tang, T.; Nathan, L.; Jaimes, J.A.; Hsu, H.-L.; Daniel, S.; Whittaker, G.R. Production of Pseudotyped Particles to Study Highly Pathogenic Coronaviruses in a Biosafety Level 2 Setting. J. Vis. Exp. 2019, 145, e59010. [Google Scholar] [CrossRef] [Green Version]
- Nie, J.; Li, Q.; Wu, J.; Zhao, C.; Hao, H.; Liu, H.; Zhang, L.; Nie, L.; Qin, H.; Wang, M.; et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microbes Infect. 2020, 9, 680–686. [Google Scholar] [CrossRef] [Green Version]
- Ju, B.; Zhang, Q.; Ge, X.; Wang, R.; Yu, J.; Shan, S.; Zhou, B.; Song, S.; Tang, X.; Yu, J.; et al. Potent human neutralizing antibodies elicited by SARS-CoV-2 infection. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Trombetta, C.M.; Remarque, E.J.; Mortier, D.; Montomoli, E. Comparison of hemagglutination inhibition, single radial hemolysis, virus neutralization assays, and ELISA to detect antibody levels against seasonal influenza viruses. Influ. Other Respir. Viruses 2018, 12, 675–686. [Google Scholar] [CrossRef]
- Manenti, A.; Maciola, A.K.; Trombetta, C.M.; Kistner, O.; Casa, E.; Hyseni, I.; Razzano, I.; Torelli, A.; Montomoli, E. Influenza Anti-Stalk Antibodies: Development of a New Method for the Evaluation of the Immune Responses to Universal Vaccine. Vaccines 2020, 8, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manenti, A.; Maggetti, M.; Casa, E.; Martinuzzi, D.; Torelli, A.; Trombetta, C.M.; Marchi, S.; Montomoli, E. Evaluation of SARS-CoV-2 neutralizing antibodies using a CPE-based colorimetric live virus micro-neutralization assay in human serum samples. J. Med Virol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.; Grayson, N.; Paton, R.; Lourenço, J.; Penman, B.; Lee, L.N.; Odon, V.; Mongkolsapaya, J.; Chinnakannan, S.; Dejnirattisai, W.; et al. Neutralising antibodies to SARS coronavirus 2 in Scottish blood donors—A pilot study of the value of serology to determine population exposure. medRxiv 2020. [Google Scholar] [CrossRef]
- Zufferey, R.; Nagy, D.; Mandel, R.J.; Naldini, L.; Trono, D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 1997, 15, 871–875. [Google Scholar] [CrossRef]
- Zufferey, R.; Dull, T.; Mandel, R.J.; Bukovsky, A.; Quiroz, D.; Naldini, L.; Trono, D. Self-Inactivating Lentivirus Vector for Safe and Efficient In Vivo Gene Delivery. J. Virol. 1998, 72, 9873–9880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Böttcher, E.; Matrosovich, T.; Beyerle, M.; Klenk, H.-D.; Garten, W.; Matrosovich, M. Proteolytic Activation of Influenza Viruses by Serine Proteases TMPRSS2 and HAT from Human Airway Epithelium. J. Virol. 2006, 80, 9896–9898. [Google Scholar] [CrossRef] [Green Version]
- Temperton, N.; Chan, P.K.; Simmons, G.; Zambon, M.C.; Tedder, R.S.; Takeuchi, Y.; Weiss, R.A. Longitudinally Profiling Neutralizing Antibody Response to SARS Coronavirus with Pseudotypes. Emerg. Infect. Dis. 2005, 11, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Grehan, K.; Ferrara, F.; Temperton, N. An optimised method for the production of MERS-CoV spike expressing viral pseudotypes. MethodsX 2015, 2, 379–384. [Google Scholar] [CrossRef]
- Stewart, S.A.; Dykxhoorn, D.M.; Palliser, D.; Mizuno, H.; Yu, E.Y.; An, N.S.; Sabatini, D.M.; Chen, I.S.; Hahn, W.C.; Sharp, P.A.; et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 2003, 9, 493–501. [Google Scholar] [CrossRef] [Green Version]
- Kundi, M. One-hit models for virus inactivation studies. Antivir. Res. 1999, 41, 145–152. [Google Scholar] [CrossRef]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shajahan, A.; Supekar, N.T.; Gleinich, A.; Azadi, P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology 2020. [Google Scholar] [CrossRef] [PubMed]
- Geraerts, M.; Willems, S.; Baekelandt, V.; Debyser, Z.; Gijsbers, R. Comparison of lentiviral vector titration methods. BMC Biotechnol. 2006, 6, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Material not intended for publication: No. Author. Affiliation, City, State. FACS analysis, 2020.
- Amanat, F.; Stadlbauer, D.; Strohmeier, S.; Nguyen, T.H.O.; Chromikova, V.; McMahon, M.; Jiang, K.; Arunkumar, G.A.; Jurczyszak, D.; Polanco, J.; et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020, 26, 1033–1036. [Google Scholar] [CrossRef] [PubMed]
- Cicchetti, D.V. Guidelines, criteria, and rules of thumb for evaluating normed and standardized assessment instruments in psychology. Psychol. Assess. 1994, 6, 284–290. [Google Scholar] [CrossRef]
- Bland, J.M.; Altman, U.G. Measuring agreement in method comparison studies. Stat. Methods Med. Res. 1999, 8, 135–160. [Google Scholar] [CrossRef]
- Bootz, A.; Karbach, A.; Spindler, J.; Kropff, B.; Reuter, N.; Sticht, H.; Winkler, T.H.; Britt, W.J.; Mach, M. Protective capacity of neutralizing and non-neutralizing antibodies against glycoprotein B of cytomegalovirus. PLoS Pathog. 2017, 13, e1006601. [Google Scholar] [CrossRef]
- Jiang, S.; Hillyer, C.; Du, L. Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends Immunol. 2020, 41, 355–359. [Google Scholar] [CrossRef]
- Milani, G.P.; Montomoli, E.; Bollati, V.; Albetti, B.; Bandi, C.; Bellini, T.; Bonzini, M.; Buscaglia, M.; Cantarella, C.; Cantone, L.; et al. SARS-CoV-2 infection among asymptomatic homebound subjects in Milan, Italy. Eur. J. Intern. Med. 2020, 78, 161–163. [Google Scholar] [CrossRef]
- Perera, R.A.; Mok, C.K.; Tsang, O.T.; Lv, H.; Ko, R.L.; Wu, N.C.; Yuan, M.; Leung, W.S.; Mc Chan, J.; Chik, T.S.; et al. Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), March 2020. Eurosurveillance 2020, 25, 2000421. [Google Scholar] [CrossRef] [Green Version]
- Seow, J.; Graham, C.; Merrick, B.; Acors, S.; Steel, K.J.; Hemmings, O.; O’Bryne, A.; Kouphou, N.; Pickering, S.; Galao, R.; et al. Longitudinal evaluation and decline of antibody responses in SARS-CoV-2 infection. MedRxiv 2020. [Google Scholar] [CrossRef]
- Molesti, E.; Cattoli, G.; Ferrara, F.; Böttcher-Friebertshäuser, E.; Terregino, C.; Temperton, N. The production and development of H7 Influenza virus pseudotypes for the study of humoral responses against avian viruses. J. Mol. Genet. Med. Int. J. Biomed. Res. 2013, 7, 315–320. [Google Scholar] [CrossRef]
- Li, Q.; Liu, Q.; Huang, W.; Li, X.; Wang, Y. Current status on the development of pseudoviruses for enveloped viruses. Rev. Med. Virol. 2018, 28, e1963. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef] [Green Version]
- Case, J.B.; Rothlauf, P.W.; Chen, R.E.; Liu, Z.; Zhao, H.; Kim, A.S.; Bloyet, L.-M.; Zeng, Q.; Tahan, S.; Droit, L.; et al. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host Microbe 2020. [Google Scholar] [CrossRef] [PubMed]
- Alberini, I.; Del Tordello, E.; Fasolo, A.; Temperton, N.; Galli, G.; Gentile, C.; Montomoli, E.; Hilbert, A.K.; Banzhoff, A.; Del Giudice, G.; et al. Pseudoparticle neutralization is a reliable assay to measure immunity and cross-reactivity to H5N1 influenza viruses. Vaccine 2009, 27, 5998–6003. [Google Scholar] [CrossRef] [PubMed]
- Molesti, E.; Wright, E.; Terregino, C.; Rahman, R.; Cattoli, G.; Temperton, N. Multiplex Evaluation of Influenza Neutralizing Antibodies with Potential Applicability to In-Field Serological Studies. J. Immunol. Res. 2014, 2014, 457932. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Butler, E.N.; Veguilla, V.; Vassell, R.; Thomas, J.T.; Moos, M.; Ye, Z.; Hancock, K.; Weiss, C.D. Establishment of retroviral pseudotypes with influenza hemagglutinins from H1, H3, and H5 subtypes for sensitive and specific detection of neutralizing antibodies. J. Virol. Methods 2008, 153, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Garcia, J.-M.; Lagarde, N.; Ma, E.S.; De Jong, M.D.; Peiris, J.M. Optimization and evaluation of an influenza A (H5) pseudotyped lentiviral particle-based serological assay. J. Clin. Virol. 2010, 47, 29–33. [Google Scholar] [CrossRef] [Green Version]
- Wright, E.; Temperton, N.; Marston, D.A.; McElhinney, L.M.; Fooks, A.R.; Weiss, R.A. Investigating antibody neutralization of lyssaviruses using lentiviral pseudotypes: A cross-species comparison. J. Gen. Virol. 2008, 89, 2204–2213. [Google Scholar] [CrossRef] [Green Version]
- Logan, N.; McMonagle, E.; Drew, A.A.; Takahashi, E.; McDonald, M.; Baron, M.; Gilbert, M.; Cleaveland, S.; Haydon, D.T.; Hosie, M.J.; et al. Efficient generation of vesicular stomatitis virus (VSV)-pseudotypes bearing morbilliviral glycoproteins and their use in quantifying virus neutralising antibodies. Vaccine 2016, 34, 814–822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The United States Pharmacopeial Convention. Design and Development of Biological Assays; The United States Pharmacopeial Convention: Rockville, MD, USA, 2010; Volume 1032, pp. 1–36. [Google Scholar]
- The United States Pharmacopeial Convention. Biological Assay Validation; The United States Pharmacopeial Convention: Rockville, MD, USA, 2010; Volume 1033, pp. 1–25. [Google Scholar]

| MNT | |||
|---|---|---|---|
| PNT | Negative | Positive | Total |
| Negative | 37 | 4 | 41 |
| Positive | 0 | 24 | 24 |
| Total | 37 | 28 | 65 |
| ID Samples | Live Virus Micro | Pseudotype Neutralisation |
|---|---|---|
| Neutralisation Titres (MNT) | Titres (PNT) | |
| 1 | 5 | <10 |
| 2 | 5 | <10 |
| 3 | 5 | <10 |
| 4 | 5 | <10 |
| 5 | 5 | <10 |
| 6 | 5 | <10 |
| 7 | 5 | <10 |
| 8 | 5 | <10 |
| 9 | 5 | <10 |
| 10 | 5 | <10 |
| 11 | 10 | 10–20 |
| 12 | 10 | <10 |
| 13 | 10 | <10 |
| 14 | 20 | 40–80 |
| 15 | 20 | 40–80 |
| 16 | 20 | <10 |
| 17 | 20 | <10 |
| 18 | 20 | 160–320 |
| 19 | 40 | 160–320 |
| 20 | 40 | 160–320 |
| 21 | 40 | 80–160 |
| 22 | 80 | 160–320 |
| 23 | 80 | 160–320 |
| 24 | 80 | 320–640 |
| 25 | 80 | 160–320 |
| 26 | 80 | 160–320 |
| 27 | 160 | 160–320 |
| 28 | 160 | 160–320 |
| 29 | 160 | >1280 |
| 30 | 320 | 640–1280 |
| 31 | 640 | >1280 |
| 32 | 640 | 20–40 |
| 33 | 640 | 320–640 |
| 34 | 640 | >1280 |
| 35 | 640 | >1280 |
| 36 | 1280 | >1280 |
| 37 | 1280 | >1280 |
| 38 | 1280 | 640–1280 |
| 39 | 5 | <10 |
| 40 | 5 | <10 |
| 41 | 5 | <10 |
| 42 | 5 | <10 |
| 43 | 5 | <10 |
| 44 | 5 | <10 |
| 45 | 5 | <10 |
| 46 | 5 | <10 |
| 47 | 5 | <10 |
| 48 | 5 | <10 |
| 49 | 5 | <10 |
| 50 | 5 | <10 |
| 51 | 5 | <10 |
| 52 | 5 | <10 |
| 53 | 5 | <10 |
| 54 | 5 | <10 |
| 55 | 5 | <10 |
| 56 | 5 | <10 |
| 57 | 5 | <10 |
| 58 | 5 | <10 |
| 59 | 5 | <10 |
| 60 | 5 | <10 |
| 61 | 5 | <10 |
| 62 | 5 | <10 |
| 63 | 5 | <10 |
| 64 | 5 | <10 |
| 65 | 5 | <10 |
| Model | one-way |
| Type | agreement |
| Subjects | 65 |
| Raters | 2 |
| ICC(1) | 0.872 |
| CI 95% | [0.799–0.92] |
| F-test | 14.7 |
| p | <0.0001 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hyseni, I.; Molesti, E.; Benincasa, L.; Piu, P.; Casa, E.; Temperton, N.J.; Manenti, A.; Montomoli, E. Characterisation of SARS-CoV-2 Lentiviral Pseudotypes and Correlation between Pseudotype-Based Neutralisation Assays and Live Virus-Based Micro Neutralisation Assays. Viruses 2020, 12, 1011. https://doi.org/10.3390/v12091011
Hyseni I, Molesti E, Benincasa L, Piu P, Casa E, Temperton NJ, Manenti A, Montomoli E. Characterisation of SARS-CoV-2 Lentiviral Pseudotypes and Correlation between Pseudotype-Based Neutralisation Assays and Live Virus-Based Micro Neutralisation Assays. Viruses. 2020; 12(9):1011. https://doi.org/10.3390/v12091011
Chicago/Turabian StyleHyseni, Inesa, Eleonora Molesti, Linda Benincasa, Pietro Piu, Elisa Casa, Nigel J Temperton, Alessandro Manenti, and Emanuele Montomoli. 2020. "Characterisation of SARS-CoV-2 Lentiviral Pseudotypes and Correlation between Pseudotype-Based Neutralisation Assays and Live Virus-Based Micro Neutralisation Assays" Viruses 12, no. 9: 1011. https://doi.org/10.3390/v12091011
APA StyleHyseni, I., Molesti, E., Benincasa, L., Piu, P., Casa, E., Temperton, N. J., Manenti, A., & Montomoli, E. (2020). Characterisation of SARS-CoV-2 Lentiviral Pseudotypes and Correlation between Pseudotype-Based Neutralisation Assays and Live Virus-Based Micro Neutralisation Assays. Viruses, 12(9), 1011. https://doi.org/10.3390/v12091011






