Expansion and Refinement of Deep Sequence-Coupled Biopanning Technology for Epitope-Specific Antibody Responses in Human Serum
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Serum Samples
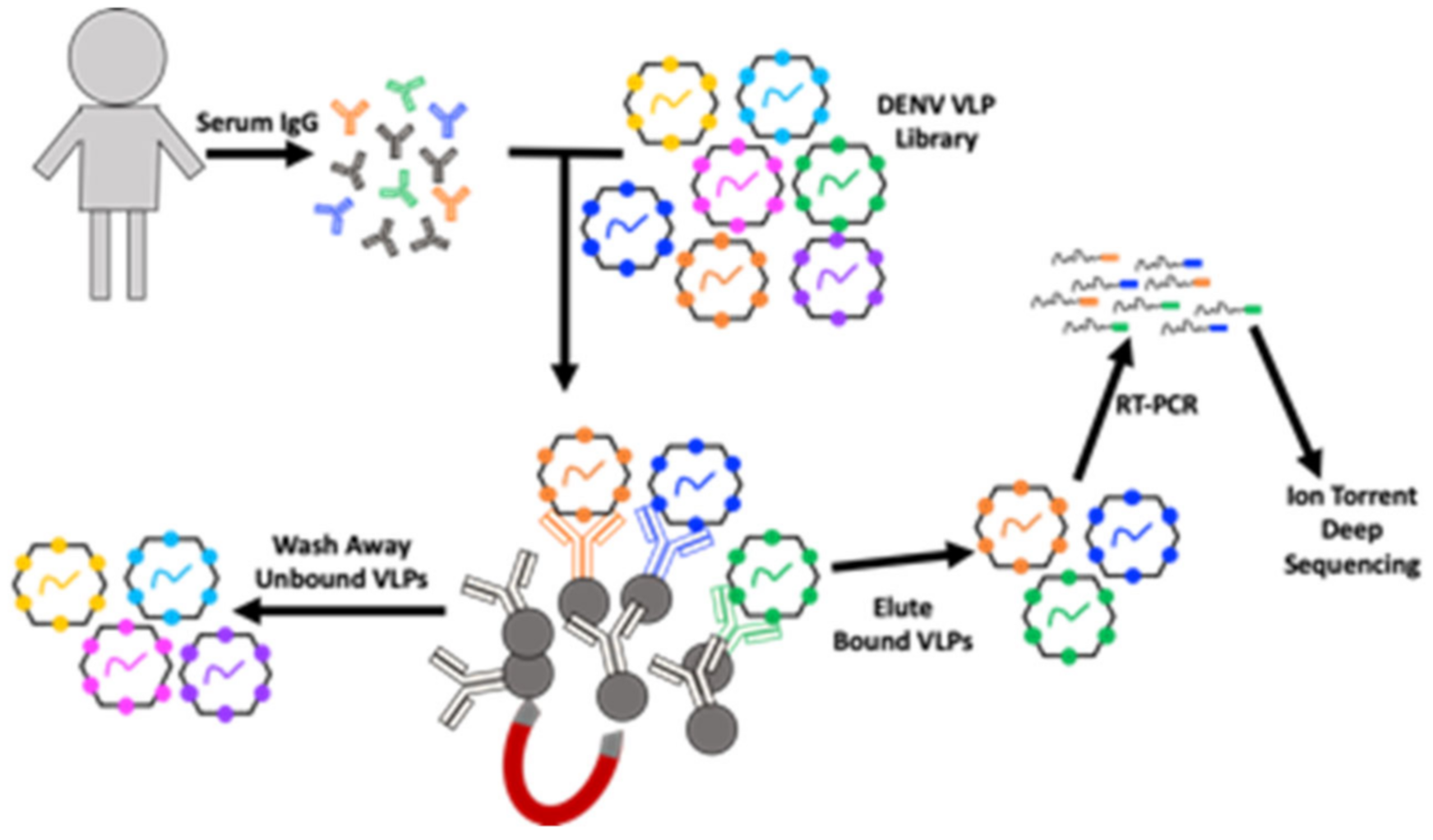
2.2. Deep Sequence-Coupled Biopanning (DSCB)
2.3. Structure and Alignments
2.4. Synthetic Peptide ELISA
2.5. MATLAB Scripts and Statistical Analysis
3. Results
3.1. Modifications to DSCB Protocol
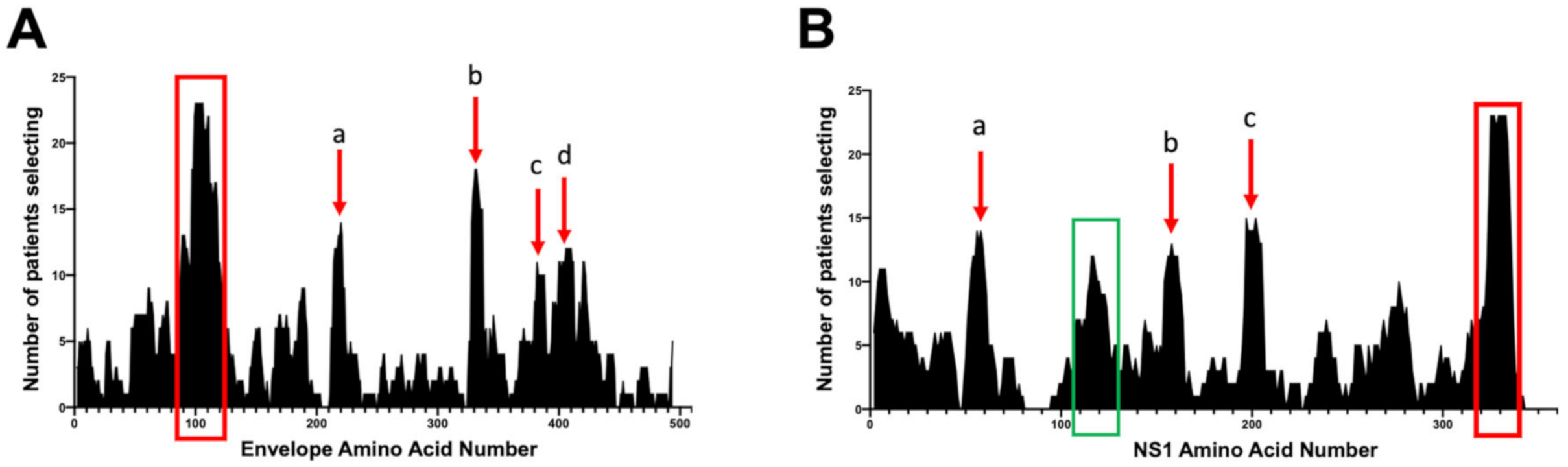
3.2. Comparison of Sera from 26 Primary DENV- Infected Individuals for Commonly Selected DENV-NS1 and Envelope Epitopes
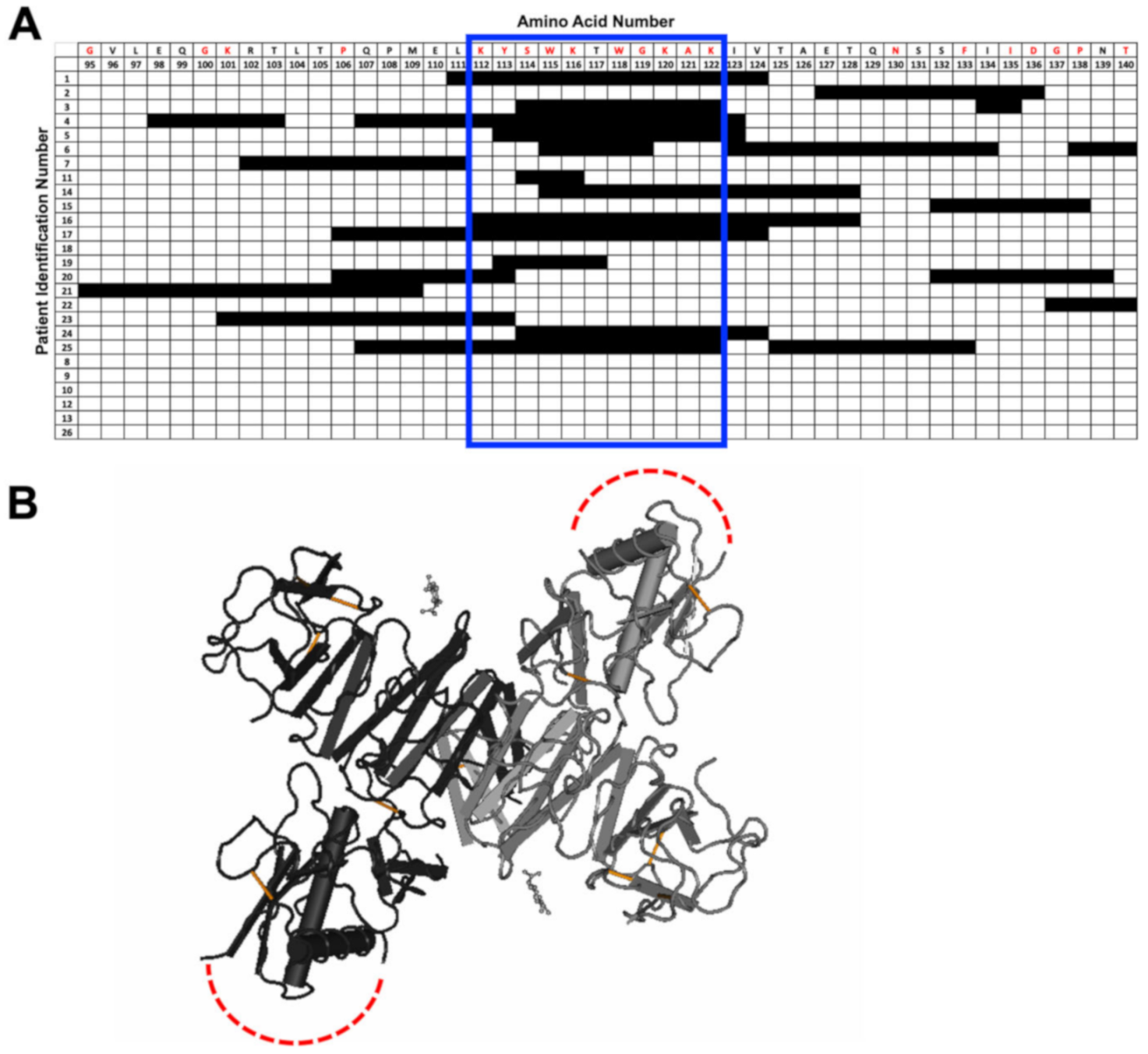
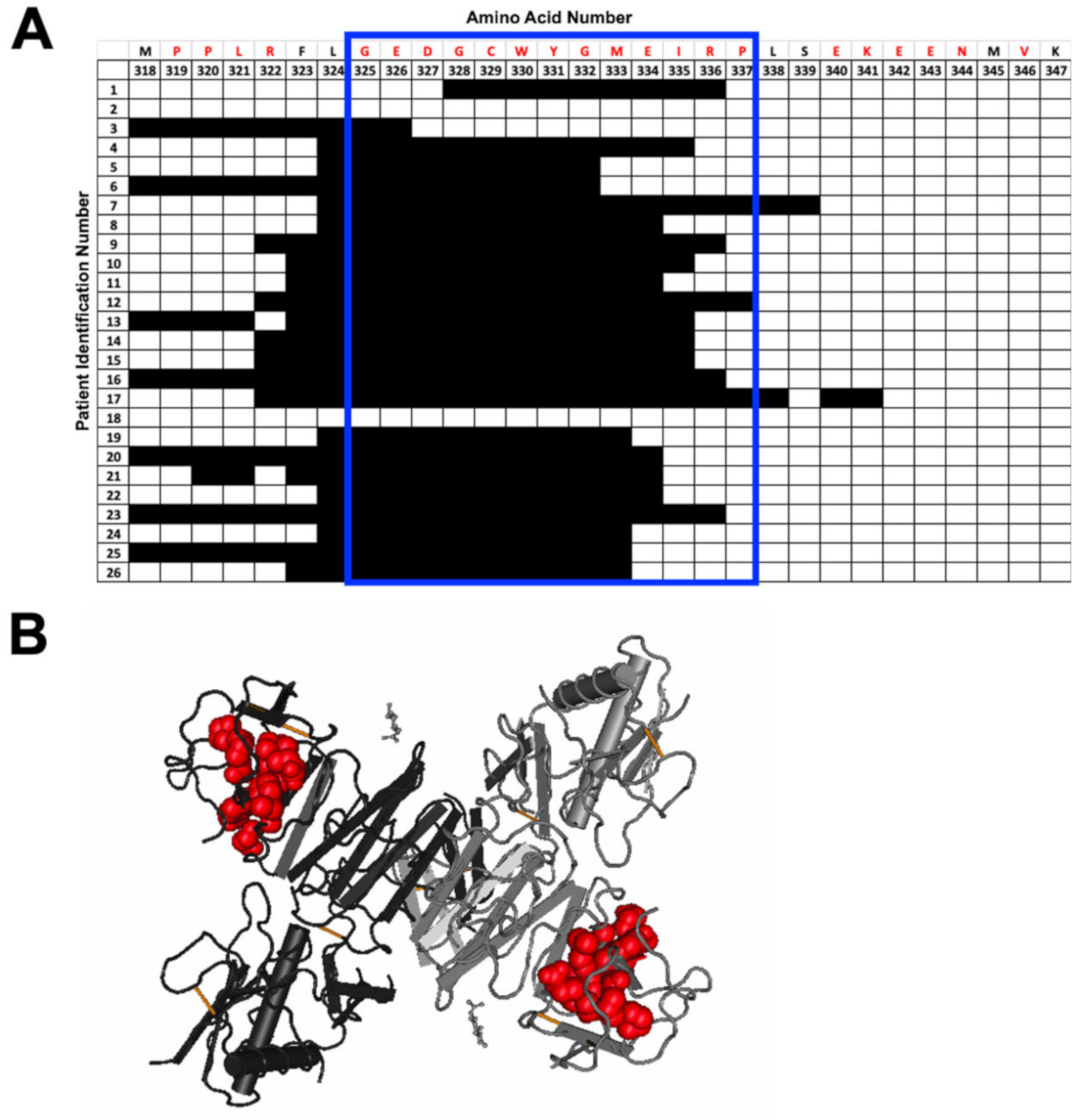
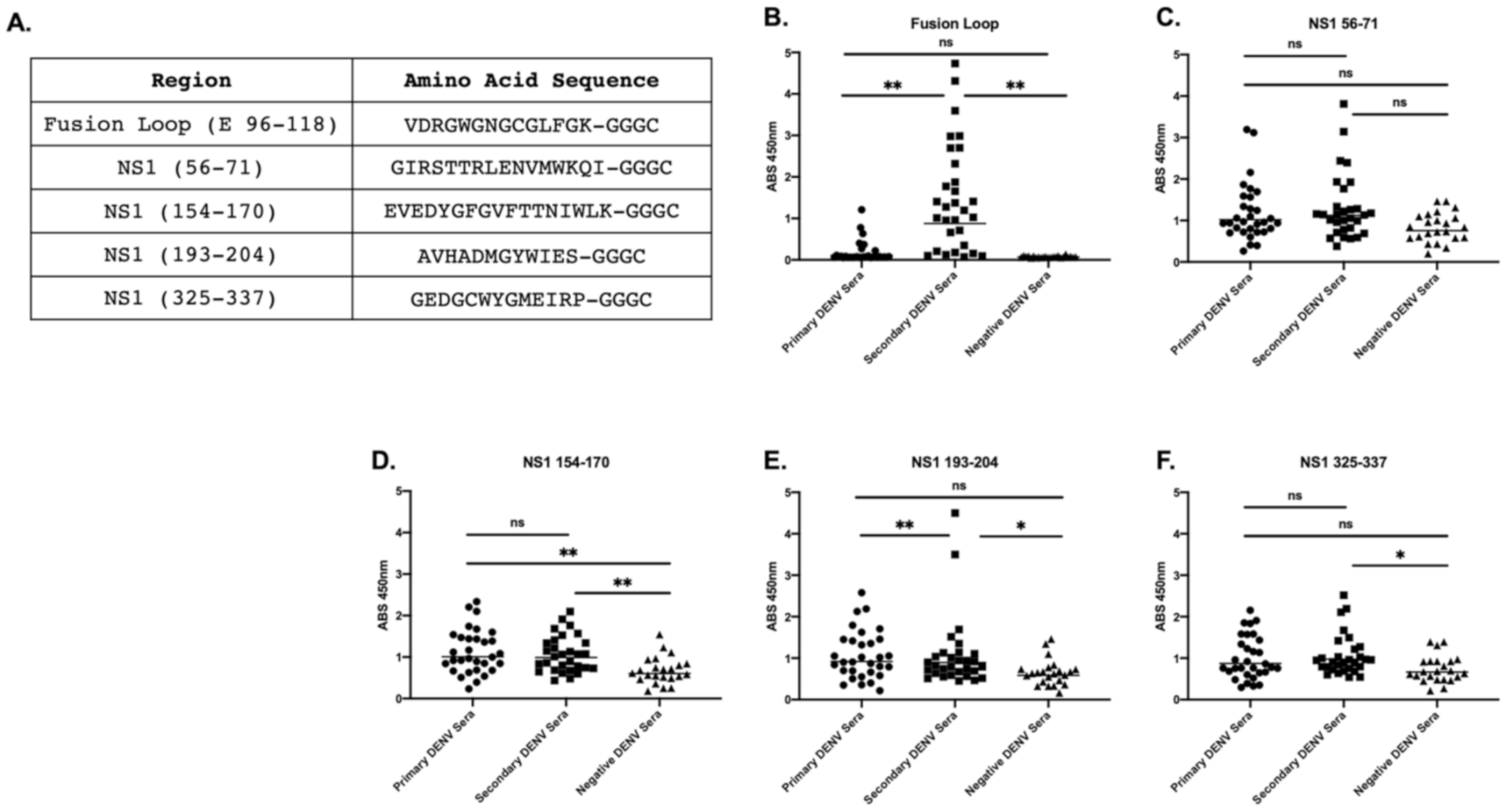
3.3. Peptide-Specific Antibody Responses to Commonly Selected Epitopes within NS1 and E
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Frietze, K.M.; Roden, R.B.S.; Lee, J.-H.; Shi, Y.; Peabody, D.S.; Chackerian, B. Identification of anti-CA125 antibody responses in ovarian cancer patients by a novel deep sequence-coupled biopanning platform. Cancer Immunol. Res. 2016, 4, 157–164. [Google Scholar] [CrossRef]
- Frietze, K.M.; Pascale, J.M.; Moreno, B.; Chackerian, B.; Peabody, D.S. Pathogen-specific deep sequence-coupled biopanning: A method for surveying human antibody responses. PLoS ONE 2017, 12, e0171511. [Google Scholar] [CrossRef] [PubMed]
- Collar, A.L.; Linville, A.C.; Core, S.B.; Wheeler, C.M.; Geisler, W.M.; Peabody, D.S.; Chackerian, B.; Frietze, K.M. Antibodies to variable domain 4 linear epitopes of the Chlamydia trachomatis major outer membrane protein are not associated with Chlamydia resolution or reinfection in women. mSphere 2020, 5, e00654-20. [Google Scholar] [CrossRef] [PubMed]
- Chackerian, B.; Caldeira, J.D.C.; Peabody, J.; Peabody, D.S. Peptide epitope identification by affinity selection on bacteriophage MS2 virus-like particles. J. Mol. Biol. 2011, 409, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Messina, J.P.; Brady, O.J.; Golding, N.; Kraemer, M.U.G.; Wint, G.R.W.; Ray, S.E.; Pigott, D.M.; Shearer, F.M.; Johnson, K.; Earl, L.; et al. The current and future global distribution and population at risk of dengue. Nat. Microbiol. 2019, 4, 1508–1515. [Google Scholar] [CrossRef]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef]
- Stanaway, J.D.; Shepard, D.S.; Undurraga, E.A.; Halasa, Y.A.; Coffeng, L.E.; Brady, O.J.; Hay, S.I.; Bedi, N.; Bensenor, I.M.; Castañeda, C.A.O.; et al. The global burden of dengue: An analysis from the global burden of disease study 2013. Lancet Infect. Dis. 2016, 16, 712–723. [Google Scholar] [CrossRef]
- Guzman, M.G.; Harris, E. Dengue. Lancet 2015, 385, 453–465. [Google Scholar] [CrossRef]
- Wahala, W.M.P.B.; de Silva, A.M. The human antibody response to dengue virus infection. Viruses 2011, 3, 2374–2395. [Google Scholar] [CrossRef]
- Halstead, S.B. Dengue antibody-dependent enhancement: Knowns and unknowns. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef]
- Modhiran, N.; Watterson, D.; Muller, D.A.; Panetta, A.K.; Sester, D.P.; Liu, L.; Hume, D.A.; Stacey, K.J.; Young, P.R. Dengue virus NS1 protein activates cells via toll-like receptor 4 and disrupts endothelial cell monolayer integrity. Sci. Transl. Med. 2015, 7, 142. [Google Scholar] [CrossRef] [PubMed]
- Modhiran, N.; Watterson, D.; Blumenthal, A.; Baxter, A.G.; Young, P.R.; Stacey, K.J. Dengue virus NS1 protein activates immune cells via TLR4 but not TLR2 or TLR6. Immunol. Cell Biol. 2017, 95, 491–495. [Google Scholar] [CrossRef]
- Glasner, D.R.; Ratnasiri, K.; Puerta-Guardo, H.; Espinosa, D.A.; Beatty, P.R.; Harris, E. Dengue virus NS1 cytokine-independent vascular leak is dependent on endothelial glycocalyx components. PLoS Pathog. 2017, 13, e1006673. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-R.; Chuang, Y.-C.; Lin, Y.-S.; Liu, H.-S.; Liu, C.-C.; Perng, G.-C.; Yeh, T.-M. Dengue Virus Nonstructural Protein 1 Induces Vascular Leakage through Macrophage Migration Inhibitory Factor and Autophagy. PLoS Negl. Trop. Dis. 2016, 10, e0004828. [Google Scholar] [CrossRef] [PubMed]
- Hertz, T.; Beatty, P.R.; MacMillen, Z.; Killingbeck, S.S.; Wang, C.; Harris, E. Antibody epitopes identified in critical regions of dengue virus nonstructural 1 protein in mouse vaccination and natural human infections. J. Immunol. 2017, 198, 4025–4035. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.-C.; Chuang, Y.-C.; Liu, C.-C.; Ho, T.-S.; Lin, Y.-S.; Anderson, R.; Yeh, T.-M. Antibodies against modified NS1 wing domain peptide protect against Dengue virus infection. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Falconar, A.K. The dengue virus nonstructural-1 protein (NS1) generates antibodies to common epitopes on human blood clotting, integrin/adhesin proteins and binds to human endothelial cells: Potential implications in haemorrhagic fever pathogenesis. Arch. Virol. 1997, 142, 897–916. [Google Scholar] [CrossRef]
- Falconar, A.K.I. Antibody responses are generated to immunodominant ELK/KLE-type motifs on the nonstructural-1 glycoprotein during live dengue virus infections in mice and humans: Implications for diagnosis, pathogenesis, and vaccine design. Clin. Vaccine Immunol. 2007, 14, 493–504. [Google Scholar] [CrossRef]
- Lin, C.-F.; Wan, S.-W.; Chen, M.-C.; Lin, S.-C.; Cheng, C.-C.; Chiu, S.-C.; Hsiao, Y.-L.; Lei, H.-Y.; Liu, H.-S.; Yeh, T.-M.; et al. Liver injury caused by antibodies against dengue virus nonstructural protein 1 in a murine model. Lab. Investig. 2008, 88, 1079–1089. [Google Scholar] [CrossRef]
- Lin, Y.-S.; Yeh, T.-M.; Lin, C.-F.; Wan, S.-W.; Chuang, Y.-C.; Hsu, T.-K.; Liu, H.-S.; Liu, C.-C.; Anderson, R.; Lei, H.Y. Molecular mimicry between virus and host and its implications for dengue disease pathogenesis. Exp. Biol. Med. 2011, 236, 515–523. [Google Scholar] [CrossRef]
- Chuang, Y.-C.; Lei, H.-Y.; Liu, H.-S.; Lin, Y.-S.; Fu, T.-F.; Yeh, T.-M. Macrophage migration inhibitory factor induced by dengue virus infection increases vascular permeability. Cytokine 2011, 54, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Chuang, Y.-C.; Wang, S.-Y.; Lin, Y.-S.; Chen, H.-R.; Yeh, T.-M. Re-evaluation of the pathogenic roles of nonstructural protein 1 and its antibodies during dengue virus infection. J. Biomed. Sci. 2013, 20, 42. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.-S.; King, C.-C.; Huang, H.-S.; Shih, Y.-L.; Lee, C.-C.; Tsai, W.-J.; Yu, C.-C.; Chang, H.H. Antiplatelet autoantibodies elicited by dengue virus non-structural protein 1 cause thrombocytopenia and mortality in mice. J. Thromb. Haemost. 2007, 5, 2291–2299. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.-J.; Lei, H.-Y.; Lin, C.-F.; Luo, Y.-H.; Wan, S.-W.; Liu, H.-S.; Yeh, T.-M.; Lin, Y.-S. Anti-dengue virus nonstructural protein 1 antibodies recognize protein disulfide isomerase on platelets and inhibit platelet aggregation. Mol. Immunol. 2009, 47, 398–406. [Google Scholar] [CrossRef]
- Chen, M.-C.; Lin, C.-F.; Lei, H.-Y.; Lin, S.-C.; Liu, H.-S.; Yeh, T.-M.; Anderson, R.; Lin, Y.-S. Deletion of the C-terminal region of dengue virus nonstructural protein 1 (NS1) abolishes anti-NS1-mediated platelet dysfunction and bleeding tendency. J. Immunol. 2009, 183, 1797–1803. [Google Scholar] [CrossRef]
- Liu, I.-J.; Chiu, C.-Y.; Chen, Y.-C.; Wu, H.-C. Molecular mimicry of human endothelial cell antigen by autoantibodies to nonstructural protein 1 of dengue virus. J. Biol. Chem. 2011, 286, 9726–9736. [Google Scholar] [CrossRef]
- Frietze, K.M.; Core, S.B.; Linville, A.; Chackerian, B.; Peabody, D.S. Assessing Antibody Specificity in Human Serum Using Deep Sequence-Coupled Biopanning. Methods Mol. Biol. 2020, 2070, 157–171. [Google Scholar] [CrossRef]
- Kunkel, T.A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 1985, 82, 488–492. [Google Scholar] [CrossRef]
- Klein, D.E.; Choi, J.L.; Harrison, S.C. Structure of a dengue virus envelope protein late-stage fusion intermediate. J. Virol. 2013, 87, 2287–2293. [Google Scholar] [CrossRef]
- Chao, C.-H.; Wu, W.-C.; Lai, Y.-C.; Tsai, P.-J.; Perng, G.-C.; Lin, Y.-S.; Yeh, T.-M. Dengue virus nonstructural protein 1 activates platelets via toll-like receptor 4, leading to thrombocytopenia and hemorrhage. PLoS Pathog. 2019, 15, e1007625. [Google Scholar] [CrossRef]
- Amexis, G.; Young, N.S. Multiple antigenic peptides as vaccine platform for the induction of humoral responses against dengue-2 virus. Viral Immunol. 2007, 20, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Peng, L.; Zhao, W.; Zhong, H.; Zhang, F.; Yan, Z.; Cao, H. Synthetic peptides containing B− and T-cell epitope of dengue virus-2 E domain III provoked B− and T-cell responses. Vaccine 2011, 29, 3695–3702. [Google Scholar] [CrossRef] [PubMed]
- Rocha, R.P.; Livonesi, M.C.; Fumagalli, M.J.; Rodrigues, N.F.; da Costa, L.C.F.; dos Santos, M.C.S.G.; de Oliveira Rocha, E.S.; Kroon, E.G.; Malaquias, L.C.C.; Coelho, L.F.L. Evaluation of tetravalent and conserved synthetic peptides vaccines derived from dengue virus envelope domain I and II. Virus Res. 2014, 188, 122–127. [Google Scholar] [CrossRef] [PubMed]
- Innis, B.L.; Thirawuth, V.; Hemachudha, C. Identification of continuous epitopes of the envelope glycoprotein of dengue type 2 virus. Am. J. Trop. Med. Hyg. 1989, 40, 676–687. [Google Scholar] [CrossRef]
- da Silva, A.N.M.R.; Nascimento, E.J.M.; Cordeiro, M.T.; Gil, L.H.V.G.; Abath, F.G.C.; Montenegro, S.M.L.; Marques, E.T.A. Identification of continuous human B-cell epitopes in the envelope glycoprotein of dengue virus type 3 (DENV-3). PLoS ONE 2009, 4, e7425. [Google Scholar] [CrossRef]
- Andrade, D.V.; Katzelnick, L.C.; Widman, D.G.; Balmaseda, A.; de Silva, A.M.; Baric, R.S.; Harris, E. Analysis of individuals from a dengue-endemic region helps define the footprint and repertoire of antibodies targeting dengue virus 3 type-specific epitopes. mBio 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.-Y.; Tsai, W.-Y.; Lin, S.-R.; Kao, C.-L.; Hu, H.-P.; King, C.-C.; Wu, H.-C.; Chang, G.-J.; Wang, W.-K. Antibodies to envelope glycoprotein of dengue virus during the natural course of infection are predominantly cross-reactive and recognize epitopes containing highly conserved residues at the fusion loop of domain II. J. Virol. 2008, 82, 6631–6643. [Google Scholar] [CrossRef]
- Crill, W.D.; Hughes, H.R.; Delorey, M.J.; Chang, G.-J.J. Humoral immune responses of dengue fever patients using epitope-specific serotype-2 virus-like particle antigens. PLoS ONE 2009, 4, e4991. [Google Scholar] [CrossRef]
- Wahala, W.M.P.B.; Kraus, A.A.; Haymore, L.B.; Accavitti-Loper, M.A.; de Silva, A.M. Dengue virus neutralization by human immune sera: Role of envelope protein domain III-reactive antibody. Virology 2009, 392, 103–113. [Google Scholar] [CrossRef]
- Lin, H.-E.; Tsai, W.-Y.; Liu, I.-J.; Li, P.-C.; Liao, M.-Y.; Tsai, J.-J.; Wu, Y.-C.; Lai, C.-Y.; Lu, C.-H.; Huang, J.-H.; et al. Analysis of epitopes on dengue virus envelope protein recognized by monoclonal antibodies and polyclonal human sera by a high throughput assay. PLoS Negl. Trop. Dis. 2012, 6, e1447. [Google Scholar] [CrossRef]
- Lai, C.-Y.; Williams, K.L.; Wu, Y.-C.; Knight, S.; Balmaseda, A.; Harris, E.; Wang, W.-K. Analysis of cross-reactive antibodies recognizing the fusion loop of envelope protein and correlation with neutralizing antibody titers in nicaraguan dengue cases. PLoS Negl. Trop. Dis. 2013, 7, e2451. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Warner, N.L.; Linville, A.C.; Core, S.B.; Moreno, B.; Pascale, J.M.; Peabody, D.S.; Chackerian, B.; Frietze, K.M. Expansion and Refinement of Deep Sequence-Coupled Biopanning Technology for Epitope-Specific Antibody Responses in Human Serum. Viruses 2020, 12, 1114. https://doi.org/10.3390/v12101114
Warner NL, Linville AC, Core SB, Moreno B, Pascale JM, Peabody DS, Chackerian B, Frietze KM. Expansion and Refinement of Deep Sequence-Coupled Biopanning Technology for Epitope-Specific Antibody Responses in Human Serum. Viruses. 2020; 12(10):1114. https://doi.org/10.3390/v12101114
Chicago/Turabian StyleWarner, Nikole L., Alexandria C. Linville, Susan B. Core, Brechla Moreno, Juan M. Pascale, David S. Peabody, Bryce Chackerian, and Kathryn M. Frietze. 2020. "Expansion and Refinement of Deep Sequence-Coupled Biopanning Technology for Epitope-Specific Antibody Responses in Human Serum" Viruses 12, no. 10: 1114. https://doi.org/10.3390/v12101114
APA StyleWarner, N. L., Linville, A. C., Core, S. B., Moreno, B., Pascale, J. M., Peabody, D. S., Chackerian, B., & Frietze, K. M. (2020). Expansion and Refinement of Deep Sequence-Coupled Biopanning Technology for Epitope-Specific Antibody Responses in Human Serum. Viruses, 12(10), 1114. https://doi.org/10.3390/v12101114





