Regulation of the Human Papillomavirus Life Cycle by DNA Damage Repair Pathways and Epigenetic Factors
Abstract
1. Introduction
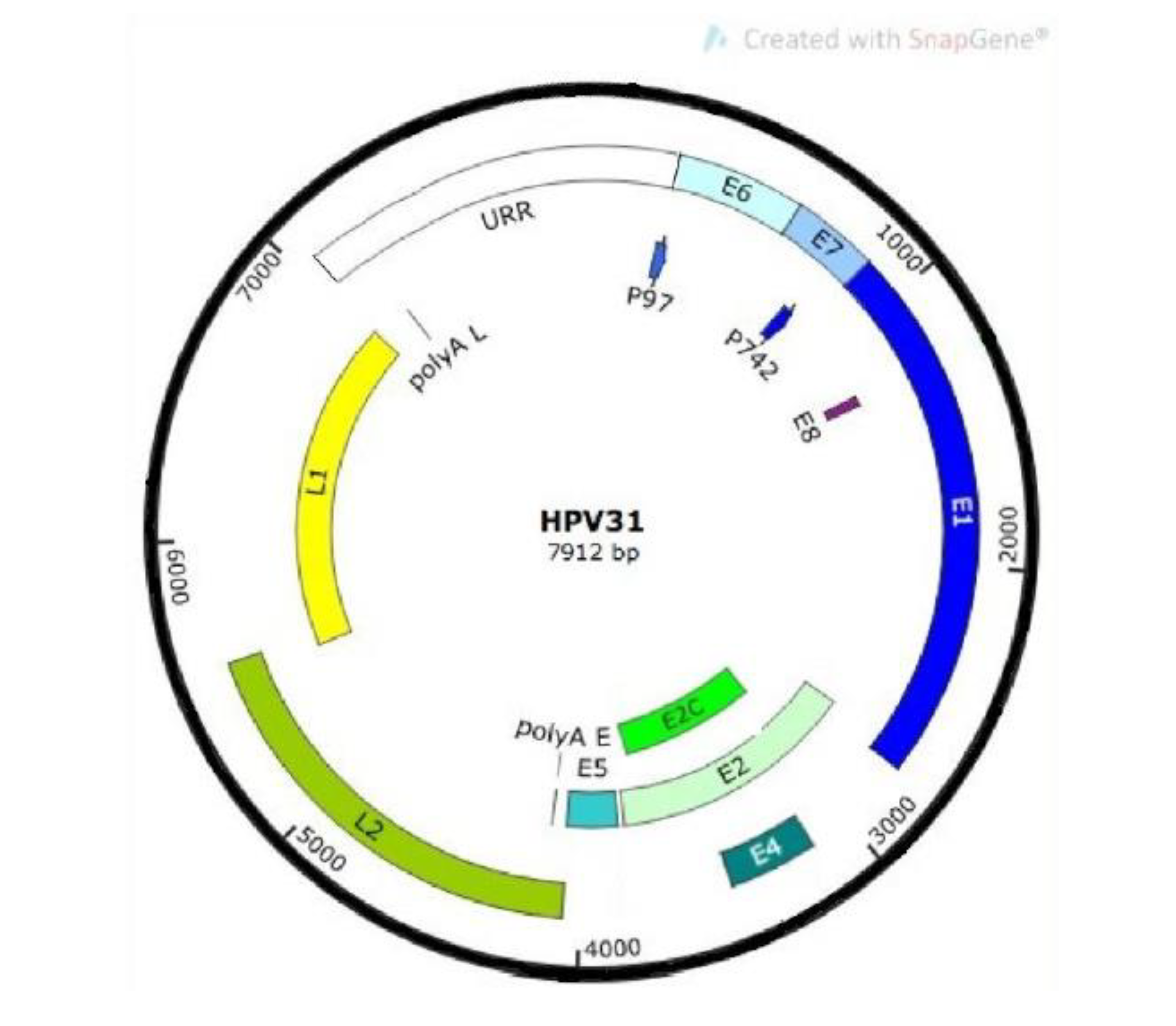
2. Human Papillomavirus Life Cycle
3. High-Risk Human Papillomavirus Integration into the Genome of the Host Keratinocyte
4. Modes of HPV Genome Replication
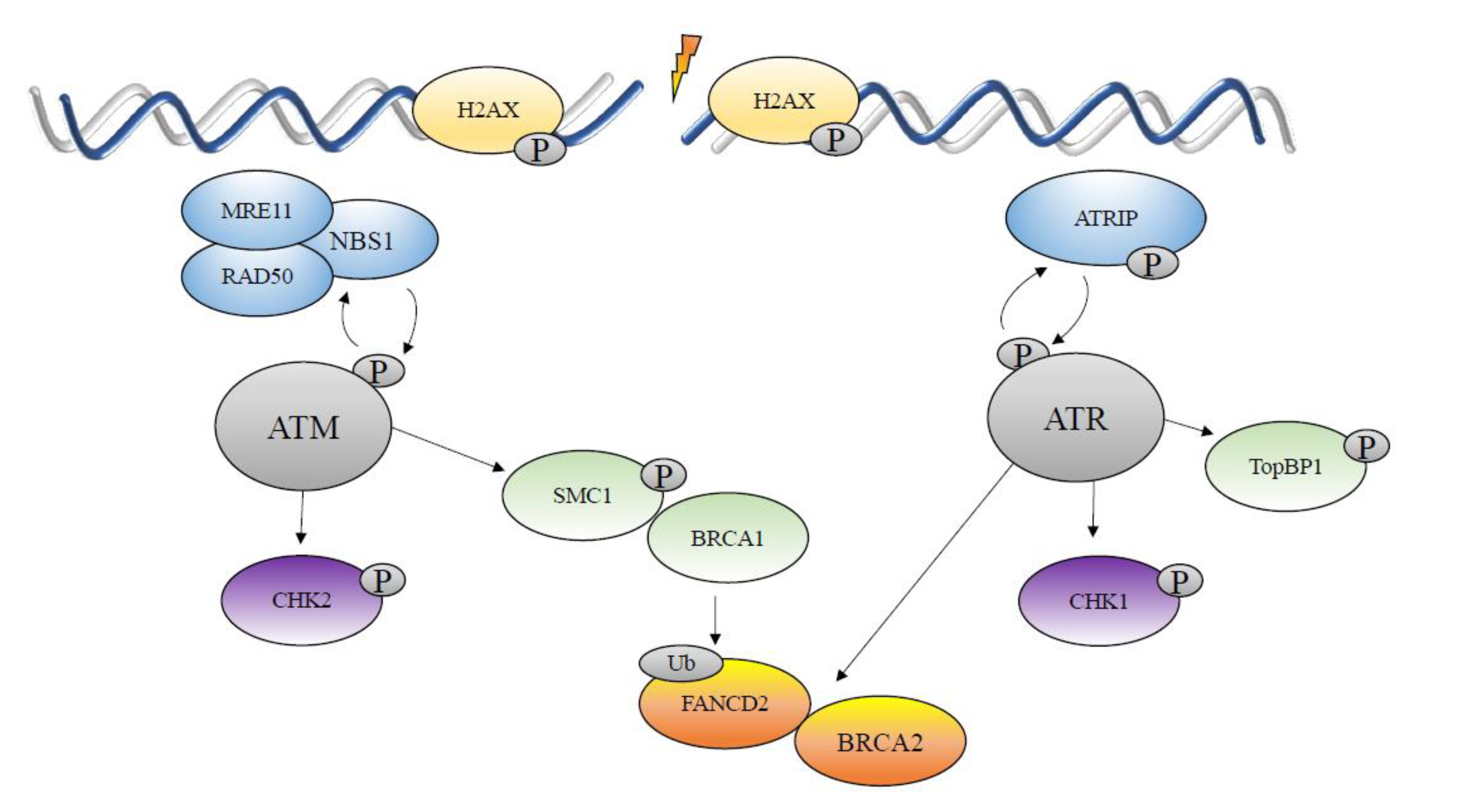
5. The DNA Damage Response
6. Double-Strand Break Repair
7. Homologous Recombination Repair
8. Stalled DNA Replication Forks
9. DNA Interstrand Cross-Links and the FANC Group of Proteins
10. DNA Damage Repair Pathways and HPV replication
11. Acetylation by Tip60
12. Sirtuin Deacetyalses
13. SIRT1 and HPV
14. Summary
Funding
Conflicts of Interest
References
- Pastrana, D.V.; Peretti, A.; Welch, N.L.; Borgogna, C.; Olivero, C.; Badolato, R.; Notarangelo, L.D.; Gariglio, M.; FitzGerald, P.C.; McIntosh, C.E.; et al. Metagenomic Discovery of 83 New Human Papillomavirus Types in Patients with Immunodeficiency. mSphere 2018, 3. [Google Scholar] [CrossRef]
- zur Hausen, H. Papillomaviruses in the causation of human cancers–a brief historical account. Virology 2009, 384, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, A.K.; Engels, E.A.; Pfeiffer, R.M.; Hernandez, B.Y.; Xiao, W.; Kim, E.; Jiang, B.; Goodman, M.T.; Sibug-Saber, M.; Cozen, W.; et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J. Clin. Oncol. 2011, 29, 4294–4301. [Google Scholar] [CrossRef] [PubMed]
- Castellsague, X.; Alemany, L.; Quer, M.; Halec, G.; Quiros, B.; Tous, S.; Clavero, O.; Alos, L.; Biegner, T.; Szafarowski, T.; et al. HPV Involvement in Head and Neck Cancers: Comprehensive Assessment of Biomarkers in 3680 Patients. J. Natl Cancer Inst. 2016, 108, djv403. [Google Scholar] [CrossRef]
- Stokley, S.; Jeyarajah, J.; Yankey, D.; Cano, M.; Gee, J.; Roark, J.; Curtis, R.C.; Markowitz, L. Immunization Services Division NCfI, Respiratory Diseases CDC, et al. Human papillomavirus vaccination coverage among adolescents, 2007-2013, and postlicensure vaccine safety monitoring, 2006-2014--United States. Mmwr Morb. Mortal. Wkly. Rep. 2014, 63, 620–624. [Google Scholar] [PubMed]
- Forhan, S.E.; Gottlieb, S.L.; Sternberg, M.R.; Xu, F.; Datta, S.D.; McQuillan, G.M.; Berman, S.M.; Markowitz, L.E. Prevalence of sexually transmitted infections among female adolescents aged 14 to 19 in the United States. Pediatrics 2009, 124, 1505–1512. [Google Scholar] [CrossRef]
- Arbyn, M.; Weiderpass, E.; Bruni, L.; de Sanjose, S.; Saraiya, M.; Ferlay, J.; Bray, F. Estimates of incidence and mortality of cervical cancer in 2018: A worldwide analysis. Lancet Glob. Health 2020, 8, e191–e203. [Google Scholar] [CrossRef]
- Moody, C.A.; Laimins, L.A. Human papillomavirus oncoproteins: Pathways to transformation. Nat. Rev. Cancer 2010, 10, 550–560. [Google Scholar] [CrossRef]
- Mittal, S.; Banks, L. Molecular mechanisms underlying human papillomavirus E6 and E7 oncoprotein-induced cell transformation. Mutat. Res. Rev. Mutat. Res. 2017, 772, 23–35. [Google Scholar] [CrossRef]
- zur Hausen, H. In the near future, our readership can look forward to reading the “editor’s choice”. Int J. Cancer 2009, 124, x. [Google Scholar] [CrossRef]
- Scheffner, M.; Huibregtse, J.M.; Vierstra, R.D.; Howley, P.M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 1993, 75, 495–505. [Google Scholar] [CrossRef]
- Martinez-Zapien, D.; Ruiz, F.X.; Poirson, J.; Mitschler, A.; Ramirez, J.; Forster, A.; Cousido-Siah, A.; Masson, M.; Vande Pol, S.; Podjarny, A.; et al. Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature 2016, 529, 541–545. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Chen, W.; Roman, A. The E7 proteins of low- and high-risk human papillomaviruses share the ability to target the pRB family member p130 for degradation. Proc. Natl. Acad. Sci. USA 2006, 103, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Heck, D.; Yee, C.; Howley, P.; Munger, K. Efficiency of binding the retinoblastoma protein correlates with the transforming capacity of the E7 oncoproteins of the human papillomaviruses. Proc. Natl. Acad. Sci. USA 1992, 89, 4442–4446. [Google Scholar] [CrossRef]
- Huang, P.; Patrick, D.; Edwards, G.; Goodhart, P.; Huber, H.; Miles, L.; Garsky, V.; Oliff, A.; Heimbrook, D. Protein domains governing interactions between E2F, the retinblastoma gene product, and human papillomavirus type 16 E7 protein. Mol. Cell. Biol. 1993, 13, 953–960. [Google Scholar] [CrossRef]
- Massimi, P.; Gammoh, N.; Thomas, M.; Banks, L. HPV E6 specifically targets different cellular pools of its PDZ domain-conatining tumor suppressor substrates for proteosome-mediated degradation. Oncogene 2004, 23, 8033–8039. [Google Scholar] [CrossRef]
- Huh, K.W.; DeMasi, J.; Ogawa, H.; Nakatani, Y.; Howley, P.M.; Munger, K. Association of the human papillomavirus type 16 E7 oncoprotein with the 600-kDa retinoblastoma protein-associated factor, p600. Proc. Natl. Acad. Sci. USA 2005, 102, 11492–11497. [Google Scholar] [CrossRef]
- Ustav, M.; Stenlund, A. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 1991, 10, 449–457. [Google Scholar] [CrossRef]
- Mohr, I.; Clark, R.; Sun, S.; Androphy, E.; MacPherson, P.; Botchan, M. Targetting the E1 replication factor to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science 1990, 250, 1694–1699. [Google Scholar] [CrossRef]
- Stubenrauch, F.; Hummel, M.; Iftner, T.; Laimins, L.A. The E8^E2C protein, a negative regulator of viral transcription and replication, is required for extrachromosoaml maintenace of HPV 31 in keratinocytes. J. Virol. 2000, 74, 1178–1186. [Google Scholar] [CrossRef]
- Doorbar, J.; Ely, S.; Sterling, J.; McLean, C.; Crawford, L. Specific interaction between HPV-16 E1-E4 and cytokeratins results in collapse of the epithelial cell intermediate filament network. Nature 1991, 352, 824–827. [Google Scholar] [CrossRef] [PubMed]
- Genther, S.M.; Sterling, S.; Duensing, S.; Munger, K.; Sattler, C.; Lambert, P. Quantitaive role of HPV 16 E5 gene during the productive stage of viral life cycle. J. Virol. 2003, 77, 2832–2842. [Google Scholar] [CrossRef] [PubMed]
- Fehrman, F.K.; Laimins, L. The E5 protein of HPV 31 acts to augment cell prolfilertion and activatin of differntiation-edependent late viral functions. J. Virol. 2003, 77, 2819–2831. [Google Scholar] [CrossRef]
- DiGiuseppe, S.; Bienkowska-Haba, M.; Guion, L.G.; Sapp, M. Cruising the cellular highways: How human papillomavirus travels from the surface to the nucleus. Virus Res. 2017, 231, 1–9. [Google Scholar] [CrossRef] [PubMed]
- McKinney, C.C.; Hussmann, K.L.; McBride, A.A. The Role of the DNA Damage Response throughout the Papillomavirus Life Cycle. Viruses 2015, 7, 2450–2469. [Google Scholar] [CrossRef]
- Durst, M.; Kleinheinz, A.; Hotz, M.; Gissman, L. The physical state of human papillomavirus type 16 DNA in benign and malignant genital tumors. J. Gen. Virol. 1985, 66, 1515–1522. [Google Scholar] [CrossRef]
- Senapati, R.; Senapati, N.N.; Dwibedi, B. Molecular mechanisms of HPV mediated neoplastic progression. Infect. Agent Cancer 2016, 11, 59. [Google Scholar] [CrossRef]
- Moody, C.A.; Laimins, L.A. Human papillomaviruses activate the ATM DNA damage pathway for viral genome amplification upon differentiation. PLoS Pathog 2009, 5, e1000605. [Google Scholar] [CrossRef]
- Orav, M.; Geimanen, J.; Sepp, E.M.; Henno, L.; Ustav, E.; Ustav, M. Initial amplification of the HPV18 genome proceeds via two distinct replication mechanisms. Sci. Rep. 2015, 5, 15952. [Google Scholar] [CrossRef]
- Flores, E.R.; Lambert, P.F. Evidence for a switch in the mode of human papillomavirus type 16 DNA replication during the viral life cycle. J. Virol. 1997, 71, 7167–7179. [Google Scholar] [CrossRef]
- Sakakibara, N.; Chen, D.; McBride, A.A. Papillomaviruses use recombination-dependent replication to vegetatively amplify their genomes in differentiated cells. PLoS Pathog 2013, 9, e1003321. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Schmidt-Grimminger, D.; Murant, T.; Broker, T.; Chow, L. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 1995, 9, 2335–2349. [Google Scholar] [CrossRef] [PubMed]
- Mehta, K.; Laimins, L. Human Papillomaviruses Preferentially Recruit DNA Repair Factors to Viral Genomes for Rapid Repair and Amplification. mBio 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, A.; Elledge, S.J. The DNA damage response: Making it safe to play with knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [PubMed]
- Gasser, S.; Raulet, D. The DNA damage response, immunity and cancer. Semin Cancer Biol. 2006, 16, 344–347. [Google Scholar] [CrossRef]
- Zhou, B.B.; Elledge, S.J. The DNA damage response: Putting checkpoints in perspective. Nature 2000, 408, 433–439. [Google Scholar] [CrossRef]
- Harper, J.W.; Elledge, S.J. The DNA damage response: Ten years after. Mol. Cell 2007, 28, 739–745. [Google Scholar] [CrossRef] [PubMed]
- Cimprich, K.A.; Cortez, D. ATR: An essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008, 9, 616–627. [Google Scholar] [CrossRef]
- Rogakou, E.P.; Pilch, D.R.; Orr, A.H.; Ivanova, V.S.; Bonner, W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998, 273, 5858–5868. [Google Scholar] [CrossRef]
- Stiff, T.; O’Driscoll, M.; Rief, N.; Iwabuchi, K.; Lobrich, M.; Jeggo, P.A. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004, 64, 2390–2396. [Google Scholar] [CrossRef]
- Fernandez-Capetillo, O.; Celeste, A.; Nussenzweig, A. Focusing on foci: H2AX and the recruitment of DNA-damage response factors. Cell Cycle 2003, 2, 426–427. [Google Scholar] [CrossRef]
- Shiloh, Y.; Ziv, Y. The ATM protein kinase: Regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 2013, 14, 197–210. [Google Scholar] [CrossRef]
- Shieh, S.; Ikeda, M.; Taya, Y.; Prives, C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 1997, 91, 325–3344. [Google Scholar] [CrossRef]
- Nakagawa, K.; Taya, Y.; Tamai, K.; Yamaizumi, M. Requirement of ATM in phosphorylation of the human p53 protein at serine 15 following DNA double-strand breaks. Mol. Cell Biol. 1999, 19, 2828–2834. [Google Scholar] [CrossRef] [PubMed]
- Hirao, A.; Kong, Y.Y.; Matsuoka, S.; Wakeham, A.; Ruland, J.; Yoshida, H.; Liu, D.; Elledge, S.J.; Mak, T.W. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 2000, 287, 1824–1827. [Google Scholar] [CrossRef] [PubMed]
- Chehab, N.H.; Malikzay, A.; Appel, M.; Halazonetis, T.D. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev. 2000, 14, 278–288. [Google Scholar] [PubMed]
- Haince, J.F.; McDonald, D.; Rodrigue, A.; Dery, U.; Masson, J.Y.; Hendzel, M.J.; Poirier, G.G. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 2008, 283, 1197–1208. [Google Scholar] [CrossRef]
- Cortez, D.; Wang, Y.; Qin, J.; Elledge, S.J. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science 1999, 286, 1162–1166. [Google Scholar] [CrossRef]
- Wold, M.S. Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997, 66, 61–92. [Google Scholar] [CrossRef]
- Morita, T.; Yoshimura, Y.; Yamamoto, A.; Murata, K.; Mori, M.; Yamamoto, H.; Matsushiro, A. A mouse homolog of the Escherichia coli recA and Saccharomyces cerevisiae RAD51 genes. Proc. Natl. Acad. Sci. USA 1993, 90, 6577–6580. [Google Scholar] [CrossRef]
- Jensen, R.B.; Carreira, A.; Kowalczykowski, S.C. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 2010, 467, 678–683. [Google Scholar] [CrossRef] [PubMed]
- West, S.C. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003, 4, 435–445. [Google Scholar] [CrossRef]
- You, Z.; Bailis, J.M. DNA damage and decisions: CtIP coordinates DNA repair and cell cycle checkpoints. Trends Cell Biol. 2010, 20, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Michaelis, C.; Ciosk, R.; Nasmyth, K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell 1997, 91, 35–45. [Google Scholar] [CrossRef]
- Kim, S.T.; Xu, B.; Kastan, M.B. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 2002, 16, 560–570. [Google Scholar] [CrossRef]
- Byun, T.S.; Pacek, M.; Yee, M.C.; Walter, J.C.; Cimprich, K.A. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005, 19, 1040–1052. [Google Scholar] [CrossRef]
- Luke-Glaser, S.; Luke, B.; Grossi, S.; Constantinou, A. FANCM regulates DNA chain elongation and is stabilized by S-phase checkpoint signalling. EMBO J. 2010, 29, 795–805. [Google Scholar] [CrossRef]
- Niraj, J.; Farkkila, A.; D’Andrea, A.D. The Fanconi Anemia Pathway in Cancer. Annu. Rev. Cancer Biol. 2019, 3, 457–478. [Google Scholar] [CrossRef]
- Hong, S.; Laimins, L.A. Regulation of the life cycle of HPVs by differentiation and the DNA damage response. Future Microbiol. 2013, 8, 1547–1557. [Google Scholar] [CrossRef]
- Edwards, T.G.; Helmus, M.J.; Koeller, K.; Bashkin, J.K.; Fisher, C. Human papillomavirus episome stability is reduced by aphidicolin and controlled by DNA damage response pathways. J. Virol. 2013, 87, 3979–3989. [Google Scholar] [CrossRef]
- Gillespie, K.A.; Mehta, K.P.; Laimins, L.A.; Moody, C.A. Human papillomaviruses recruit cellular DNA repair and homologous recombination factors to viral replication centers. J. Virol. 2013, 86, 9520–9526. [Google Scholar] [CrossRef]
- Anacker, D.C.; Aloor, H.L.; Shepard, C.N.; Lenzi, G.M.; Johnson, B.A.; Kim, B.; Moody, C.A. HPV31 utilizes the ATR-Chk1 pathway to maintain elevated RRM2 levels and a replication-competent environment in differentiating Keratinocytes. Virology 2016, 499, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Laimins, L.A. Manipulation of the innate immune response by human papillomaviruses. Virus Res. 2017, 231, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Wallace, N.A.; Khanal, S.; Robinson, K.L.; Wendel, S.O.; Messer, J.J.; Galloway, D.A. High-Risk Alphapapillomavirus Oncogenes Impair the Homologous Recombination Pathway. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Hong, S.; Cheng, S.; Lovane, A.; Laimins, L.A. STAT-5 regulates transcription of the topoisomerase IIβ-binding protein 1 (TopBP1) gene to activate the ATR pathway and promote HPV replication. mBio 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Anacker, D.C.; Moody, C.A. Modulation of the DNA damage response during the life cycle of human papillomaviruses. Virus Res. 2017, 231, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Sitz, J.; Blanchet, S.A.; Gameiro, S.F.; Biquand, E.; Morgan, T.M.; Galloy, M.; Dessapt, J.; Lavoie, E.G.; Blondeau, A.; Smith, B.C.; et al. Human papillomavirus E7 oncoprotein targets RNF168 to hijack the host DNA damage response. Proc. Natl. Acad. Sci. USA 2019, 116, 19552–19562. [Google Scholar] [CrossRef] [PubMed]
- Fradet-Turcotte, A.; Moody, C.; Laimins, L.A.; Archambault, J. Nuclear export of human papillomavirus type 31 E1 is regulated by Cdk2 phosphorylation and required for viral genome maintenance. J. Virol. 2010, 84, 11747–11760. [Google Scholar] [CrossRef] [PubMed]
- Mehta, K.; Gunasekharan, V.; Satsuka, A.; Laimins, L.A. Human papillomaviruses activate and recruit SMC1 cohesin proteins for the differentiation-dependent life cycle through association with CTCF insulators. PLoS Pathog 2015, 11, e1004763. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, T.; Garcia-Higuera, I.; Xu, B.; Andreassen, P.R.; Gregory, R.C.; Kim, S.T.; Lane, W.S.; Kastan, M.B. D’Andrea AD: Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell 2002, 109, 459–472. [Google Scholar] [CrossRef]
- Park, D.J.; Wilczynski, S.P.; Paquette, R.L.; Miller, C.W.; Koeffler, H.P. p53 mutations in HPV-negative cervical carcinoma. Oncogene 1993, 9, 205–210. [Google Scholar]
- Park, J.W.; Shin, M.K.; Pitot, H.C.; Lambert, P.F. High incidence of HPV-associated head and neck cancers in FA deficient mice is associated with E7′s induction of DNA damage through its inactivation of pocket proteins. PLoS ONE 2013, 8, e75056. [Google Scholar]
- Hoskins, E.E.; Morreale, R.J.; Werner, S.P.; Higginbotham, J.M.; Laimins, L.A.; Lambert, P.F.; Brown, D.R.; Gillison, M.L.; Nuovo, G.J.; Witte, D.P.; et al. The fanconi anemia pathway limits human papillomavirus replication. J. Virol. 2012, 86, 8131–8138. [Google Scholar] [CrossRef]
- Spriggs, C.C.; Laimins, L.A. FANCD2 Binds Human Papillomavirus Genomes and Associates with a Distinct Set of DNA Repair Proteins to Regulate Viral Replication. MBio 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Khanal, S.; Galloway, D.A. High-risk human papillomavirus oncogenes disrupt the Fanconi anemia DNA repair pathway by impairing localization and de-ubiquitination of FancD2. PLoS Pathog 2019, 15, e1007442. [Google Scholar] [CrossRef]
- Sun, Y.; Jiang, X.; Chen, S.; Fernandes, N.; Price, B.D. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. USA 2005, 102, 13182–13187. [Google Scholar] [CrossRef]
- Hong, S.; Dutta, A.; Laimins, L.A. The acetyltransferase Tip60 is a critical regulator of the differentiation-dependent amplification of human papillomaviruses. J. Virol. 2015, 89, 4668–4675. [Google Scholar] [CrossRef]
- Rine, J.; Herskowitz, I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics 1987, 116, 9–22. [Google Scholar]
- Houtkooper, R.H.; Pirinen, E.; Auwerx, J. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 2012, 13, 225–238. [Google Scholar] [CrossRef]
- Imai, S.; Johnson, F.B.; Marciniak, R.A.; McVey, M.; Park, P.U.; Guarente, L. Sir2: An NAD-dependent histone deacetylase that connects chromatin silencing, metabolism, and aging. Cold Spring Harb Symp. Quant. Biol. 2000, 65, 297–302. [Google Scholar] [CrossRef]
- Toiber, D.; Sebastian, C.; Mostoslavsky, R. Characterization of nuclear sirtuins: Molecular mechanisms and physiological relevance. Handb. Exp. Pharm. 2011, 206, 189–224. [Google Scholar]
- Bonkowski, M.S.; Sinclair, D.A. Slowing ageing by design: The rise of NAD(+) and sirtuin-activating compounds. Nat. Rev. Mol. Cell Biol. 2016, 17, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Alves-Fernandes, D.K.; Jasiulionis, M.G. The Role of SIRT1 on DNA Damage Response and Epigenetic Alterations in Cancer. Int J. Mol. Sci 2019, 20. [Google Scholar]
- Wang, R.H.; Zheng, Y.; Kim, H.S.; Xu, X.; Cao, L.; Luhasen, T.; Lee, M.H.; Xiao, C.; Vassilopoulos, A.; Chen, W.; et al. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated tumorigenesis. Mol. Cell 2008, 32, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Mostoslavsky, R.; Chua, K.F.; Lombard, D.B.; Pang, W.W.; Fischer, M.R.; Gellon, L.; Liu, P.; Mostoslavsky, G.; Franco, S.; Murphy, M.M.; et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 2006, 124, 315–329. [Google Scholar] [CrossRef]
- Michishita, E.; McCord, R.A.; Berber, E.; Kioi, M.; Padilla-Nash, H.; Damian, M.; Cheung, P.; Kusumoto, R.; Kawahara, T.L.; Barrett, J.C.; et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 2008, 452, 492–496. [Google Scholar] [CrossRef]
- Mao, Z.; Hine, C.; Tian, X.; Van Meter, M.; Au, M.; Vaidya, A.; Seluanov, A.; Gorbunova, V. SIRT6 promotes DNA repair under stress by activating PARP1. Science 2011, 332, 1443–1446. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, J.; Hong, T.; Chen, X.; Cui, L. SIRT2: Controversy and multiple roles in disease and physiology. Ageing Res. Rev. 2019, 55, 100961. [Google Scholar] [CrossRef]
- Zhang, X.; Brachner, A.; Kukolj, E.; Slade, D.; Wang, Y. SIRT2 deacetylates GRASP55 to facilitate post-mitotic Golgi assembly. J. Cell Sci 2019, 132. [Google Scholar]
- Langsfeld, E.S.; Bodily, J.M.; Laimins, L.A. The Deacetylase Sirtuin 1 Regulates Human Papillomavirus Replication by Modulating Histone Acetylation and Recruitment of DNA Damage Factors NBS1 and Rad51 to Viral Genomes. PLoS Pathog 2015, 11, e1005181. [Google Scholar] [CrossRef]
- Das, D.; Smith, N.; Wang, X. Morgan IM: The Deacetylase SIRT1 Regulates the Replication Properties of Human Papillomavirus 16 E1 and E2. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]

| DDR proteins affected by E6 and E7 Header | Proteins |
|---|---|
| A. Increased total levels of DDR proteins | ATR |
| CHK1 | |
| TOPBP1 | |
| FANCD2 | |
| RAD51 | |
| NBS1 | |
| BRCA1 | |
| 53BP1 | |
| RNF168 | |
| Tip60 | |
| B. Increased levels of phosphorylated forms | pATM |
| pCHK2 | |
| pNBS1 | |
| pBRCA1 | |
| γH2AX | |
| pATR | |
| pCHK1 | |
| pTOPBP1 | |
| pSMC1 | |
| pMRE11 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albert, E.; Laimins, L. Regulation of the Human Papillomavirus Life Cycle by DNA Damage Repair Pathways and Epigenetic Factors. Viruses 2020, 12, 744. https://doi.org/10.3390/v12070744
Albert E, Laimins L. Regulation of the Human Papillomavirus Life Cycle by DNA Damage Repair Pathways and Epigenetic Factors. Viruses. 2020; 12(7):744. https://doi.org/10.3390/v12070744
Chicago/Turabian StyleAlbert, Ekaterina, and Laimonis Laimins. 2020. "Regulation of the Human Papillomavirus Life Cycle by DNA Damage Repair Pathways and Epigenetic Factors" Viruses 12, no. 7: 744. https://doi.org/10.3390/v12070744
APA StyleAlbert, E., & Laimins, L. (2020). Regulation of the Human Papillomavirus Life Cycle by DNA Damage Repair Pathways and Epigenetic Factors. Viruses, 12(7), 744. https://doi.org/10.3390/v12070744





