A Well-Defined H9N2 Avian Influenza Virus Genotype with High Adaption in Mammals was Prevalent in Chinese Poultry Between 2016 to 2019
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Viruses, Cells, and Animals
2.3. Experimental Design
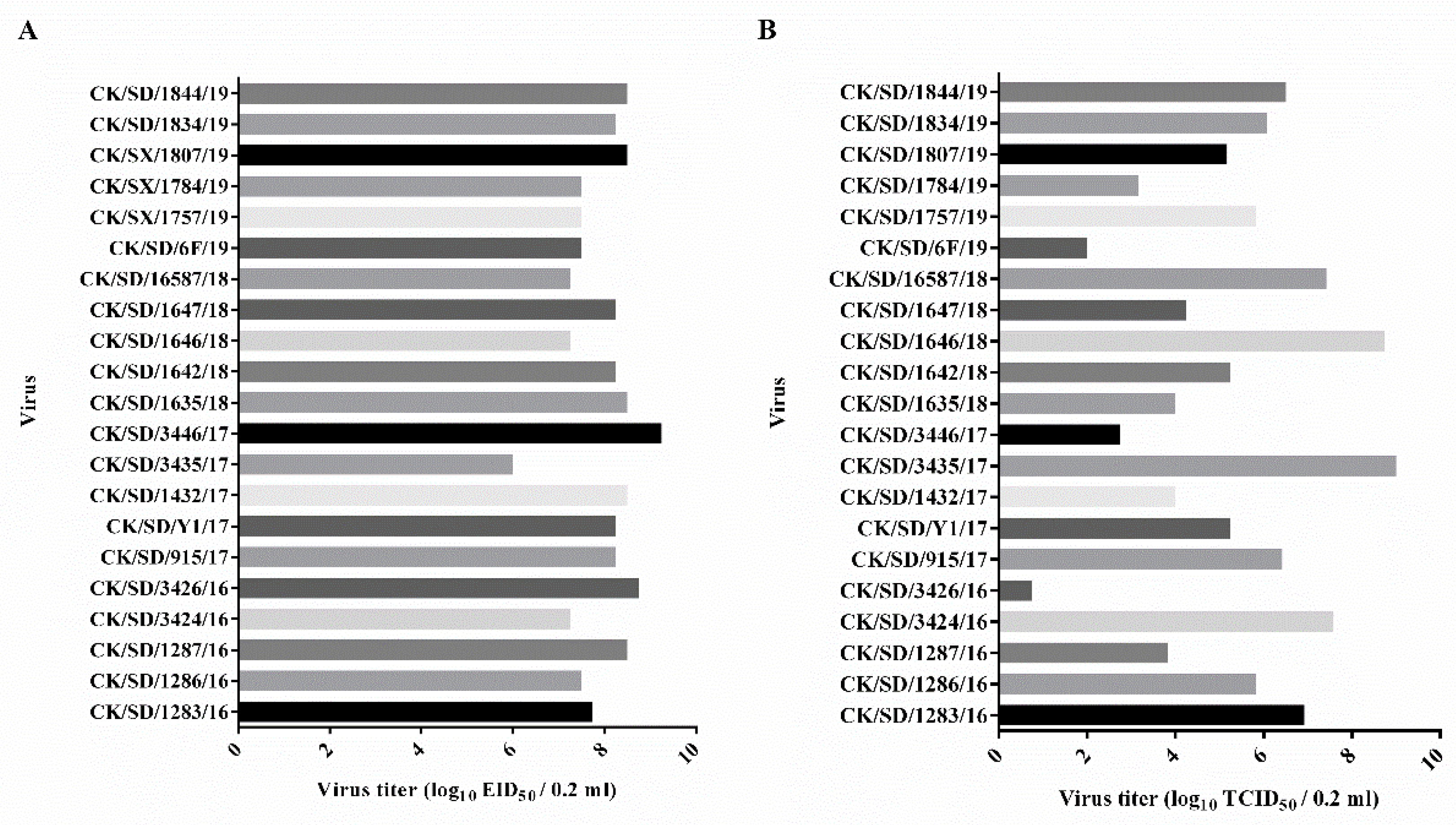
2.4. 50% Egg Infectious Dose (EID50) Determination
2.5. 50% Tissue Infectious Dose (TCID50) Determination
2.6. Sample Collection
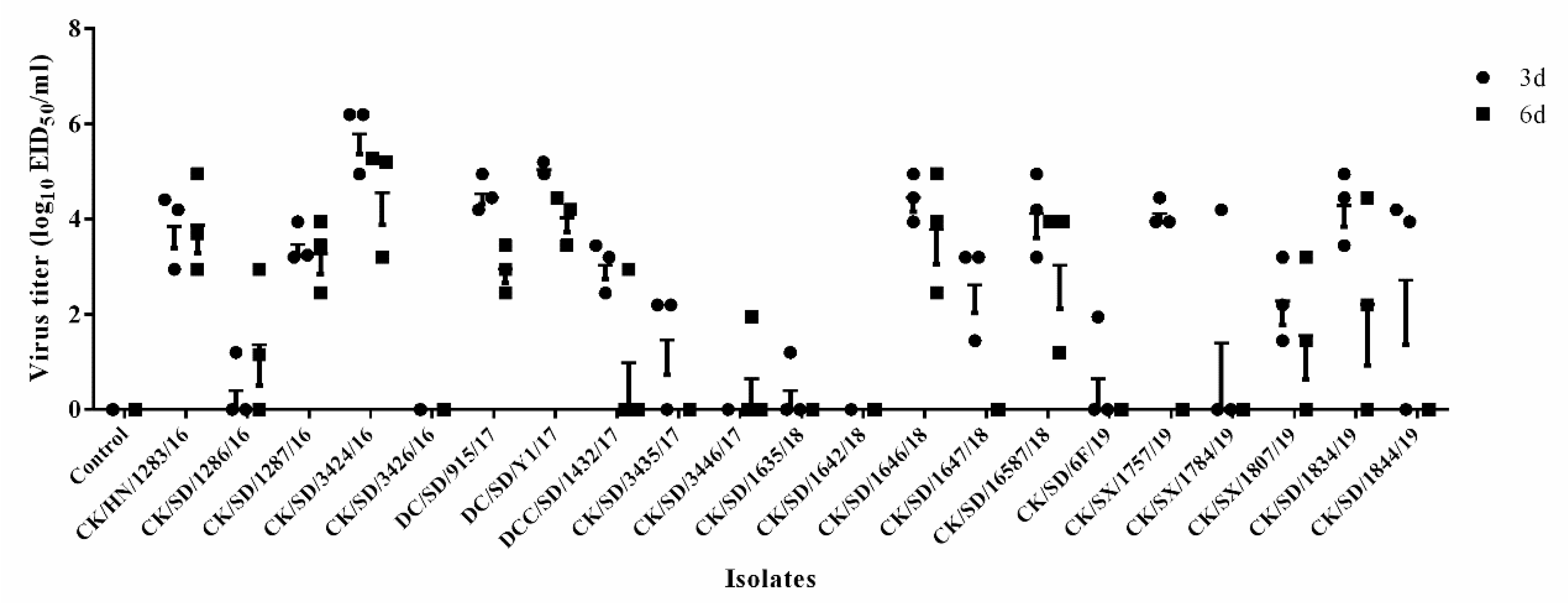
2.7. Determination of Viral Titers in Tissues of Mice
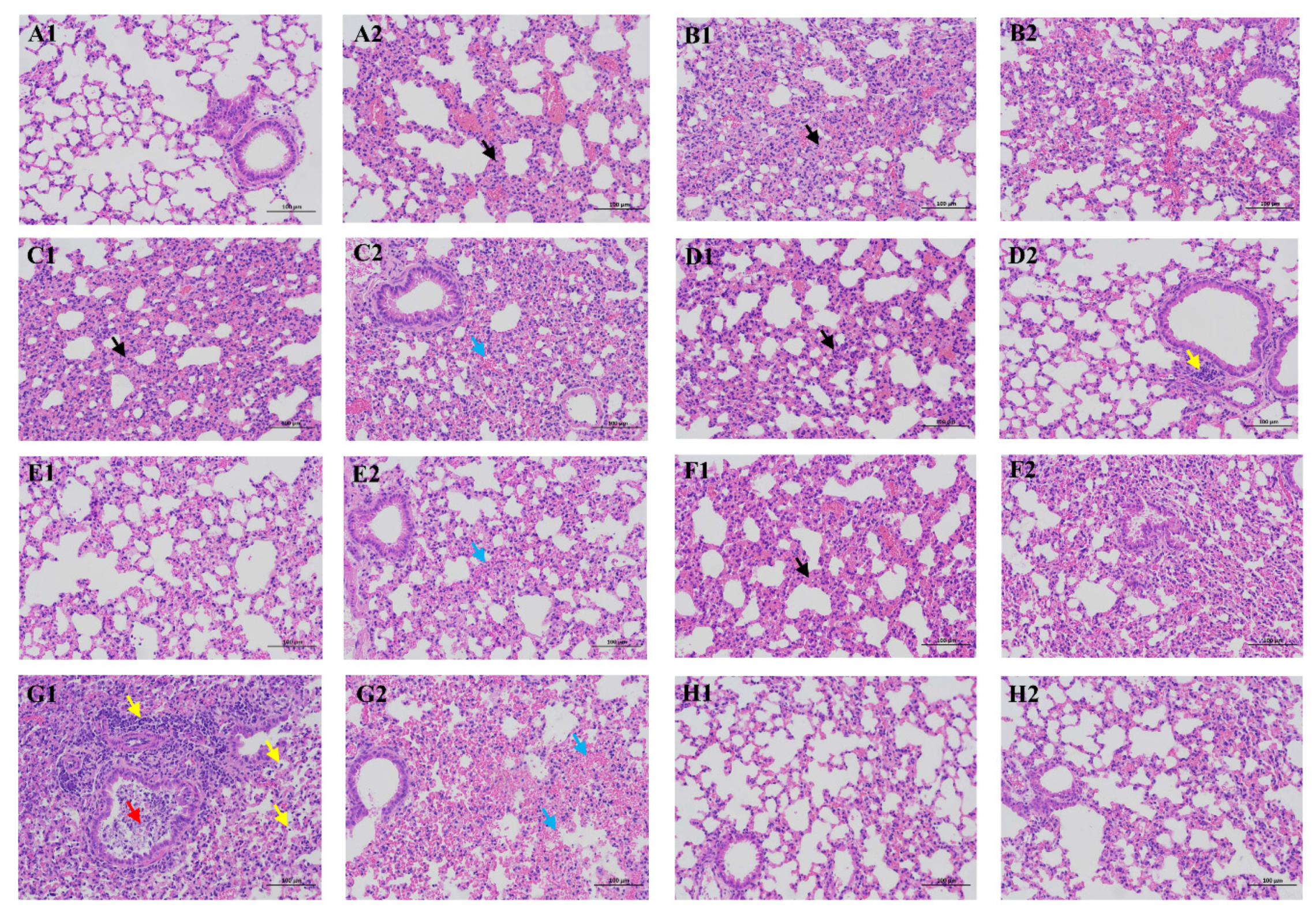
2.8. Pathology Study
2.9. Genotype and Mutation Definition
2.10. Statistical Analysis
3. Results
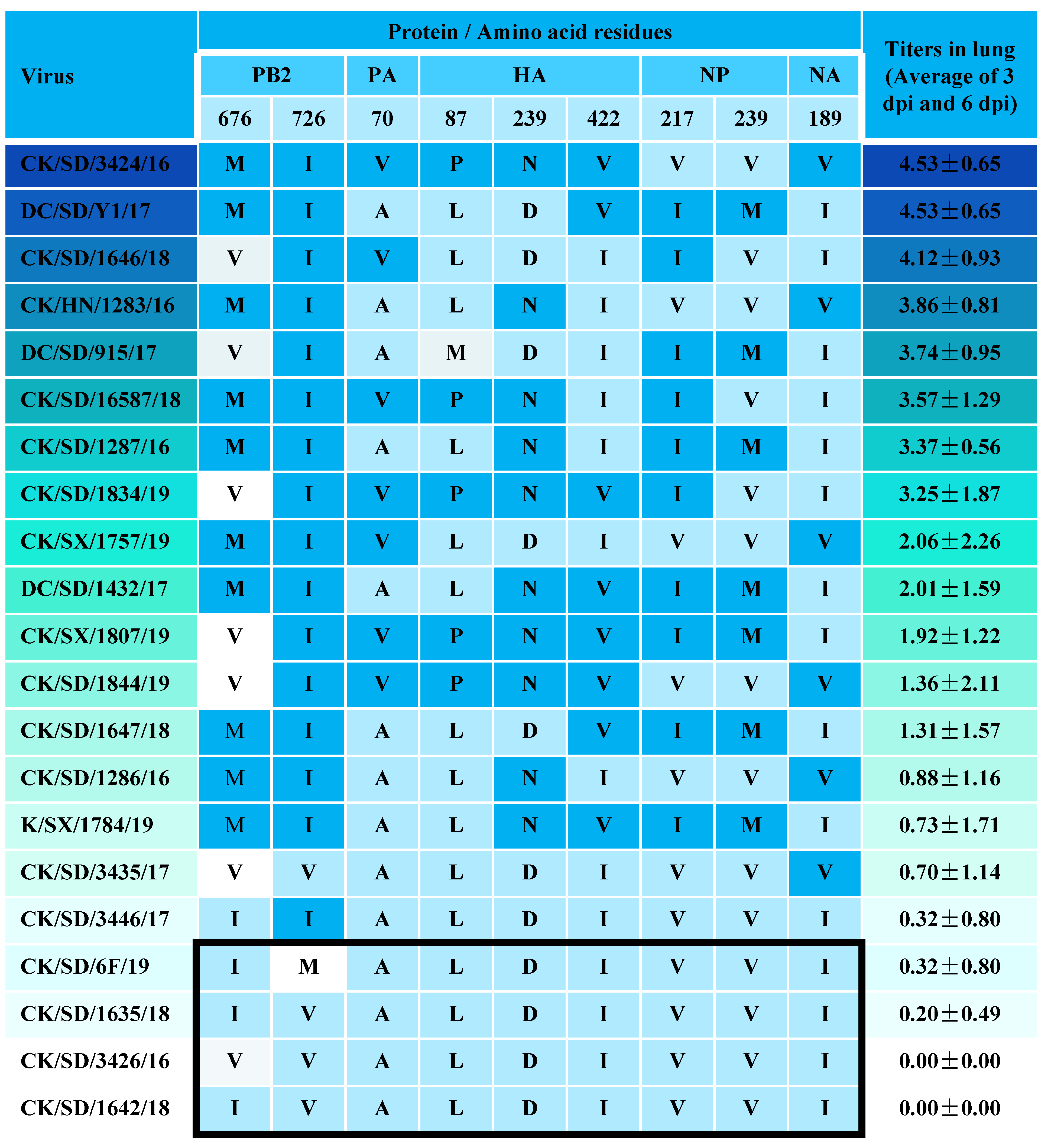
3.1. Determination of Viral Replication Capacities
3.2. Viral Titer in the Lungs of Mice
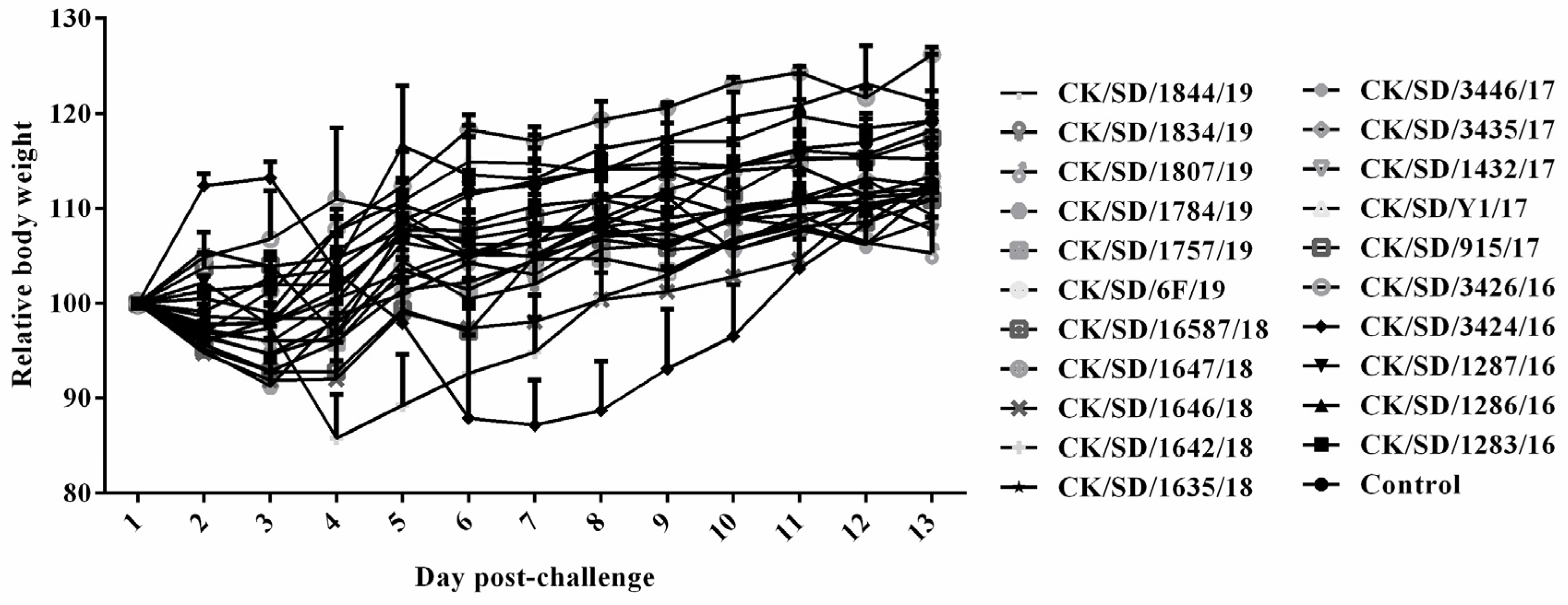
3.3. Body Weight of Mice
3.4. Pathology of Infected Mice
3.5. Genotype
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Homme, P.J.; Easterday, B.C. Avian influenza virus infection. I. Characterization of influenza A/turkey/Wisconsin/1966 virus in turkey. Avian Dis. 1970, 14, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.L.; Zhang, Z.J.; Chen, W.B. Isolation and identification of avian influenza virus. Chin. J. Vet. Med. 1994, 10, 3–5. [Google Scholar]
- Li, C.; Yu, K.; Tian, G.; Yu, D.; Liu, L.; Jing, B.; Ping, J.; Chen, H. Evolution of H9N2 influenza viruses from domestic poultry in Mainland China. Virology 2005, 340, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Liu, A.; Zhang, F.; Ling, Y.; Ou, C.; Hou, N.; He, C. Co-infection of broilers with Ornithobacterium rhinotracheale and H9N2 avian influenza virus. BMC Vet. Res. 2012, 8, 104. [Google Scholar] [CrossRef]
- Brown, I.H.; Banks, J.; Manvell, R.J.; Essen, S.C.; Alexander, D.J. Recent epidemiology and ecology of influenza A viruses in avian species in Europe and the Middle East. Dev. Biol. 2006, 124, 45–50. [Google Scholar]
- Peacock, T.P.; Joe, J.; Joshua, E.S.; Munir, I. A Global Perspective on H9N2 Avian Influenza Virus. Viruses 2019, 11, 620. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, J. H9N2 influenza virus in China: A cause of concern. Protein Cell 2015, 6, 18–25. [Google Scholar] [CrossRef]
- Peiris, M.; Yam, W.C.; Chan, K.H.; Ghose, P.; Shortridge, K.F. Influenza A H9N2: Aspects of laboratory diagnosis. J. Clin. Microbiol. 1999, 37, 3426–3427. [Google Scholar] [CrossRef]
- Guo, Y.J.; Li, J.W.; Cheng, I. Discovery of humans infected by avian influenza A (H9N2) virus. Chin. J. Exp. Clin. Virol. 1999, 15, 105–108. [Google Scholar]
- Peiris, M.; Yuen, K.Y.; Leung, C.W.; Chan, K.H.; Ip, P.L.; Lai, R.W.; Orr, W.K.; Shortridge, K.F. Human infection with influenza H9N2. Lancet 1999, 354, 916–917. [Google Scholar] [CrossRef]
- Lin, Y.P.; Shaw, M.; Gregory, V.; Cameron, K.; Lim, W.; Klimov, A.; Subbarao, K.; Guan, Y.; Krauss, S.; Shortridge, K.; et al. Avian-to-human transmission of H9N2 subtype influenza A viruses: Relationship between H9N2 and H5N1 human isolates. Proc. Natl. Acad. Sci. USA 2000, 97, 9654–9658. [Google Scholar] [CrossRef] [PubMed]
- Matrosovich, M.N.; Krauss, S.; Webster, R.G. H9N2 Influenza A Viruses from Poultry in Asia Have Human Virus-like Receptor Specificity. Virology 2001, 281, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Li, K.S.; Xu, K.M.; Peiris, J.S.M.; Poon, L.L.M.; Yu, K.Z.; Yuen, K.Y.; Short- ridge, K.F.; Webster, R.G.; Guan, Y. Characterization of H9 subtype influenza viruses from the ducks of southern China: A candidate for the next influenza pandemic in humans? J. Virol. 2003, 77, 6988–6994. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Perez, D.R. Amino acid 226 in the hemagglutinin of H9N2 influenza viruses determines cell tropism and replication in human airway epithelial cells. J. Virol. 2007, 81, 5181–5191. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Shortridge, K.; Krauss, S.; Webster, R.G. Molecular characterization of H9N2 influenza viruses: Were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc. Natl. Acad. Sci. USA 1999, 96, 9363–9367. [Google Scholar] [CrossRef]
- Liu, D.; Shi, W.; Gao, G.F. Poultry carrying H9N2 act as incubators for novel human avian influenza viruses. Lancet 2014, 383, 869. [Google Scholar] [CrossRef]
- Cui, L.; Liu, D.; Shi, W.; Pan, J.; Qi, X.; Li, X.; Guo, X.; Zhou, M.; Li, W.; Li, J.; et al. Dynamic reassortments and genetic heterogeneity of the human-infecting influenza A (H7N9) virus. Nat. Commun. 2014, 5, 3142. [Google Scholar] [CrossRef]
- Nfon, C.; Berhane, Y.; Pasick, J.; Kobinger, G.; Kobasa, D.; Babiuk, S. Prior infection of chickens with H1N1 avian influenza virus elicits heterologous protection against highly pathogenic H5N2. Vaccine 2012, 30, 7187–7192. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A Simple Method of Estimating Fifty Percent Endo-Points. Am. J. Hyg. 1938, 27, 493–797. [Google Scholar]
- Pu, J.; Wang, S.; Yin, Y.; Zhang, G.Z.; Carter, R.A.; Wang, J.L.; Xu, G.L.; Sun, H.L.; Wang, M.; Wen, C.; et al. Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus. Proc. Natl. Acad. Sci. USA 2015, 112, 548–553. [Google Scholar] [CrossRef]
- Hinshaw, V.S.; Bean, W.J.; Webster, R.G.; Sriram, G. Genetic reassortment of influenza A viruses in the intestinal tract of ducks. Virology 1980, 102, 412–419. [Google Scholar] [CrossRef]
- Guo, Y.J.; Krauss, S.; Senne, D.A.; Mo, I.P.; Lo, K.S.; Xiong, X.P.; Norwood, M.; Shortridge, K.F.; Webster, R.G.; Guan, Y. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology 2000, 267, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.K.; Ozaki, H.; Webby, R.J.; Webster, R.G.; Peiris, J.S.; Poon, L.; Butt, C.; Leung, Y.H.C.; Guan, Y. Continuing evolution of H9N2 influenza viruses in southeastern China. J. Virol. 2004, 78, 8609–8614. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, S.; Bing, G.; Carter, R.A.; Wang, Z.; Wang, J.; Wang, C.; Wang, L.; Wu, J.; Webster, R.G.; et al. Genetic evolution of influenza H9N2 viruses isolated from various hosts in china from 1994 to 2013. Emerg. Microbes Infect. 2017, 6, e106. [Google Scholar] [CrossRef]
- Li, X.; Shi, J.; Guo, J.; Deng, G.; Zhang, Q.; Wang, J.; He, X.; Wang, K.; Chen, J.; Li, Y.; et al. Genetics, receptor binding property, and transmissibility in mammals of naturally isolated H9N2 avian influenza viruses. PLoS Pathog. 2014, 10, e1004508. [Google Scholar] [CrossRef]
- Liu, Q.; Liu, Y.; Yang, J.; Huang, X.; Han, K.; Zhao, D.; Bi, K.; Li, Y. Two Genetically Similar H9N2 Influenza A Viruses Show Different Pathogenicity in Mice. Front. Microbiol. 2016, 7, 1737. [Google Scholar] [CrossRef]
- Teng, Q.; Xu, D.; Shen, W.; Liu, Q.; Rong, G.; Li, X.; Yan, L.; Yang, J.; Chen, H.; Yu, H.; et al. A Single Mutation at Position 190 in Hemagglutinin Enhances Binding Affinity for Human Type Sialic Acid Receptor and Replication of H9N2 Avian Influenza Virus in Mice. J. Virol. 2016, 90, 9806–9825. [Google Scholar] [CrossRef]
- Smith, G.J.; Vijaykrishna, D.; Bahl, J.; Lycett, S.J.; Worobey, M.; Pybus, O.G.; Ma, S.K.; Cheung, C.L.; Raghwani, J.; Bhatt, S.; et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 2009, 459, 1122–1125. [Google Scholar] [CrossRef]
- Neumann, G.; Kawaoka, Y. Host range restriction and pathogenicity in the context of influenza pandemic. Emerg. Infect. Dis. 2006, 12, 881–886. [Google Scholar] [CrossRef]
- Mehle, A.; Dugan, V.G.; Taubenberger, J.K.; Doudna, J.A. Reassortment and mutation of the avian influenza virus polymerase PA subunit overcome species barriers. J. Virol. 2012, 86, 1750–1757. [Google Scholar] [CrossRef]
- Ince, W.L.; Gueye-Mbaye, A.; Bennink, J.R.; Yewdell, J.W. Reassortment Complements Spontaneous Mutation in Influenza A Virus NP and M1 Genes To Accelerate Adaptation to a New Host. J. Virol. 2013, 87, 4330–4338. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Lu, L.; Li, J.; Yin, Y.; Zhang, Y.; Gao, H.; Qin, Z.; Zeshan, B.; Liu, J.; Sun, L.; et al. Novel genetic reassortants in H9N2 influenza A viruses and their diverse pathogenicity to mice. Virol. J. 2011, 8, 505. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ishaq, M.; Prudence, M.; Xi, X.; Hu, T.; Liu, Q.; Guo, D. Single mutation at the amino acid position 627 of PB2 that leads to increased virulence of an H5N1 avian influenza virus during adaptation in mice can be compensated by multiple mutations at other sites of PB2. Virus Res. 2009, 144, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Tan, Y.; Wei, K.; Sun, H.; Shi, Y.; Pu, J.; Yang, H.; Gao, G.F.; Yin, Y.; Feng, W.; et al. Amino acid 316 of hemagglutinin and the neuraminidase stalk length influence virulence of H9N2 influenza virus in chickens and mice. J. Virol. 2013, 87, 2963–2968. [Google Scholar] [CrossRef] [PubMed]
- Steel, J.; Lowen, A.C.; Mubareka, S.; Palese, P. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. 2009, 5, e1000252. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Zu, Z.; Liu, J.; Song, J.; Wang, X.; Wang, C.; Liu, L.; Tong, Q.; Wang, M.; Sun, H. Prevailing I292V PB2 mutation in avian influenza H9N2 virus increases viral polymerase function and attenuates IFN-β induction in human cells. J. Gen. Virol. 2019, 100, 1273–1281. [Google Scholar] [CrossRef]
- Xiao, C.; Ma, W.; Sun, N.; Huang, L.; Li, Y.; Zeng, Z.; Wen, Y.; Zhang, Z.; Li, H.; Li, Q.; et al. PB2-588 V promotes the mammalian adaptation of H10N8, H7N9 and H9N2 avian influenza viruses. Sci. Rep. 2016, 6, 19474. [Google Scholar] [CrossRef]
- Gabriel, G.; Dauber, B.; Wolff, T.; Planz, O.; Klenk, H.D.; Stech, J. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc. Natl. Acad. Sci. USA 2005, 102, 18590–18595. [Google Scholar] [CrossRef]
- Bussey, K.A.; Desmet, E.A.; Mattiacio, J.L.; Hamilton, A.; Bradel-Tretheway, B.; Bussey, H.E.; Kim, B.; Dewhurst, S.; Takimoto, T. PA residues in the 2009 H1N1 pandemic influenza virus enhance avian influenza virus polymerase activity in mammalian cells. J. Virol. 2011, 85, 7020–7028. [Google Scholar] [CrossRef]
- Liedmann, S.; Hrincius, E.R.; Guy, C.; Anhlan, D.; Dierkes, R.; Carter, R.; Wu, G.; Staeheli, P.; Green, D.R.; Wolff, T.; et al. Viral suppressors of the RIG-I-mediated interferon response are pre-packaged in influenza virions. Nat. Commun. 2014, 5, 5645. [Google Scholar] [CrossRef]
- Xu, G.; Zhang, X.; Gao, W.; Wang, C.; Wang, J.; Sun, H.; Sun, Y.; Guo, L.; Zhang, Z.; Chang, K.; et al. Prevailing PA mutation K356R in avian influenza H9N2 virus increases mammalian replication and pathogenicity. J. Virol. 2016, 90, 8105–8114. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Ibrahim, M.S.; Ellakany, H.F.; Ellakany, N.; Norihito, K.; Rika, M.; Hiroaki, H.; Nogluk, S.; Tatsuya, T.; Yasuo, S.; et al. Acquisition of Human-Type Receptor Binding Specificity by New H5N1 Influenza Virus Sublineages during Their Emergence in Birds in Egypt. PLoS Pathog. 2011, 7, e1002068. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Deng, G.; Song, J.; Tian, G.; Suo, Y.; Jiang, Y.; Guan, Y.; Bu, Z.; Kawaoka, Y.; Chen, H. Two amino acid residues in the matrix protein M1 contribute to the virulence difference of H5N1 avian influenza viruses in mice. Virology 2009, 384, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Li, K.S.; Guan, Y.; Wang, J.; Smith, G.J.D.; Xu, K.M.; Duan, L.; Rahardjo, A.P.; Puthavathana, P.; Buranathai, C.; Nguyen, T.D.; et al. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 2004, 430, 209–213. [Google Scholar] [CrossRef]
- Labadie, K.; Dos Santos Afonso, E.; Rameix-Welti, M.A.; Van der Werf, S.; Naffakh, N. Host-range determinants on the PB2 protein of influenza A viruses control the interaction between the viral polymerase and nucleoprotein in human cells. Virology 2007, 362, 271–282. [Google Scholar] [CrossRef]
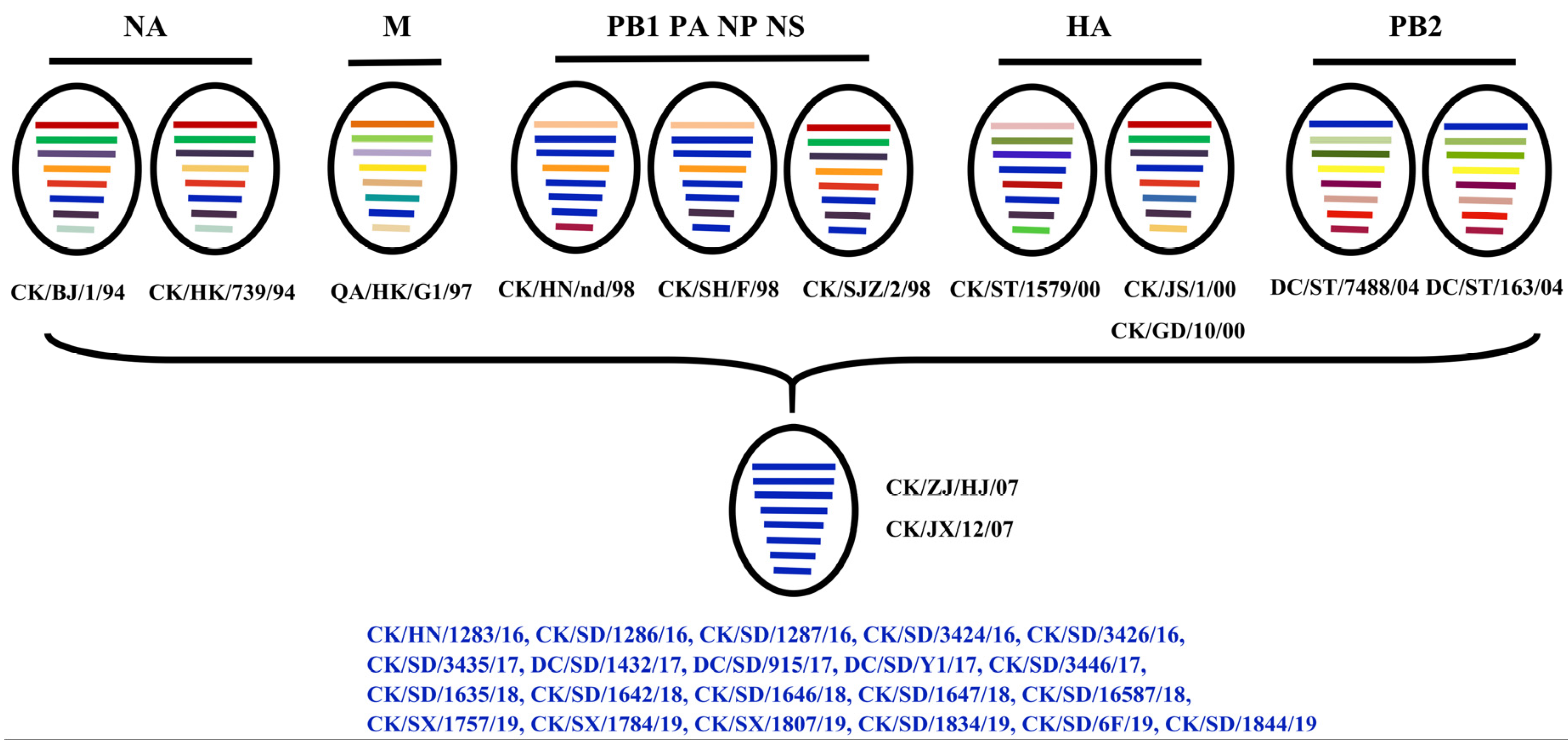
| Isolate | Abbreviation | Host | Date of Isolation | Genbank Accession No. |
|---|---|---|---|---|
| A/Chicken/Henan/1283/2016 | CK/HN/1283/16 | Chicken | February 2016 | MH375347-MH375354 |
| A/Chicken/Shandong/1286/2016 | CK/SD/1286/16 | Chicken | April 2016 | MH375359-MH375366 |
| A/Chicken/Shandong/1287/2016 | CK/SD/1287/16 | Chicken | April 2016 | MH375368-MH375375 |
| A/Chicken/Shandong/3424/2016 | CK/SD/3424/16 | Chicken | June 2016 | MH667569-MH667576 |
| A/Chicken/Shandong/3426/2016 | CK/SD/3426/16 | Chicken | July 2016 | MH667582-MH667589 |
| A/Chicken/Shandong/3435/2017 | CK/SD/3435/17 | Chicken | January 2017 | MH667590-MH667597 |
| A/Duck/Shandong/1432/2017 | DC/SD/1432/17 | Duck | April 2017 | MH375444-MH375451 |
| A/Duck/Shandong/915/2017 | DC/SD/915/17 | Duck | May 2017 | MH375378-MH375385 |
| A/Duck/Shandong/Y1/2017 | DC/SD/Y1/17 | Duck | May 2017 | MH375416-MH375423 |
| A/Chicken/Shandong/3446/2017 | CK/SD/3446/17 | Chicken | May 2017 | MH889082-MH889089 |
| A/Chicken/Shandong/1635/2018 | CK/SD/1635/18 | Chicken | April 2018 | MK367676-MK367683 |
| A/Chicken/Shandong/1642/2018 | CK/SD/1642/18 | Chicken | April 2018 | MK367685-MK367692 |
| A/Chicken/Shandong/1646/2018 | CK/SD/1646/18 | Chicken | May 2018 | MK367659-MK367666 |
| A/Chicken/Shandong/1647/2018 | CK/SD/1647/18 | Chicken | May 2018 | MK367667-MK367674 |
| A/Chicken/Shandong/16587/2018 | CK/SD/16587/18 | Chicken | May 2018 | MK367643-MK367650 |
| A/Chicken/ShanXi/1757/2019 | CK/SX/1757/19 | Chicken | January 2019 | MN780494-MN780501 |
| A/Chicken/ShanXi/1784/2019 | CK/SX/1784/19 | Chicken | March 2019 | MN780586-MN780593 |
| A/Chicken/ShanXi/1807/2019 | CK/SX/1807/19 | Chicken | April 2019 | MN780832-MN780839 |
| A/Chicken/Shandong/1834/2019 | CK/SD/1834/19 | Chicken | May 2019 | MN765112-MN765119 |
| A/Chicken/Shandong/6F/2019 | CK/SD/6F/19 | Chicken | May 2019 | MN759632-MN759639 |
| A/Chicken/Shandong/1844/2019 | CK/SD/1844/19 | Chicken | June 2019 | MN765144-MN765151 |
| Protein | Site | H9N2 AIV Strain | References | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3424 | 915 | 1283 | 1646 | 16587 | 1834 | 1287 | 1757 | 1432 | 1844 | 1807 | 1647 | 1286 | 3435 | 1784 | 6F | 3446 | 1635 | 3426 | 1642 | |||
| PB2 | 89V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | [33] |
| 292V | V | V | I | V | V | V | V | V | V | V | V | V | V | V | V | I | V | I | V | V | [36] | |
| 309D | D | D | D | D | D | D | D | D | D | D | D | D | D | D | D | D | D | D | D | D | [33] | |
| 477G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | [33] | |
| 588V | V | V | V | V | V | V | V | V | A | V | V | A | V | V | A | V | V | V | V | A | [37] | |
| PB1 | 13P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | [38] |
| PA | 224S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | [39] |
| 351E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | [40] | |
| 356R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | [41] | |
| HA a | 165N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | [25] |
| 174H | H | H | H | Q | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | [42] | |
| 226L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | [14] | |
| 316S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | [34] | |
| NA | 62-64 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | [34] |
| M1 | 30D | D | D | D | D | D | D | D | D | D | D | D | D | D | D | D | D | D | D | D | D | [43] |
| 215A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | [43] | |
| NS1 | 149A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | [44] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; Huang, Q.; Yang, S.; Su, S.; Li, B.; Cui, N.; Xu, C. A Well-Defined H9N2 Avian Influenza Virus Genotype with High Adaption in Mammals was Prevalent in Chinese Poultry Between 2016 to 2019. Viruses 2020, 12, 432. https://doi.org/10.3390/v12040432
Chen Z, Huang Q, Yang S, Su S, Li B, Cui N, Xu C. A Well-Defined H9N2 Avian Influenza Virus Genotype with High Adaption in Mammals was Prevalent in Chinese Poultry Between 2016 to 2019. Viruses. 2020; 12(4):432. https://doi.org/10.3390/v12040432
Chicago/Turabian StyleChen, Zhaokun, Qinghua Huang, Shaohua Yang, Shuai Su, Baoquan Li, Ning Cui, and Chuantian Xu. 2020. "A Well-Defined H9N2 Avian Influenza Virus Genotype with High Adaption in Mammals was Prevalent in Chinese Poultry Between 2016 to 2019" Viruses 12, no. 4: 432. https://doi.org/10.3390/v12040432
APA StyleChen, Z., Huang, Q., Yang, S., Su, S., Li, B., Cui, N., & Xu, C. (2020). A Well-Defined H9N2 Avian Influenza Virus Genotype with High Adaption in Mammals was Prevalent in Chinese Poultry Between 2016 to 2019. Viruses, 12(4), 432. https://doi.org/10.3390/v12040432




