Investigating the Diversity of Marine Bacteriophage in Contrasting Water Masses Associated with the East Australian Current (EAC) System
Abstract
1. Introduction
2. Materials and Methods
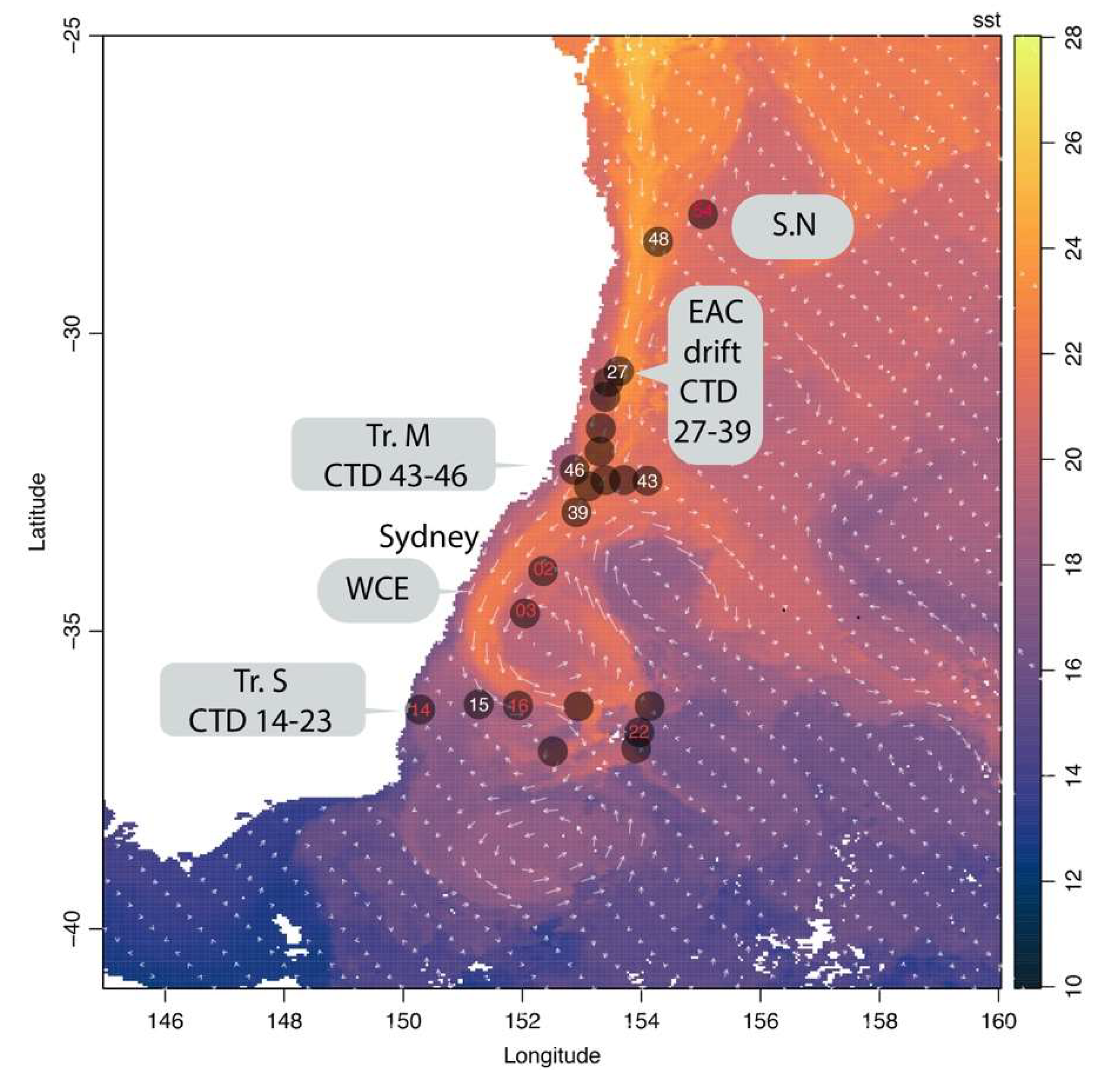
2.1. Sample Collection
2.2. DNA Extraction
2.3. Metagenome Analysis
2.4. Taxonomy of Viral and Prokaryotics Contigs
2.5. Identification of Bacteriophage Contigs
2.6. Analysis of Functional Gene Profiles for Each Site and Bacteriophage Lifestyle
2.7. Statistical Analysis
3. Results
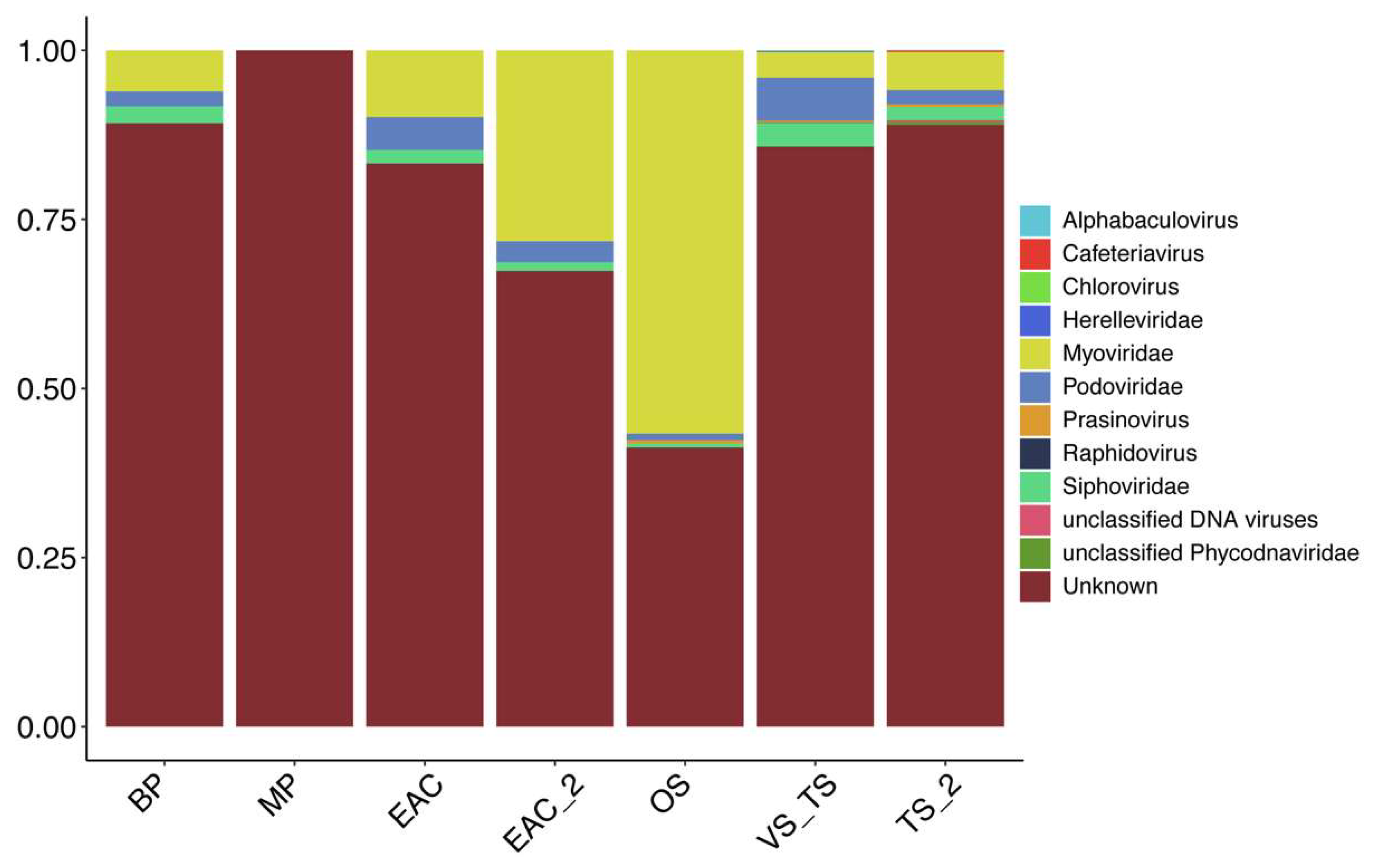
3.1. Viral Similarity to Previously Published Databases
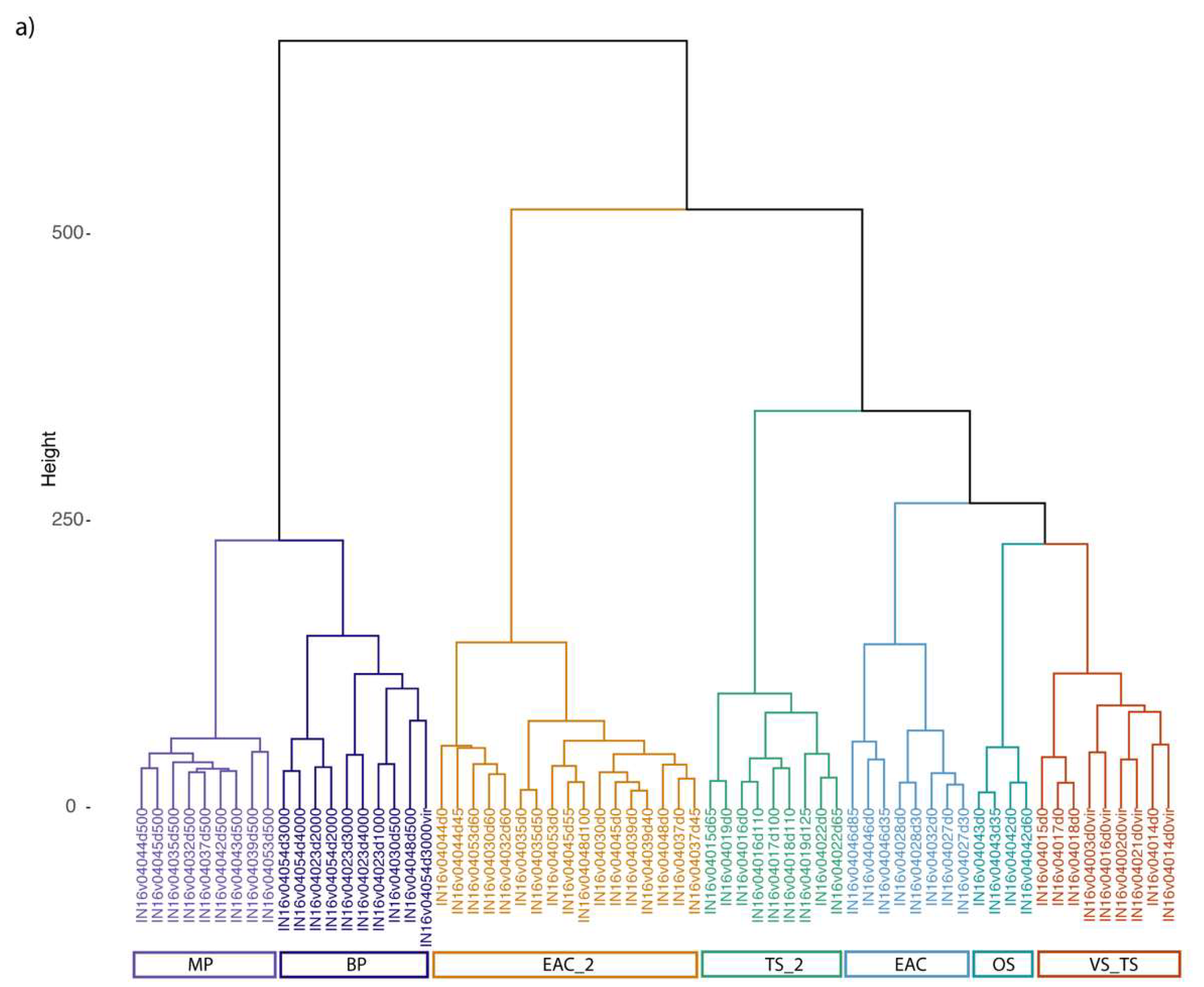
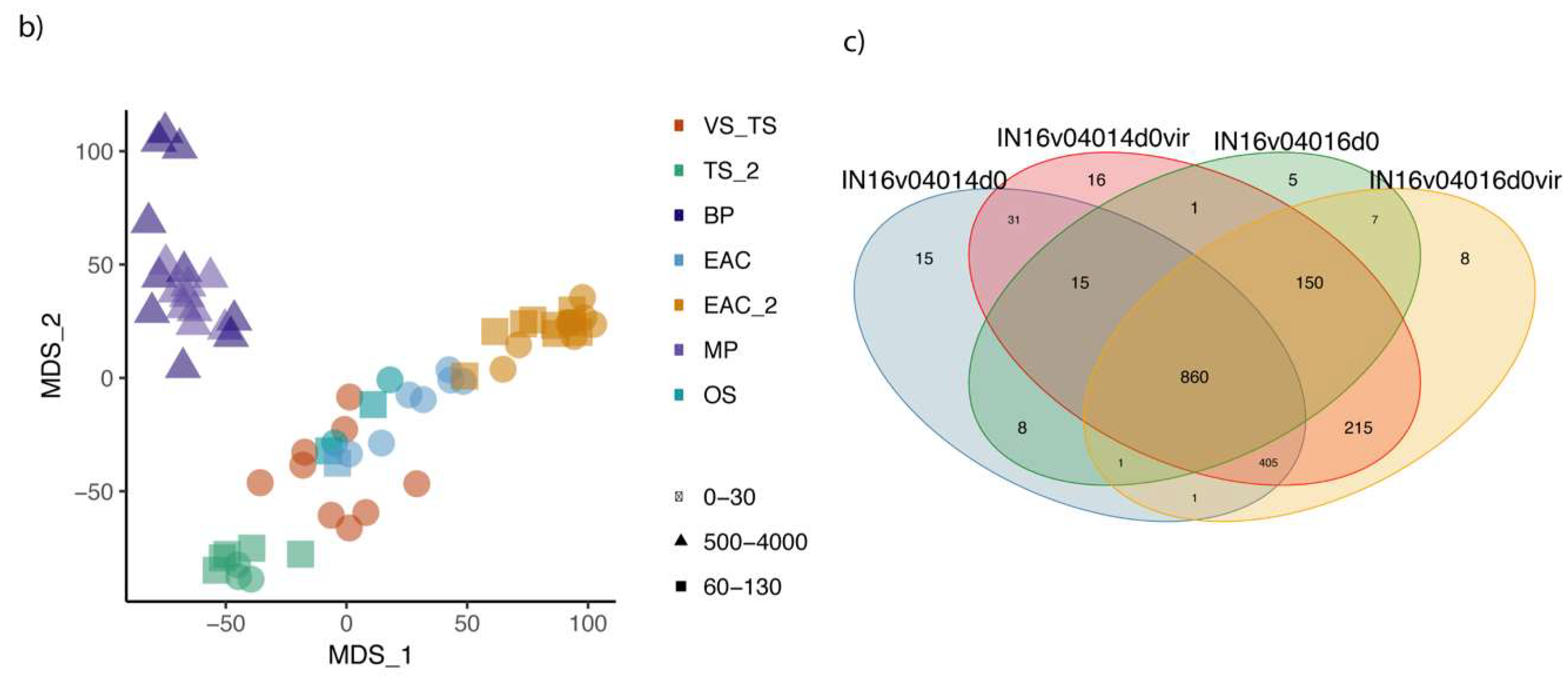
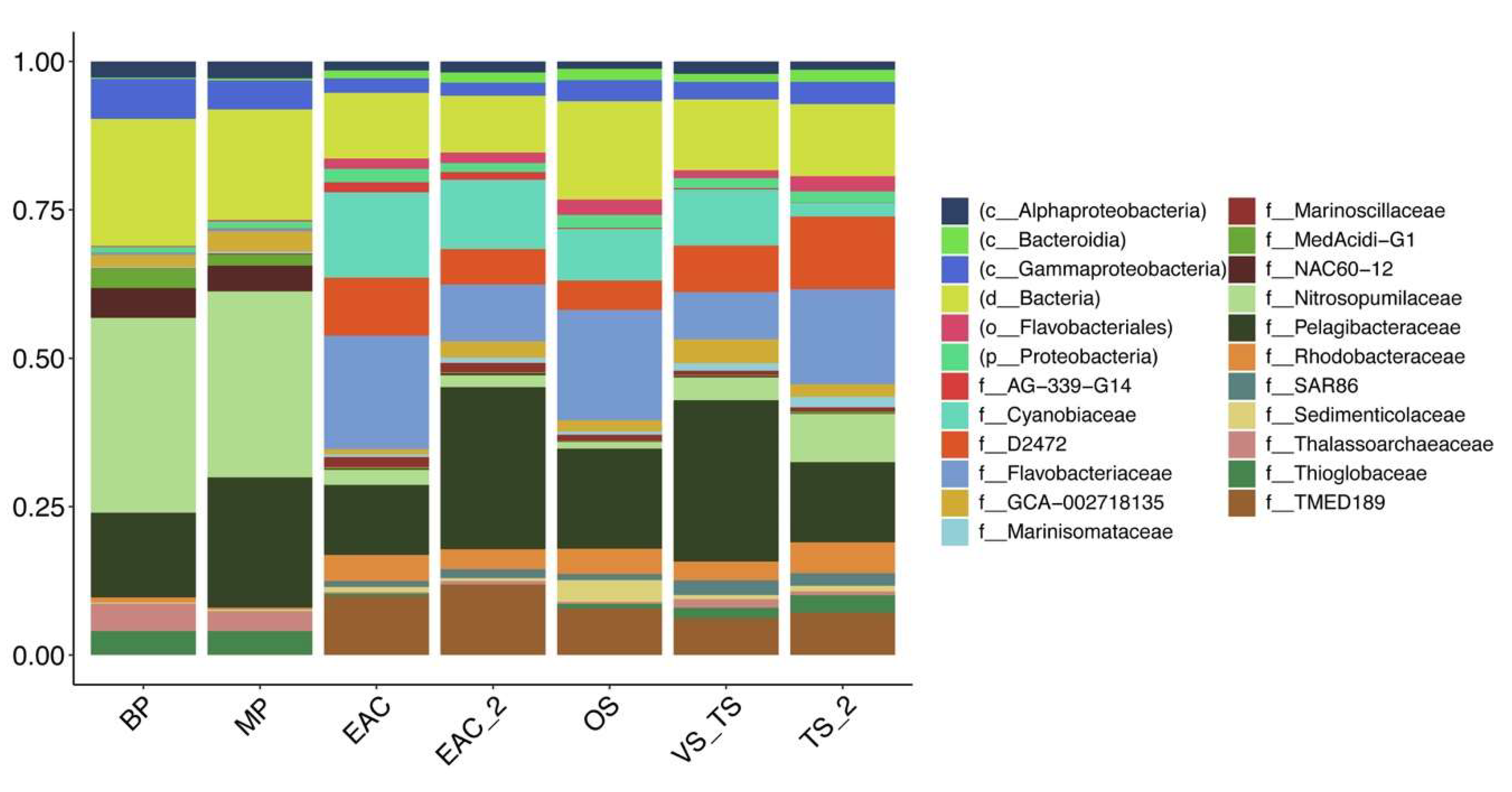
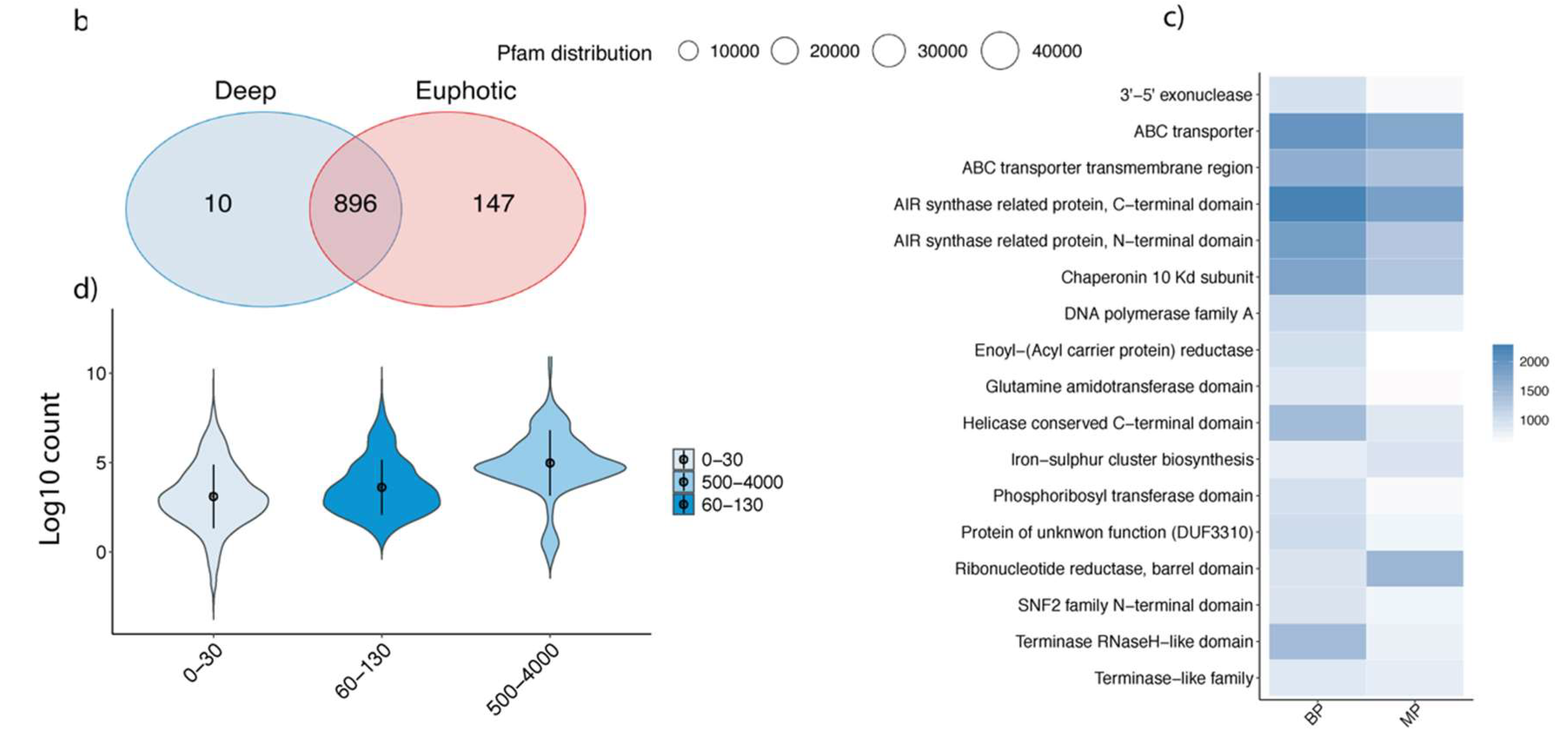
3.2. Viral Diversity in the Different Oceanic Provinces, Depth Distribution and Comparison between Size Fractions
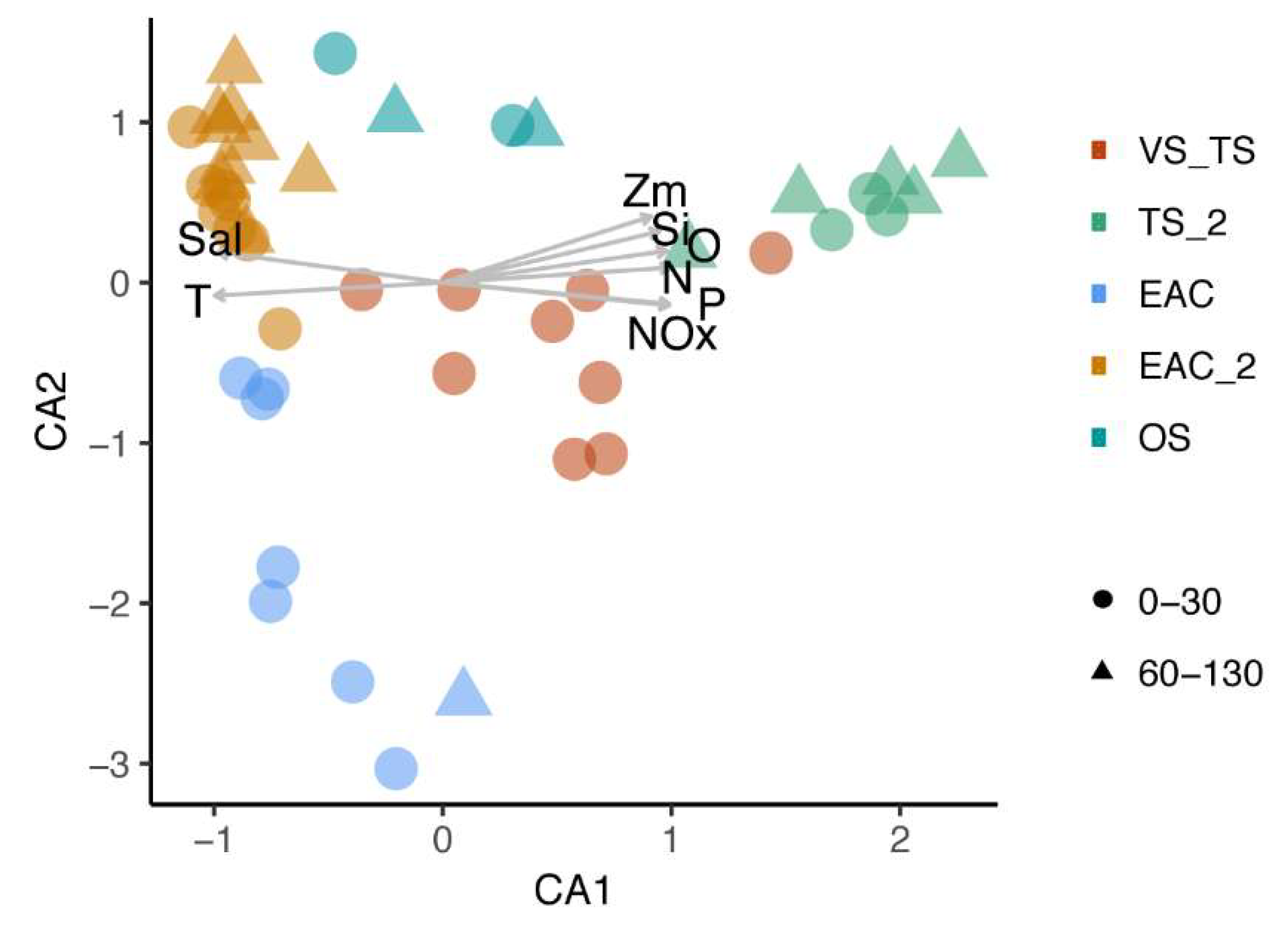
3.3. EAC and Tasman Sea Presents Different Viral Communities
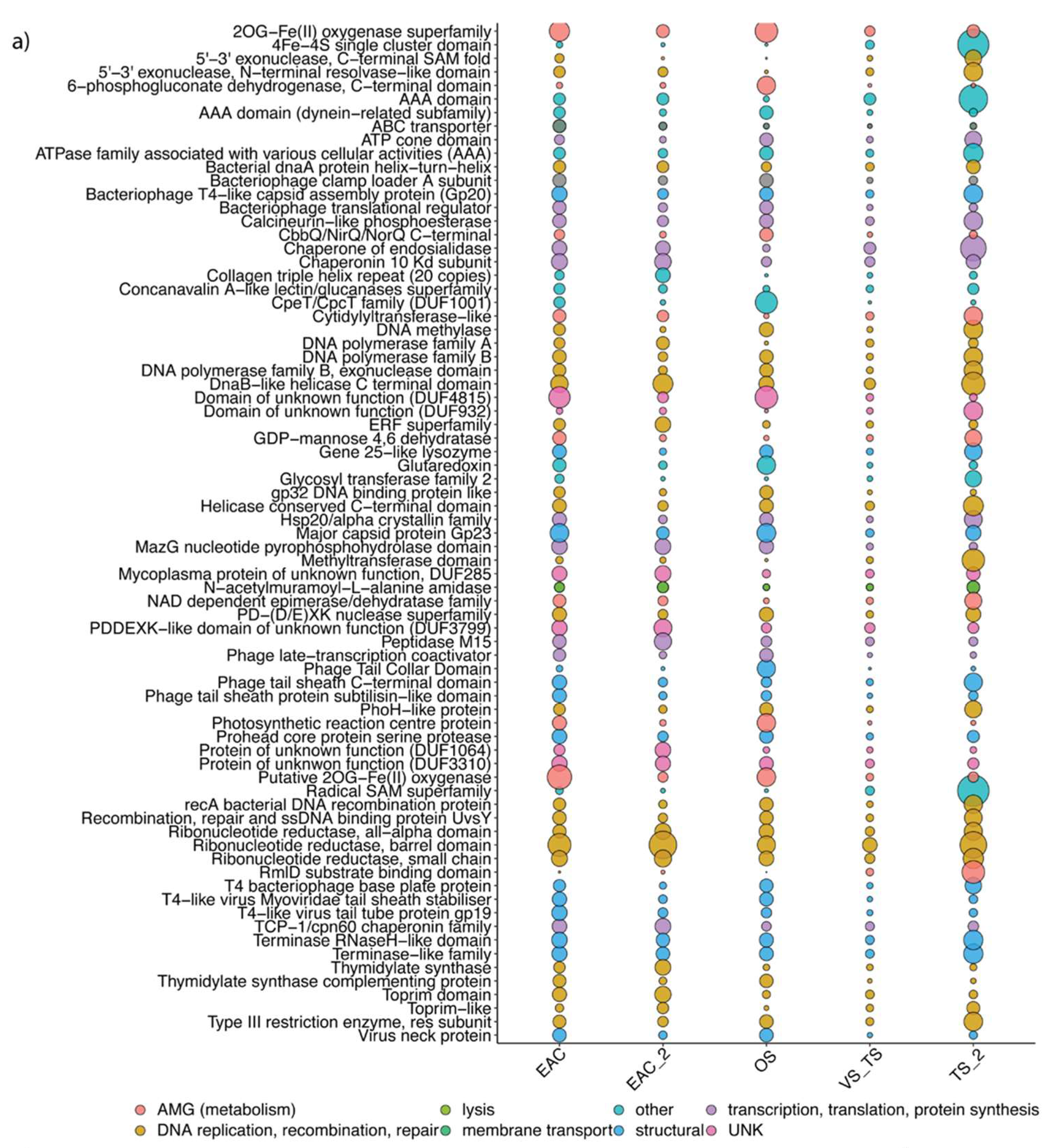
3.4. Functional Profile and Main AMGs in the Different Regions
3.5. Influence of Environmental Parameters on Viral Communities
4. Discussion
4.1. Comparisons between Different Size Fractions: How Much Diversity Are We Losing?
4.2. Ecology of Mesopelagic and Bathypelagic Phages
4.3. Phage Adapt Their Genomic Content Based on Host and Environmental Conditions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fuhrman, J.A. Marine viruses and their biogeochemical and ecological effects. Nature 1999, 399, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Wommack, K.E.; Colwell, R.R. Virioplankton: Viruses in Aquatic Ecosystems. Microbiol. Mol. Biol. Rev. 2000, 64, 69–114. [Google Scholar] [CrossRef] [PubMed]
- Suttle, C.A. Marine viruses-Major players in the global ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Breitbart, M.; Bonnain, C.; Malki, K.; Sawaya, N.A. Phage puppet masters of the marine microbial realm. Nat. Microbiol. 2018, 3, 754–766. [Google Scholar] [CrossRef]
- Lara, E.; Boras, J.A.; Gomes, A.; Borrull, E.; Teira, E.; Pernice, M.C.; Garcia, F.C.; Forn, I.; Castillo, Y.M.; Salazar, G.; et al. Unveiling the role and life strategies of viruses from the surface to the dark ocean. Sci. Adv. 2017, 3, e1602565. [Google Scholar] [CrossRef]
- Danovaro, R.; Corinaldesi, C.; Dell’Anno, A.; Fuhrman, J.A.; Middelburg, J.J.; Noble, R.T.; Suttle, C.A. Marine viruses and global climate change. FEMS Microbiol. Rev. 2011, 35, 993–1034. [Google Scholar] [CrossRef]
- Jover, L.F.; Effler, T.C.; Buchan, A.; Wilhelm, S.W.; Weitz, J.S. The elemental composition of virus particles: Implications for marine biogeochemical cycles. Nat. Rev. Microbiol. 2014, 12, 519–528. [Google Scholar] [CrossRef]
- Guidi, L.; Chaffron, S.; Bittner, L.; Eveillard, D.; Larhlimi, A.; Roux, S.; Darzi, Y.; Audic, S.; Berline, L.; Brum, J.R.; et al. Plankton networks driving carbon export in the oligotrophic ocean. Nature 2016, 532, 465–470. [Google Scholar] [CrossRef]
- Weinbauer, M.G. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 2004, 28, 127–181. [Google Scholar] [CrossRef]
- Lindell, D.; Sullivan, M.B.; Johnson, Z.I.; Tolonen, A.C.; Rohwer, F.; Chisholm, S.W. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc. Natl. Acad. Sci. USA 2004, 101, 11013–11018. [Google Scholar] [CrossRef]
- Hurwitz, B.L.; U’Ren, J.M. Viral metabolic reprogramming in marine ecosystems. Curr. Opin. Microbiol. 2016, 31, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Mann, N.H.; Cook, A.; Bailey, S.; Clokie, M.; Amanullah, A.; Azam, N.; Balliet, A.; Hollander, C.; Hoffman, B.; Jr, A.F.; et al. Bacterial photosynthesis genes in a virus. Nature 2003, 424, 741–742. [Google Scholar] [CrossRef] [PubMed]
- Puxty, R.J.; Millard, A.D.; Evans, D.J.; Scanlan, D.J. Shedding new light on viral photosynthesis. Photosynth. Res. 2015, 126, 71–97. [Google Scholar] [CrossRef] [PubMed]
- Puxty, R.J.; Evans, D.J.; Millard, A.D.; Scanlan, D.J. Energy limitation of cyanophage development: Implications for marine carbon cycling. ISME J. 2018, 12, 1273–1286. [Google Scholar] [CrossRef] [PubMed]
- Millard, A.D.; Zwirglmaier, K.; Downey, M.J.; Mann, N.H.; Scanlan, D.J. Comparative genomics of marine cyanomyoviruses reveals the widespread occurrence of Synechococcus host genes localized to a hyperplastic region: Implications for mechanisms of cyanophage evolution. Environ. Microbiol. 2009, 11, 2370–2387. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.B.; Huang, K.H.; Ignacio-Espinoza, J.C.; Berlin, A.M.; Kelly, L.; Weigele, P.R.; DeFrancesco, A.S.; Kern, S.E.; Thompson, L.R.; Young, S.; et al. Genomic analysis of oceanic cyanobacterial myoviruses compared with T4-Like myoviruses from diverse hosts and environments. Environ. Microbiol. 2010, 12, 3035–3056. [Google Scholar] [CrossRef]
- Thompson, L.R.; Zeng, Q.; Kelly, L.; Huang, K.H.; Singer, A.U.; Stubbe, J.; Chisholm, S.W. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc. Natl. Acad. Sci. USA 2011, 108, E757–E764. [Google Scholar] [CrossRef]
- Wu, L.; Cai, W.; Zhang, L.; Nakamura, H.; Timmermann, A.; Joyce, T.; McPhaden, M.J.; Alexander, M.; Qiu, B.; Visbeck, M.; et al. Enhanced warming over the global subtropical western boundary currents. Nat. Clim. Chang. 2012, 2, 161–166. [Google Scholar] [CrossRef]
- Hobday, A.J.; Pecl, G.T. Identification of global marine hotspots: Sentinels for change and vanguards for adaptation action. Rev. Fish Biol. Fish. 2014, 24, 415–425. [Google Scholar] [CrossRef]
- Pecl, G.T.; Hobday, A.J.; Frusher, S.; Sauer, W.H.H.; Bates, A.E. Ocean warming hotspots provide early warning laboratories for climate change impacts. Rev. Fish Biol. Fish. 2014, 24, 409–413. [Google Scholar] [CrossRef]
- Ridgway, K.R. Long-Term trend and decadal variability of the southward penetration of the East Australian Current. Geophys. Res. Lett. 2007, 34, 1–5. [Google Scholar] [CrossRef]
- Suthers, I.M.; Young, J.W.; Baird, M.E.; Roughan, M.; Everett, J.D.; Brassington, G.B.; Byrne, M.; Condie, S.A.; Hartog, J.R.; Hassler, C.S.; et al. The strengthening East Australian Current, its eddies and biological effects-An introduction and overview. Deep. Res. Part II Top. Stud. Oceanogr. 2011, 58, 538–546. [Google Scholar] [CrossRef]
- Johnson, C.R.; Banks, S.C.; Barrett, N.S.; Cazassus, F.; Dunstan, P.K.; Edgar, G.J.; Frusher, S.D.; Gardner, C.; Haddon, M.; Helidoniotis, F.; et al. Climate change cascades: Shifts in oceanography, species’ ranges and subtidal marine community dynamics in eastern Tasmania. J. Exp. Mar. Biol. Ecol. 2011, 400, 17–32. [Google Scholar] [CrossRef]
- Brown, M.V.; Van De Kamp, J.; Ostrowski, M.; Seymour, J.R.; Ingleton, T.; Messer, L.F.; Jeffries, T.; Siboni, N.; Laverock, B.; Bibiloni-Isaksson, J.; et al. Data Descriptor: Systematic, continental scale temporal monitoring of marine pelagic microbiota by the Australian Marine Microbial Biodiversity Initiative. Sci. Data 2018, 5, 1–10. [Google Scholar] [CrossRef]
- Seymour, J.R.; Doblin, M.A.; Jeffries, T.C.; Brown, M.V.; Newton, K.; Ralph, P.J.; Baird, M.; Mitchell, J.G. Contrasting microbial assemblages in adjacent water masses associated with the East Australian Current. Environ. Microbiol. Rep. 2012, 4, 548–555. [Google Scholar] [CrossRef]
- John, S.G.; Mendez, C.B.; Deng, L.; Poulos, B.; Kauffman, A.K.M.; Kern, S.; Brum, J.; Polz, M.F.; Boyle, E.A.; Sullivan, M.B. A simple and efficient method for concentration of ocean viruses by chemical flocculation. Environ. Microbiol. Rep. 2011, 3, 195–202. [Google Scholar] [CrossRef]
- Hurwitz, B.L.; Deng, L.; Poulos, B.T.; Sullivan, M.B. Evaluation of methods to concentrate and purify ocean virus communities through comparative, replicated metagenomics. Environ. Microbiol. 2013, 15, 1428–1440. [Google Scholar] [CrossRef]
- Andrews, S.; Krueger, F. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 27 February 2020).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.P.P. metaSPAdes: A New Versatile Metagenomic Assembler. Genome Res. 2017, 1, 30–47. [Google Scholar] [CrossRef]
- Nishimura, Y.; Watai, H.; Honda, T.; Mihara, T.; Omae, K.; Roux, S.; Blanc-Mathieu, R.; Yamamoto, K.; Hingamp, P.; Sako, Y.; et al. Environmental viral genomes shed new light on virus-host interactions in the ocean. mSphere 2017, 2, e00359-16. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Enault, F.; Hurwitz, B.L.; Sullivan, M.B. VirSorter: Mining viral signal from microbial genomic data. PeerJ 2015, 3, e985. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Lomsadze, A.; Borodovsky, M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010, 38, e132. [Google Scholar] [CrossRef] [PubMed]
- Menzel, P.; Ng, K.L.; Krogh, A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 2016, 7, 11257. [Google Scholar] [CrossRef]
- Paez-Espino, D.; Chen, I.M.A.; Palaniappan, K.; Ratner, A.; Chu, K.; Szeto, E.; Pillay, M.; Huang, J.; Markowitz, V.M.; Nielsen, T.; et al. IMG/VR: A database of cultured and uncultured DNA viruses and retroviruses. Nucleic Acids Res. 2017, 45, D457–D465. [Google Scholar] [CrossRef]
- Gruber-Vodicka, H.R.; Seah, B.K.B.; Pruesse, E. phyloFlash–Rapid SSU rRNA profiling and targeted assembly from metagenomes. bioRxiv 2019, 521922. [Google Scholar] [CrossRef]
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.-A.; Hugenholtz, P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018, 36, 996. [Google Scholar] [CrossRef]
- Kauffman, K.M.; Hussain, F.A.; Yang, J.; Arevalo, P.; Brown, J.M.; Chang, W.K.; Vaninsberghe, D.; Elsherbini, J.; Sharma, R.S.; Cutler, M.B.; et al. A major lineage of non-Tailed dsDNA viruses as unrecognized killers of marine bacteria. Nature 2018, 554, 118–122. [Google Scholar] [CrossRef]
- Castillo, D.; Kauffman, K.; Hussain, F.; Kalatzis, P.; Rørbo, N.; Polz, M.F.; Middelboe, M. Widespread distribution of prophage-Encoded virulence factors in marine Vibrio communities. Sci. Rep. 2018, 8, 9973. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Langmead Bowtie2. Nat. Methods 2013, 9, 357–359. [Google Scholar] [CrossRef]
- Coutinho, F.H.; Silveira, C.B.; Gregoracci, G.B.; Thompson, C.C.; Edwards, R.A.; Brussaard, C.P.D.; Dutilh, B.E.; Thompson, F.L. Marine viruses discovered via metagenomics shed light on viral strategies throughout the oceans. Nat. Commun. 2017, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, 2018. Available online: https://www.R-project.org/ (acceseed on 27 February 2020).
- Clarke, K.R.; Somerfield, P.J.; Gorley, R.N. Testing of null hypotheses in exploratory community analyses: Similarity profiles and biota-Environment linkage. J. Exp. Mar. Biol. Ecol. 2008, 366, 56–69. [Google Scholar] [CrossRef]
- Oksanen, A.J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; Hara, R.B.O.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. vegan: Community Ecology Package; 2019, R Package Version 2.5-6. Available online: https://CRAN.R-project.org/package=vegan (acceseed on 27 February 2020).
- Paez-Espino, D.; Pavlopoulos, G.A.; Ivanova, N.N.; Kyrpides, N.C. Nontargeted virus sequence discovery pipeline and virus clustering for metagenomic data. Nat. Protoc. 2017, 12, 1673–1682. [Google Scholar] [CrossRef] [PubMed]
- Brum, J.R.; Ignacio-espinoza, J.C.; Roux, S.; Doulcier, G.; Acinas, S.G.; Alberti, A.; Chaffron, S.; Cruaud, C.; De Vargas, C.; Gasol, J.M.; et al. Patterns and ecological drivers of ocean viral communities. Science (80-.) 2016, 348. [Google Scholar] [CrossRef]
- Roux, S.; Brum, J.R.; Dutilh, B.E.; Sunagawa, S.; Duhaime, M.B.; Loy, A.; Poulos, B.T.; Solonenko, N.; Lara, E.; Poulain, J.; et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 2016, 537, 689–693. [Google Scholar] [CrossRef]
- Williamson, S.J.; Rusch, D.B.; Yooseph, S.; Halpern, A.L.; Heidelberg, K.B.; Glass, J.I.; Andrews, C.; Fadrosh, D.; Miller, C.S.; Sutton, G.; et al. The Sorcerer II Global Ocean Sampling Expedition: Metagenomic Characterization of Viruses within Aquatic Microbial Samples. PLoS ONE 2008, 3, e1456. [Google Scholar] [CrossRef]
- Brum, J.R.; Schenck, R.O.; Sullivan, M.B. Global morphological analysis of marine viruses shows minimal regional variation and dominance of non-Tailed viruses. ISME J. 2013, 7, 1738–1751. [Google Scholar] [CrossRef]
- Rusch, D.B.; Halpern, A.L.; Sutton, G.; Heidelberg, K.B.; Williamson, S.; Yooseph, S.; Wu, D.; Eisen, J.A.; Hoffman, J.M.; Remington, K.; et al. The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007, 5, 0398–0431. [Google Scholar] [CrossRef]
- Hurwitz, B.L.; Brum, J.R.; Sullivan, M.B. Depth-Stratified functional and taxonomic niche specialization in the ‘ core ’ and ‘ flexible ’ Pacific Ocean Virome. ISME J. 2015, 472–484. [Google Scholar] [CrossRef]
- Gregory, A.C.; Zayed, A.A.; Conceição-Neto, N.; Temperton, B.; Bolduc, B.; Alberti, A.; Ardyna, M.; Arkhipova, K.; Carmichael, M.; Cruaud, C.; et al. Marine DNA Viral Macro- and Microdiversity from Pole to Pole. Cell 2019, 177, 1–15. [Google Scholar] [CrossRef]
- Touchon, M.; Bernheim, A.; Rocha, E.P.C. Genetic and life-History traits associated with the distribution of prophages in bacteria. ISME J. 2016, 10, 2744–2754. [Google Scholar] [CrossRef] [PubMed]
- Luo, E.; Aylward, F.O.; Mende, D.R.; DeLong, E.F. Bacteriophage Distributions and Temporal Variability in the Ocean’s Interior. MBio 2017, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mende, D.R.; Bryant, J.A.; Aylward, F.O.; Eppley, J.M.; Nielsen, T.; Karl, D.M.; Delong, E.F. Environmental drivers of a microbial genomic transition zone in the ocean’s interior. Nat. Microbiol. 2017, 2, 1367–1373. [Google Scholar] [CrossRef]
- Ahlgren, N.A.; Perelman, J.N.; Yeh, Y.-C.; Fuhrman, J.A. Multi-Year dynamics of fine-scale marine cyanobacterial populations are more strongly explained by phage interactions than abiotic, bottom-Up factors. Environ. Microbiol. 2019, 21, 2948–2963. [Google Scholar] [CrossRef] [PubMed]
- Gasper, R.; Schwach, J.; Hartmann, J.; Holtkamp, A.; Wiethaus, J.; Riedel, N.; Hofmann, E.; Frankenberg-Dinkel, N. Distinct features of cyanophage-Encoded T-Type phycobiliprotein lyase ΦCpeT: The role of auxiliary metabolic genes. J. Biol. Chem. 2017, 292, 3089–3098. [Google Scholar] [CrossRef] [PubMed]
- Behrenfeld, M.J.; O’Malley, R.T.; Siegel, D.A.; McClain, C.R.; Sarmiento, J.L.; Feldman, G.C.; Milligan, A.J.; Falkowski, P.G.; Letelier, R.M.; Boss, E.S. Climate-Driven trends in contemporary ocean productivity. Nature 2006, 444, 752–755. [Google Scholar] [CrossRef]
- Gross, M.; Marianovsky, I.; Glaser, G. MazG—A regulator of programmed cell death in Escherichia coli. Mol. Microbiol. 2006, 59, 590–601. [Google Scholar] [CrossRef]
- Warwick-Dugdale, J.; Buchholz, H.H.; Allen, M.J.; Temperton, B. Host-Hijacking and planktonic piracy: How phages command the microbial high seas. Virol. J. 2019, 16, 15. [Google Scholar] [CrossRef]
- Thompson, P.A.; Bonham, P.; Waite, A.M.; Clementson, L.A.; Cherukuru, N.; Hassler, C.; Doblin, M.A. Contrasting oceanographic conditions and phytoplankton communities on the east and west coasts of Australia. Deep. Res. Part II Top. Stud. Oceanogr. 2011, 58, 645–663. [Google Scholar] [CrossRef]
- Dobler, I.W.; Biebl, H. Environmental Biology of the Marine Roseobacter Lineage. Annu. Rev. Microbiol. 2006, 60, 255–280. [Google Scholar] [CrossRef]
- Wang, S.C.; Frey, P.A. S-Adenosylmethionine as an oxidant: The radical SAM superfamily. Trends Biochem. Sci. 2007, 32, 209. [Google Scholar] [CrossRef] [PubMed]
- Markine-Goriaynoff, N.; Gillet, L.; Van Etten, J.L.; Korres, H.; Verma, N.; Vanderplasschen, A. Glycosyltransferases encoded by viruses. J. Gen. Virol. 2004, 85, 2741–2754. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Golding, I.; Sawai, S.; Guo, L.; Cox, E.C. Population fitness and the regulation of Escherichia coli genes by bacterial viruses. PLoS Biol. 2005, 3, e299. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Focardi, A.; Ostrowski, M.; Goossen, K.; Brown, M.V.; Paulsen, I. Investigating the Diversity of Marine Bacteriophage in Contrasting Water Masses Associated with the East Australian Current (EAC) System. Viruses 2020, 12, 317. https://doi.org/10.3390/v12030317
Focardi A, Ostrowski M, Goossen K, Brown MV, Paulsen I. Investigating the Diversity of Marine Bacteriophage in Contrasting Water Masses Associated with the East Australian Current (EAC) System. Viruses. 2020; 12(3):317. https://doi.org/10.3390/v12030317
Chicago/Turabian StyleFocardi, Amaranta, Martin Ostrowski, Kirianne Goossen, Mark V. Brown, and Ian Paulsen. 2020. "Investigating the Diversity of Marine Bacteriophage in Contrasting Water Masses Associated with the East Australian Current (EAC) System" Viruses 12, no. 3: 317. https://doi.org/10.3390/v12030317
APA StyleFocardi, A., Ostrowski, M., Goossen, K., Brown, M. V., & Paulsen, I. (2020). Investigating the Diversity of Marine Bacteriophage in Contrasting Water Masses Associated with the East Australian Current (EAC) System. Viruses, 12(3), 317. https://doi.org/10.3390/v12030317





