Common and Strain-Specific Post-Translational Modifications of the Potyvirus Plum pox virus Coat Protein in Different Hosts
Abstract
1. Introduction
2. Materials and Methods
2.1. Viral cDNA Clones and other Plasmids
2.2. Plant Growth Conditions and Viral Inoculation
2.3. Assessment of Viral Infection and Virion Purification
2.4. Fractionation by Centrifugation and Immunoprecipitation of CP
2.5. Proteomic Approaches
2.5.1. Matrix-assisted Laser Desorption Ionization–Time of Flight (MALDI-TOF)
2.5.2. Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS)
3. Results
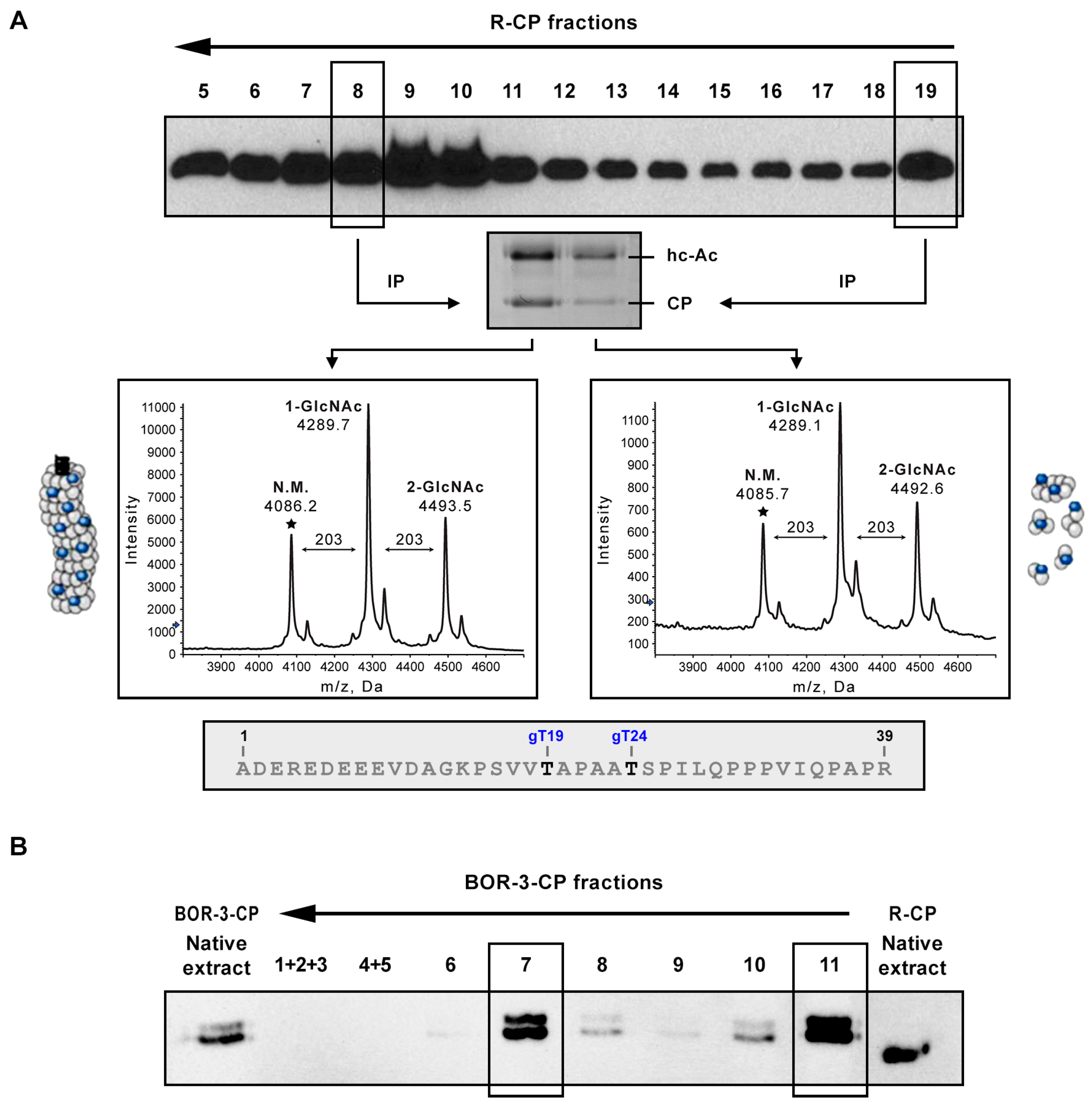
3.1. O-GlcNAcylation and Phosphorylation of PPV CP also Affect Protein not Assembled in Mature Viral Particles
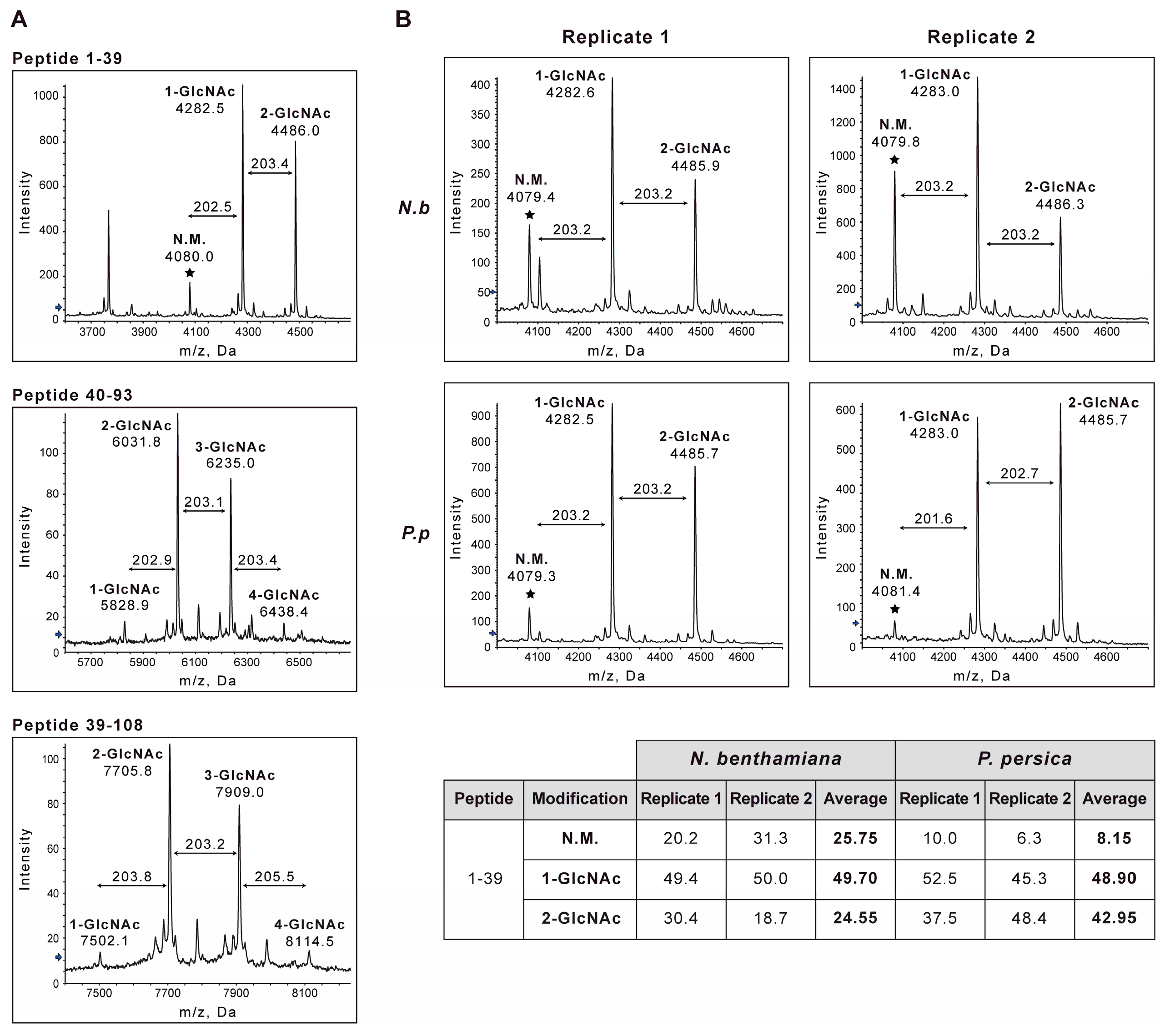
3.2. Post-Translational Modifications Affecting PPV CP Take Place in the PPV Natural Host Prunus Persica
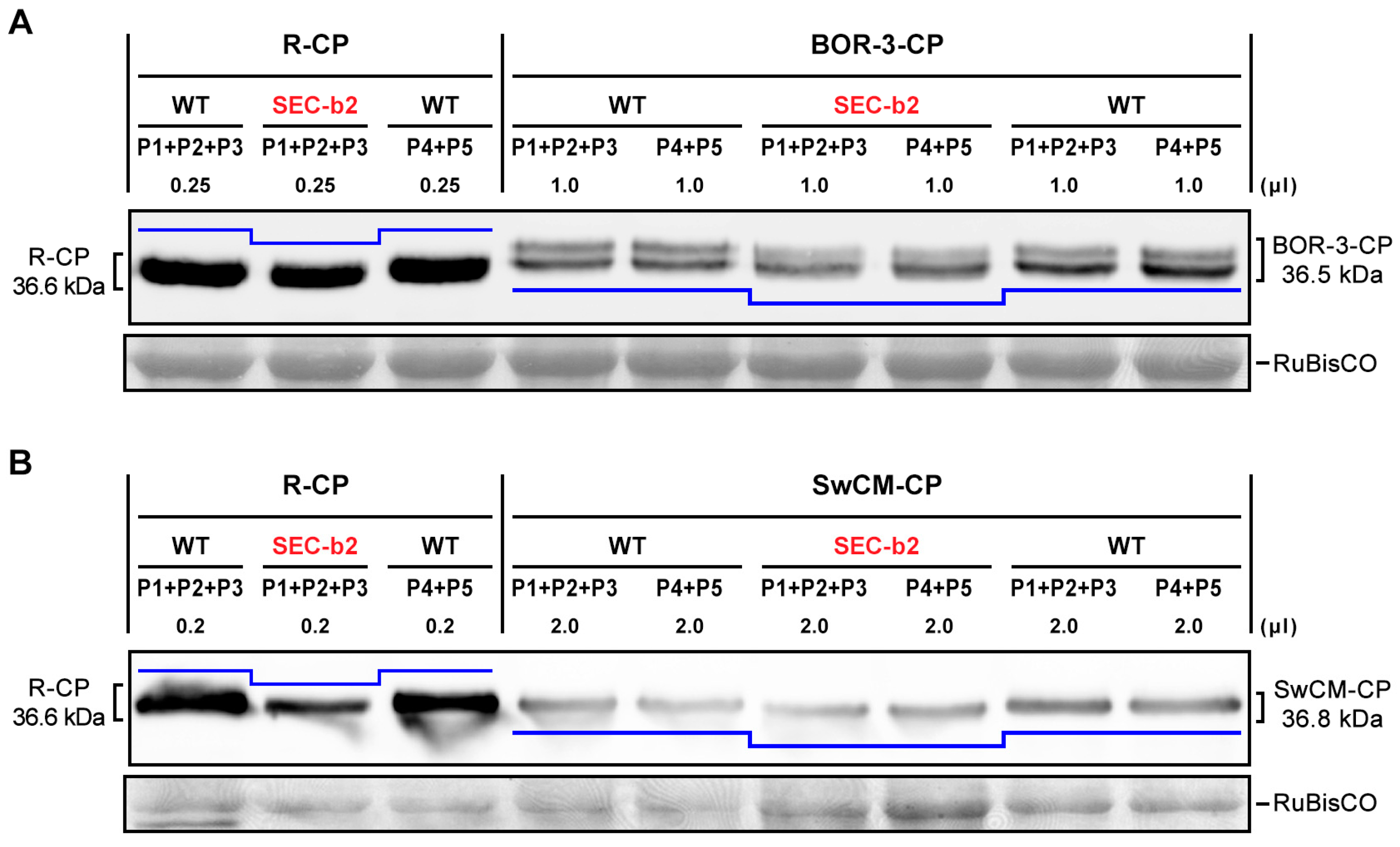
3.3. Post-Translational Modifications Affect the CP of Different PPV Strains
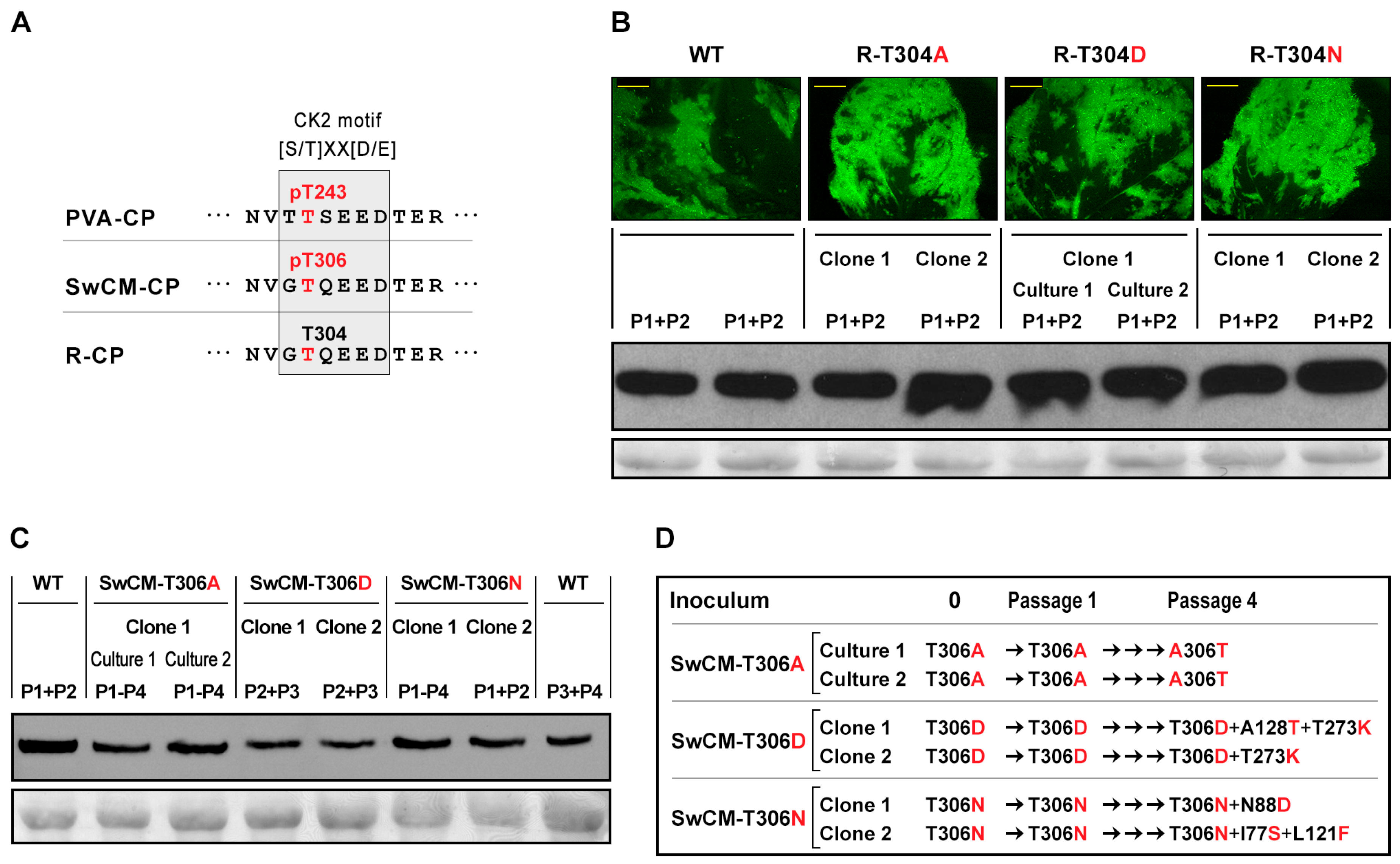
3.4. The Relevance of Phosphorylation at Conserved CK2 Motif of CP Differs among Potyviruses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bretana, N.A.; Lu, C.T.; Chiang, C.Y.; Su, M.G.; Huang, K.Y.; Lee, T.Y.; Weng, S.L. Identifying protein phosphorylation sites with kinase substrate specificity on human viruses. PLoS ONE 2012, 7, e40694. [Google Scholar] [CrossRef] [PubMed]
- Criglar, J.M.; Anish, R.; Hu, L.; Crawford, S.E.; Sankaran, B.; Prasad, B.V.V.; Estes, M.K. Phosphorylation cascade regulates the formation and maturation of rotaviral replication factories. Proc. Natl. Acad. Sci. USA 2018, 115, E12015–E12023. [Google Scholar] [CrossRef] [PubMed]
- Gil-Ranedo, J.; Hernando, E.; Valle, N.; Riolobos, L.; Maroto, B.; Almendral, J.M. Differential phosphorylation and n-terminal configuration of capsid subunits in parvovirus assembly and viral trafficking. Virology 2018, 518, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sun, L.; Zheng, W.; Madina, M.; Li, J.; Bi, Y.; Wang, H.; Liu, W.; Luo, T.R. Phosphorylation and dephosphorylation of threonine 188 in nucleoprotein is crucial for the replication of influenza A virus. Virology 2018, 520, 30–38. [Google Scholar] [CrossRef]
- Zhang, K.; Brownlie, R.; Snider, M.; van Drunen Littel-van den Hurk, S. Phosphorylation of bovine herpesvirus 1 VP8 plays a role in viral DNA encapsidation and is essential for its cytoplasmic localization and optimal virion incorporation. J. Virol. 2016, 90, 4427. [Google Scholar] [CrossRef]
- Citovsky, V.; MaLean, B.G.; Zupan, J.R.; Zambryski, P. Phosphorylation of tobacco mosaic virus cell-to-cell movement protein by a developmentally regulated plant cell wall-associated protein kinase. Genes Dev. 1993, 7, 904–910. [Google Scholar] [CrossRef]
- Kim, S.H.; Palukaitis, P.; Park, Y.I. Phosphorylation of cucumber mosaic virus RNA polymerase 2a protein inhibits formation of replicase complex. EMBO J. 2002, 21, 2292–2300. [Google Scholar] [CrossRef]
- Jakubiec, A.; Tournier, V.; Drugeon, G.; Pflieger, S.; Camborde, L.; Vinh, J.; Héricourt, F.; Redeker, V.; Jupin, I. Phosphorylation of viral RNA-dependent RNA polymerase and its role in replication of a plus-strand RNA virus. J. Biol. Chem. 2006, 281, 21236–21249. [Google Scholar] [CrossRef]
- Módena, N.A.; Zelada, A.M.; Conte, F.; Mentaberry, A. Phosphorylation of the TGBp1 movement protein of Potato virus X by a Nicotiana tabacum CK2-like activity. Virus Res. 2008, 137, 16–23. [Google Scholar] [CrossRef]
- Hu, Y.; Li, Z.; Yuan, C.; Jin, X.; Yan, L.; Zhao, X.; Zhang, Y.; Jackson, A.O.; Wang, X.; Han, C.; et al. Phosphorylation of TGB1 by protein kinase CK2 promotes barley stripe mosaic virus movement in monocots and dicots. J. Exp. Bot. 2015, 66, 4733–4747. [Google Scholar] [CrossRef]
- Nemes, K.; Gellért, Á.; Almási, A.; Vági, P.; Sáray, R.; Kádár, K.; Salánki, K. Phosphorylation regulates the subcellular localization of Cucumber Mosaic Virus 2b protein. Sci. Rep. 2017, 7, 13444. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Dong, K.; Xu, K.; Zhang, K.; Jin, X.; Yang, M.; Zhang, Y.; Wang, X.; Han, C.; Yu, J.; et al. Barley stripe mosaic virus infection requires PKA-mediated phosphorylation of γb for suppression of both RNA silencing and the host cell death response. New Phytol. 2018, 218, 1570–1585. [Google Scholar] [CrossRef] [PubMed]
- Atabekov, J.G.; Rodionova, N.P.; Karpova, O.V.; Kozlovsky, S.V.; Novikov, V.K.; Arkhipenko, M.V. Translational activation of encapsidated potato virus X RNA by coat protein phosphorylation. Virology 2001, 286, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.J.; Huang, Y.W.; Liou, M.R.; Lee, Y.C.; Lin, N.S.; Meng, M.; Tsai, C.H.; Hu, C.C.; Hsu, Y.H. Phosphorylation of coat protein by protein kinase CK2 regulates cell-to-cell movement of Bamboo mosaic virus through modulating RNA binding. Mol. Plant Microbe Interact. 2014, 27, 1211–1225. [Google Scholar] [CrossRef] [PubMed]
- Hoover, H.S.; Wang, J.C.; Middleton, S.; Ni, P.; Zlotnick, A.; Vaughan, R.C.; Kao, C.C. Phosphorylation of the brome mosaic virus capsid regulates the timing of viral infection. J. Virol. 2016, 90, 7748–7760. [Google Scholar] [CrossRef][Green Version]
- Zhao, X.; Wang, X.; Dong, K.; Zhang, Y.; Hu, Y.; Zhang, X.; Chen, Y.; Wang, X.; Han, C.; Yu, J.; et al. Phosphorylation of Beet black scorch virus coat protein by PKA is required for assembly and stability of virus particles. Sci. Rep. 2015, 5, 11585. [Google Scholar] [CrossRef]
- Nemes, K.; Gellért, Á.; Bóka, K.; Vági, P.; Salánki, K. Symptom recovery is affected by Cucumber mosaic virus coat protein phosphorylation. Virology 2019, 536, 68–77. [Google Scholar] [CrossRef]
- Valli, A.; García, J.A.; López-Moya, J.J. Potyviruses (Potyviridae). In Encyclopedia of Virology, 4th ed.; Bamford, D., Zuckerman, M., Eds.; Elsevier: Oxford, UK, 2020; in press. [Google Scholar]
- Revers, F.; García, J.A. Molecular biology of potyviruses. Adv. Virus Res. 2015, 92, 101–199. [Google Scholar]
- Ivanov, K.I.; Eskelin, K.; Lõhmus, A.; Mäkinen, K. Molecular and cellular mechanisms underlying potyvirus infection. J. Gen. Virol. 2014, 95, 1415–1429. [Google Scholar] [CrossRef]
- Chung, B.Y.W.; Miller, W.A.; Atkins, J.F.; Firth, A.E. An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. USA 2008, 105, 5897–5902. [Google Scholar] [CrossRef]
- Rodamilans, B.; Valli, A.; Mingot, A.; San León, D.; Baulcombe, D.; López-Moya, J.J.; García, J.A. RNA polymerase slippage as a mechanism for the production of frameshift gene products in plant viruses of the Potyviridae family. J. Virol. 2015, 89, 6965–6967. [Google Scholar] [CrossRef] [PubMed]
- Olspert, A.; Chung, B.Y.; Atkins, J.F.; Carr, J.P.; Firth, A.E. Transcriptional slippage in the positive-sense RNA virus family Potyviridae. EMBO Rep. 2015, 16, 995–1004. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, K.I.; Puustinen, P.; Gabrenaite, R.; Vihinen, H.; Rönnstrand, L.; Valmu, L.; Kalkkinen, N.; Mäkinen, K. Phosphorylation of the potyvirus capsid protein by protein kinase CK2 and its relevance for virus infection. Plant Cell 2003, 15, 2124–2139. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, K.I.; Puustinen, P.; Merits, A.; Saarma, M.; Mäkinen, K. Phosphorylation down-regulates the RNA binding function of the coat protein of potato virus A. J. Biol. Chem. 2001, 276, 13530–13540. [Google Scholar] [CrossRef] [PubMed]
- Lõhmus, A.; Hafrén, A.; Mäkinen, K. Coat protein regulation by CK2, CPIP, HSP70 and CHIP is required for Potato virus A replication and coat protein accumulation. J. Virol. 2017, 91, e01316. [Google Scholar] [CrossRef] [PubMed]
- Hafrén, A.; Hofius, D.; Ronnholm, G.; Sonnewald, U.; Mäkinen, K. HSP70 and its cochaperone CPIP promote potyvirus infection in Nicotiana benthamiana by regulating viral coat protein functions. Plant Cell 2010, 22, 523–535. [Google Scholar] [CrossRef]
- Fernández-Fernández, M.R.; Camafeita, E.; Bonay, P.; Méndez, E.; Albar, J.P.; García, J.A. The capsid protein of a plant single-stranded RNA virus is modified by O-linked N-acetylglucosamine. J. Biol. Chem. 2002, 277, 135–140. [Google Scholar] [CrossRef]
- Šubr, Z.; Ryšlavá, H.; Kollerová, E. Electrophoretic mobility of the capsid protein of the Plum pox virus strain PPV-Rec indicates its partial phosphorylation. Acta Virol. 2007, 51, 135–138. [Google Scholar]
- Chen, D.; Juárez, S.; Hartweck, L.; Alamillo, J.M.; Simón-Mateo, C.; Pérez, J.J.; Fernández-Fernández, M.R.; Olszewski, N.E.; García, J.A. Identification of secret agent as the O-GlcNAc transferase that participates in Plum pox virus infection. J. Virol. 2005, 79, 9381–9387. [Google Scholar] [CrossRef][Green Version]
- van der Laarse, S.A.M.; Leney, A.C.; Heck, A.J.R. Crosstalk between phosphorylation and O-GlcNAcylation: Friend or foe. FEBS J. 2018, 285, 3152–3167. [Google Scholar] [CrossRef]
- Angelova, M.; Ortiz-Meoz, R.F.; Walker, S.; Knipe, D.M. Inhibition of O-Linked N-Acetylglucosamine transferase reduces replication of herpes simplex virus and human cytomegalovirus. J. Virol. 2015, 89, 8474–8483. [Google Scholar] [CrossRef] [PubMed]
- Groussaud, D.; Khair, M.; Tollenaere, A.I.; Waast, L.; Kuo, M.S.; Mangeney, M.; Martella, C.; Fardini, Y.; Coste, S.; Souidi, M.; et al. Hijacking of the O-GlcNAcZYME complex by the HTLV-1 Tax oncoprotein facilitates viral transcription. PLoS Pathog. 2017, 13, e1006518. [Google Scholar] [CrossRef] [PubMed]
- Jochmann, R.; Pfannstiel, J.; Chudasama, P.; Kuhn, E.; Konrad, A.; Stürzl, M. O-GlcNAc transferase inhibits KSHV propagation and modifies replication relevant viral proteins as detected by systematic O-GlcNAcylation analysis. Glycobiology 2013, 23, 1114–1130. [Google Scholar] [CrossRef] [PubMed]
- Herzog, K.; Bandiera, S.; Pernot, S.; Fauvelle, C.; Juhling, F.; Weiss, A.; Bull, A.; Durand, S.C.; Chane-Woon-Ming, B.; Pfeffer, S.; et al. Functional microRNA screen uncovers O-linked N-acetylglucosamine transferase as a host factor modulating hepatitis C virus morphogenesis and infectivity. Gut 2019, 69, 380–392. [Google Scholar] [CrossRef]
- Kim, Y.C.; Udeshi, N.D.; Balsbaugh, J.L.; Shabanowitz, J.; Hunt, D.F.; Olszewski, N.E. O-GlcNAcylation of the Plum pox virus capsid protein catalyzed by SECRET AGENT: Characterization of O-GlcNAc sites by electron transfer dissociation mass spectrometry. Amino Acids 2011, 40, 869–876. [Google Scholar] [CrossRef]
- Pérez, J.J.; Udeshi, N.D.; Shabanowitz, J.; Ciordia, S.; Juárez, S.; Scott, C.L.; Olszewski, N.E.; Hunt, D.F.; García, J.A. O-GlcNAc modification of the coat protein of the potyvirus Plum pox virus enhances viral infection. Virology 2013, 442, 122–131. [Google Scholar] [CrossRef]
- Martínez-Turiño, S.; Pérez, J.J.; Hervás, M.; Navajas, R.; Ciordia, S.; Udeshi, N.D.; Shabanowitz, J.; Hunt, D.F.; García, J.A. Phosphorylation coexists with O-GlcNAcylation in a plant virus protein and influences viral infection. Mol. Plant Pathol. 2018, 19, 1427–1443. [Google Scholar] [CrossRef]
- Hervás, M.; Navajas, R.; Chagoyen, M.; Garcia, J.A.; Martinez-Turiño, S. Phosphorylation-related cross-talk between distant regions of the core region of the coat protein contributes to virion assembly of Plum pox virus. Mol. Plant Microbe Interact. 2020. [Google Scholar] [CrossRef]
- García, J.A.; Glasa, M.; Cambra, M.; Candresse, T. Plum pox virus and sharka: A model potyvirus and a major disease. Mol. Plant Pathol. 2014, 15, 226–241. [Google Scholar] [CrossRef]
- Šubr, Z.; Glasa, M. Unfolding the secrets of plum pox virus: From epidemiology to genomics. Acta Virol. 2013, 57, 217–228. [Google Scholar] [CrossRef]
- Rodamilans, B.; Valli, A.; García, J.A. Molecular plant-plum pox virus interactions. Mol. Plant Microbe Interact. 2020, 33, 6–17. [Google Scholar] [CrossRef]
- Hajizadeh, M.; Gibbs, A.J.; Amirnia, F.; Glasa, M. The global phylogeny of Plum pox virus is emerging. J. Gen. Virol. 2019, 100, 1457–1468. [Google Scholar] [CrossRef] [PubMed]
- Kollerová, E.; Glasa, M.; Šubr, Z.W. Western blotting analysis of the Plum pox virus capsid protein. J. Plant Pathol. 2008, 90, 19–22. [Google Scholar]
- Pasin, F.; García, J.A. Centro Nacional de Biotecnology, CNB-CSIC, Madrid Spain. cDNA sequence of the chimeric virus PPV-5′BD-GFP cloned in a pSN- vector. 2014. [Google Scholar]
- Salvador, B.; Delgadillo, M.O.; Saénz, P.; García, J.A.; Simón-Mateo, C. Identification of Plum pox virus pathogenicity determinants in herbaceous and woody hosts. Mol. Plant Microbe Interact. 2008, 21, 20–29. [Google Scholar] [CrossRef]
- Pasin, F.; Simón-Mateo, C.; García, J.A. The hypervariable amino-terminus of P1 protease modulates potyviral replication and host defense responses. PLoS Pathog. 2014, 10, e1003985. [Google Scholar] [CrossRef] [PubMed]
- Calvo, M.; Martínez-Turiño, S.; García, J.A. Resistance to Plum pox virus strain C in Arabidopsis thaliana and Chenopodium foetidum involves genome-linked viral protein and other viral determinants and might depend on compatibility with host translation initiation factors. Mol. Plant Microbe Interact. 2014, 27, 1291–1301. [Google Scholar] [CrossRef]
- Ho, S.N.; Hunt, H.D.; Horton, R.M.; Pullen, J.K.; Pease, L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 1989, 77, 51–59. [Google Scholar] [CrossRef]
- Gallo, A.; Valli, A.; Calvo, M.; García, J.A. A functional link between RNA replication and virion assembly in the potyvirus Plum pox virus. J. Virol. 2018, 92, e02179-17. [Google Scholar] [CrossRef]
- Laín, S.; Riechmann, J.L.; Méndez, E.; García, J.A. Nucleotide sequence of the 3’ terminal region of plum pox potyvirus RNA. Virus Res. 1988, 10, 325–342. [Google Scholar] [CrossRef]
- Sheveleva, A.A.; Nikitin, N.; Trifonova, E.A.; Zakubanskiy, A.V.; Chirkov, S. Improved method of purification of Plum pox virus and serological analysis of the coat protein. Agric. Biol. 2016, 51, 385–391. [Google Scholar] [CrossRef]
- Valli, A.; Gallo, A.; Calvo, M.; Pérez, J.J.; García, J.A. A novel role of the potyviral helper component proteinase contributes to enhance the yield of viral particles. J. Virol. 2014, 88, 9808–9818. [Google Scholar] [CrossRef] [PubMed]
- Taurino, M.; Abelenda, J.A.; Río-Alvarez, I.; Navarro, C.; Vicedo, B.; Farmaki, T.; Jiménez, P.; García-Agustín, P.; López-Solanilla, E.; Prat, S.; et al. Jasmonate-dependent modifications of the pectin matrix during potato development function as a defense mechanism targeted by Dickeya dadantii virulence factors. Plant J. 2014, 77, 418–429. [Google Scholar] [CrossRef] [PubMed]
- Navajas, R.; Paradela, A.; Albar, J.P. Immobilized metal affinity chromatography/reversed-phase enrichment of phosphopeptides and analysis by CID/ETD tandem mass spectrometry. Methods Mol. Biol. 2011, 681, 337–348. [Google Scholar] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Calvo, M. Identification of Pathogenicity Determinants Involved in the Adaptation of Plum pox virus Strain C to Prunus avium and Herbaceous Hosts; Universidad Autónoma de Madrid: Madrid, Spain, 2014. [Google Scholar]
- Pérez, J.J. Caracterización de la Modificación por O-GlcNAc de la Proteína de la Cápside del Potyvirus Plum pox virus y su Relevancia par la Infección Viral; Universidad Autónoma de Madrid: Madrid, Spain, 2014. [Google Scholar]
- Šubr, Z.W.; Kamencayová, M.; Nováková, S.; Nagyová, A.; Nosek, J.; Glasa, M. A single amino acid mutation alters the capsid protein electrophoretic double-band phenotype of the Plum pox virus strain PPV-Rec. Arch. Virol. 2010, 155, 1151–1155. [Google Scholar] [CrossRef]
- Dolja, V.V.; Haldeman, R.; Robertson, N.L.; Dougherty, W.G.; Carrington, J.C. Distinct functions of capsid protein in assembly and movement of tobacco etch potyvirus in plants. EMBO J. 1994, 13, 1482–1491. [Google Scholar] [CrossRef]
- Nicaise, V.; Candresse, T. Plum pox virus capsid protein suppresses plant pathogen-associated molecular pattern (PAMP)-triggered immunity. Mol. Plant Pathol. 2017, 18, 878–886. [Google Scholar] [CrossRef]
- Tatineni, S. Wheat streak mosaic virus coat protein is a host-specific long-distance transport determinant in oat. Virus Res. 2017, 242, 37–42. [Google Scholar] [CrossRef]
- Decroocq, V.; Salvador, B.; Sicard, O.; Glasa, M.; Cosson, P.; Svanella-Dumas, L.; Revers, F.; García, J.A.; Candresse, T. The determinant of potyvirus ability to overcome the RTM resistance of Arabidopsis thaliana maps to the N-terminal region of the coat protein. Mol. Plant Microbe Interact. 2009, 22, 1302–1311. [Google Scholar] [CrossRef]
- Carbonell, A.; Maliogka, V.I.; Pérez, J.D.J.; Salvador, B.; San León, D.; García, J.A.; Simón-Mateo, C. Diverse amino acid changes at specific positions in the N-terminal region of the coat protein allow Plum pox virus to adapt to new hosts. Mol. Plant Microbe Interact. 2013, 26, 1211–1224. [Google Scholar] [CrossRef]
- Desbiez, C.; Chandeysson, C.; Lecoq, H. A short motif in the N-terminal part of the coat protein is a host-specific determinant of systemic infectivity for two potyviruses. Mol. Plant Pathol. 2014, 15, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Gelens, L.; Saurin, A.T. Exploring the function of dynamic phosphorylation-dephosphorylation cycles. Dev. Cell 2018, 44, 659–663. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Fernández, M.R.; Martínez-Torrecuadrada, J.L.; Roncal, F.; Domínguez, E.; García, J.A. Identification of immunogenic hot spots within plum pox potyvirus capsid protein for efficient antigen presentation. J. Virol. 2002, 76, 12646–12653. [Google Scholar] [CrossRef] [PubMed]
- Pérez, J.J.; Juárez, S.; Chen, D.; Scott, C.L.; Hartweck, L.M.; Olszewski, N.E.; García, J.A. Mapping of two O-GlcNAc modification sites in the capsid protein of the potyvirus Plum pox virus. FEBS Lett. 2006, 580, 5822–5828. [Google Scholar] [CrossRef] [PubMed]
- James, D.; Varga, A.; Sanderson, D. Genetic diversity of Plum pox virus: Strains, disease and related challenges for control. Can. J. Plant Pathol. 2013, 35, 431–441. [Google Scholar] [CrossRef]
| Viral Isolate | Peptide Sequence | Start-End | Modification Site | z | m/z | Search Engine |
|---|---|---|---|---|---|---|
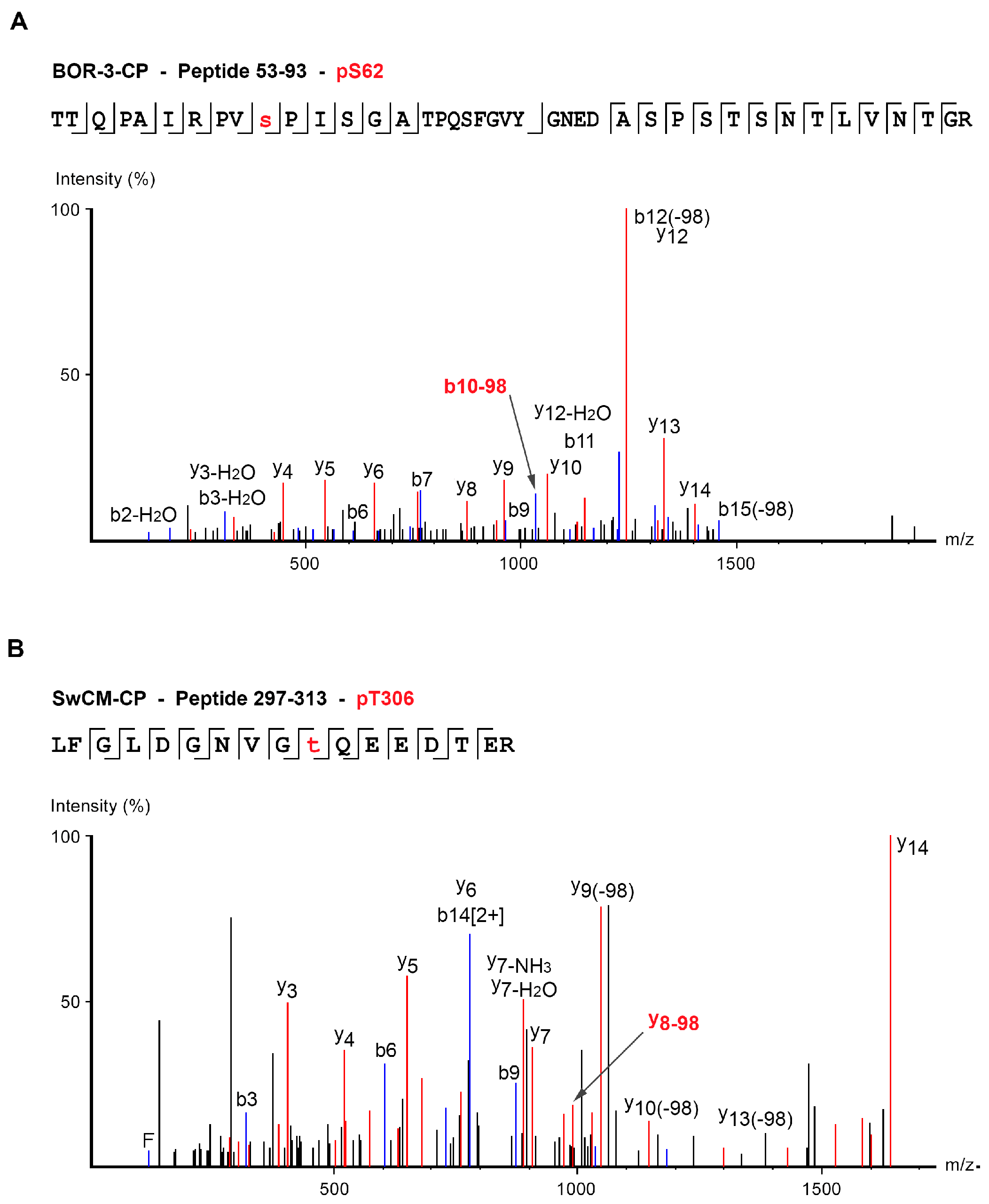
| BOR-3 | TTQPAIRPVpSPISGATPQSFGVYGNEDASPSTSNTLVNTGR | 53–93 | S62 | 4 | 1064.51 | Peaks, score > 50 |
| SFGVYGNEDApSPSTSNTLVNTGR | 71–93 | S81 | 3 | 818.35 | Mascot, score > 50 Peaks, score > 50 | |
| GpSIGTFAVPR | 100–109 | S101 | 2 | 542.76 | Mascot, score > 50 Peaks, score > 50 | |
| TMTSKLpSLPK | 112–121 | S118 | 2 | 593.30 | Mascot, score > 40 Peaks, score > 50 | |
| NLpTDYSLAR | 252–260 | T254 | 2 | 566.75 | Mascot, score > 30 Peaks, score > 50 | |
| SwCM | QNVpTPSSSNALVNTR | 80–94 | T83 | 2 | 834.39 | Mascot, score > 50 Peaks, score > 50 |
| SMTSKLpSLPK | 114–123 | S120 | 2 | 586.29 | Mascot, score > 25 Peaks, score > 50 | |
| NLpTDYSLAR | 254–262 | T256 | 2 | 566.75 | Mascot, score > 35 Peaks, score > 50 | |
| LFGLDGNVGpTQEEDTER | 297–313 | T306 | 3 | 653.95 | Mascot, score > 30 Peaks, score > 50 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hervás, M.; Ciordia, S.; Navajas, R.; García, J.A.; Martínez-Turiño, S. Common and Strain-Specific Post-Translational Modifications of the Potyvirus Plum pox virus Coat Protein in Different Hosts. Viruses 2020, 12, 308. https://doi.org/10.3390/v12030308
Hervás M, Ciordia S, Navajas R, García JA, Martínez-Turiño S. Common and Strain-Specific Post-Translational Modifications of the Potyvirus Plum pox virus Coat Protein in Different Hosts. Viruses. 2020; 12(3):308. https://doi.org/10.3390/v12030308
Chicago/Turabian StyleHervás, Marta, Sergio Ciordia, Rosana Navajas, Juan Antonio García, and Sandra Martínez-Turiño. 2020. "Common and Strain-Specific Post-Translational Modifications of the Potyvirus Plum pox virus Coat Protein in Different Hosts" Viruses 12, no. 3: 308. https://doi.org/10.3390/v12030308
APA StyleHervás, M., Ciordia, S., Navajas, R., García, J. A., & Martínez-Turiño, S. (2020). Common and Strain-Specific Post-Translational Modifications of the Potyvirus Plum pox virus Coat Protein in Different Hosts. Viruses, 12(3), 308. https://doi.org/10.3390/v12030308





