A Kayvirus Distant Homolog of Staphylococcal Virulence Determinants and VISA Biomarker Is a Phage Lytic Enzyme
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Bacteriophage and Bacteriophage Propagation
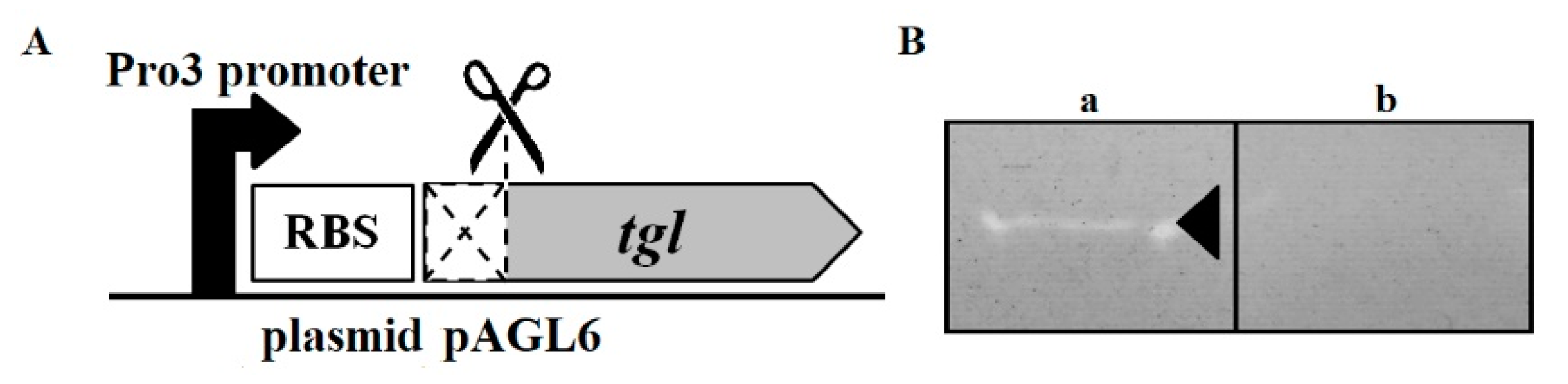
2.3. Plasmids
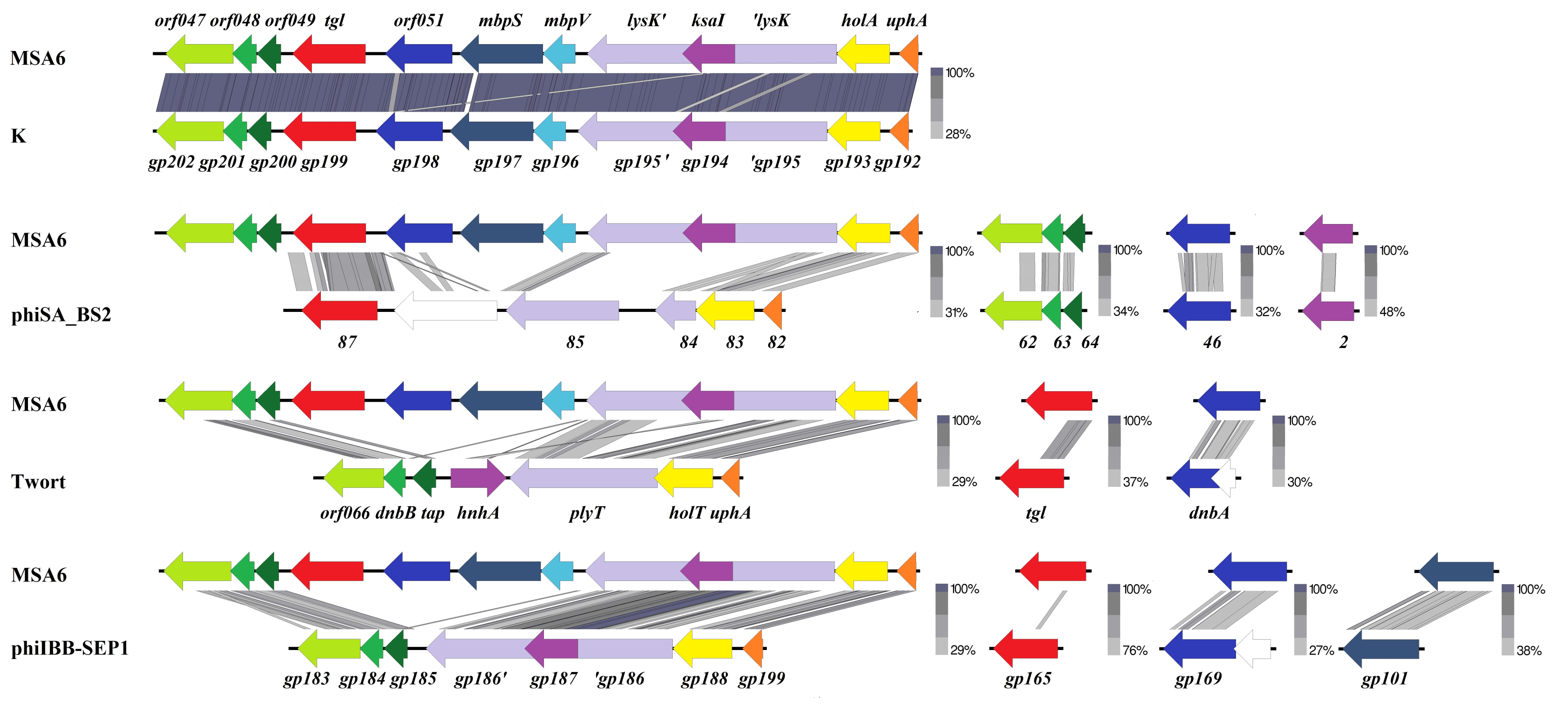
2.4. Bioinformatics Analysis
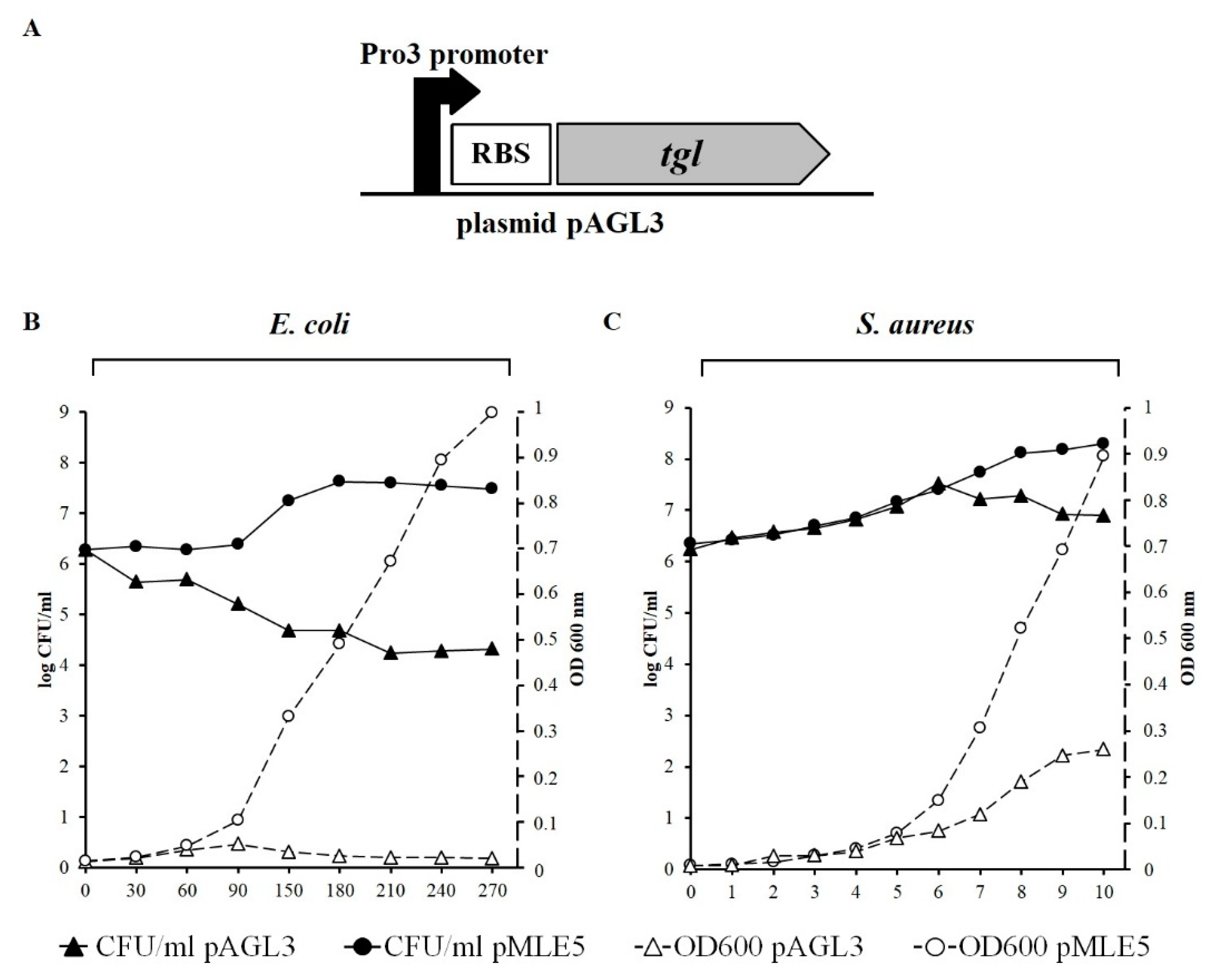
2.5. Testing the Influence of Intracellular Tgl Protein on the Growth and Survivability of E. coli and S. aureus Cells
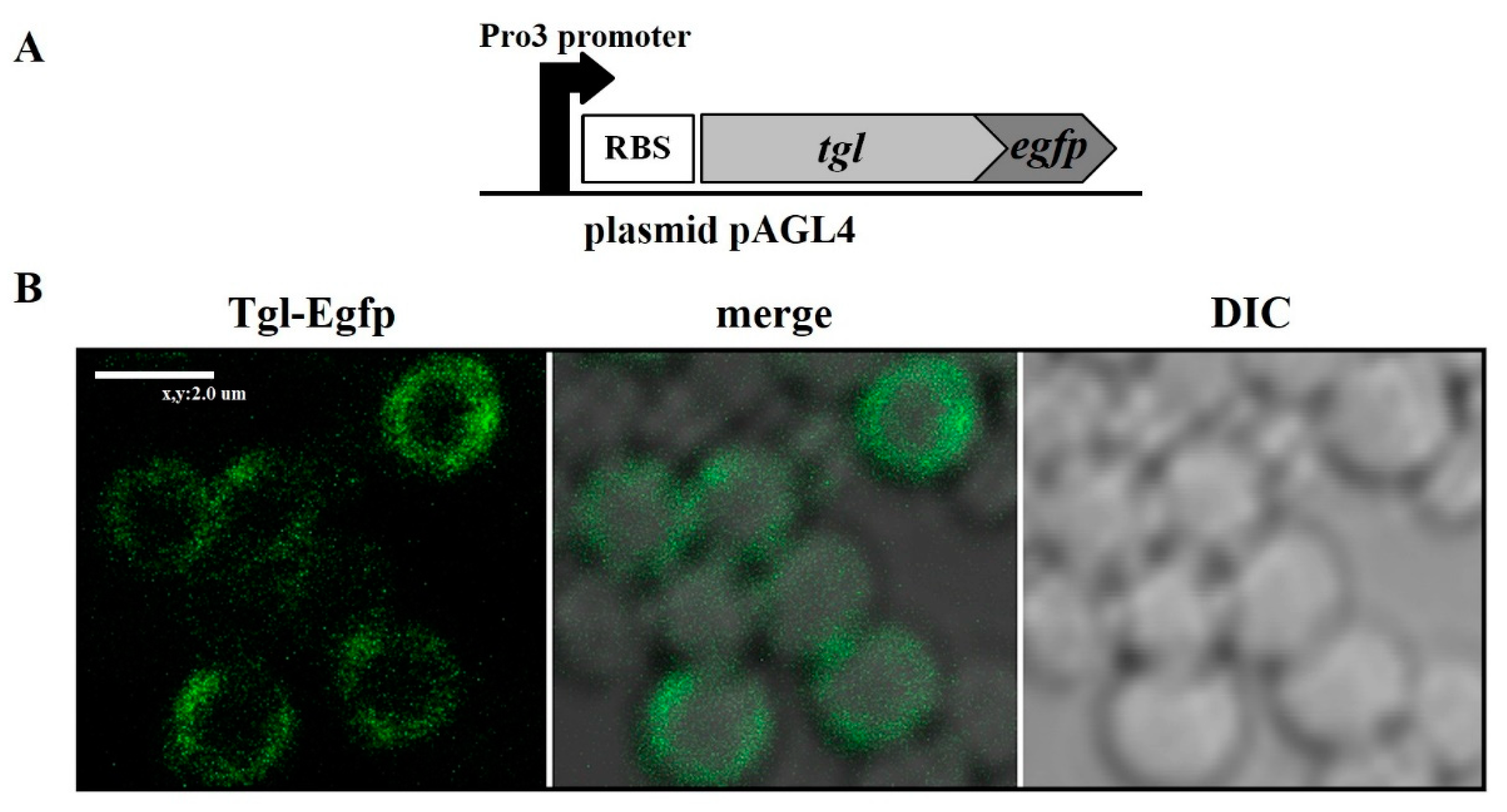
2.6. Determination of the Intracellular Localization of Tgl-Egfp Protein in S. aureus Cells
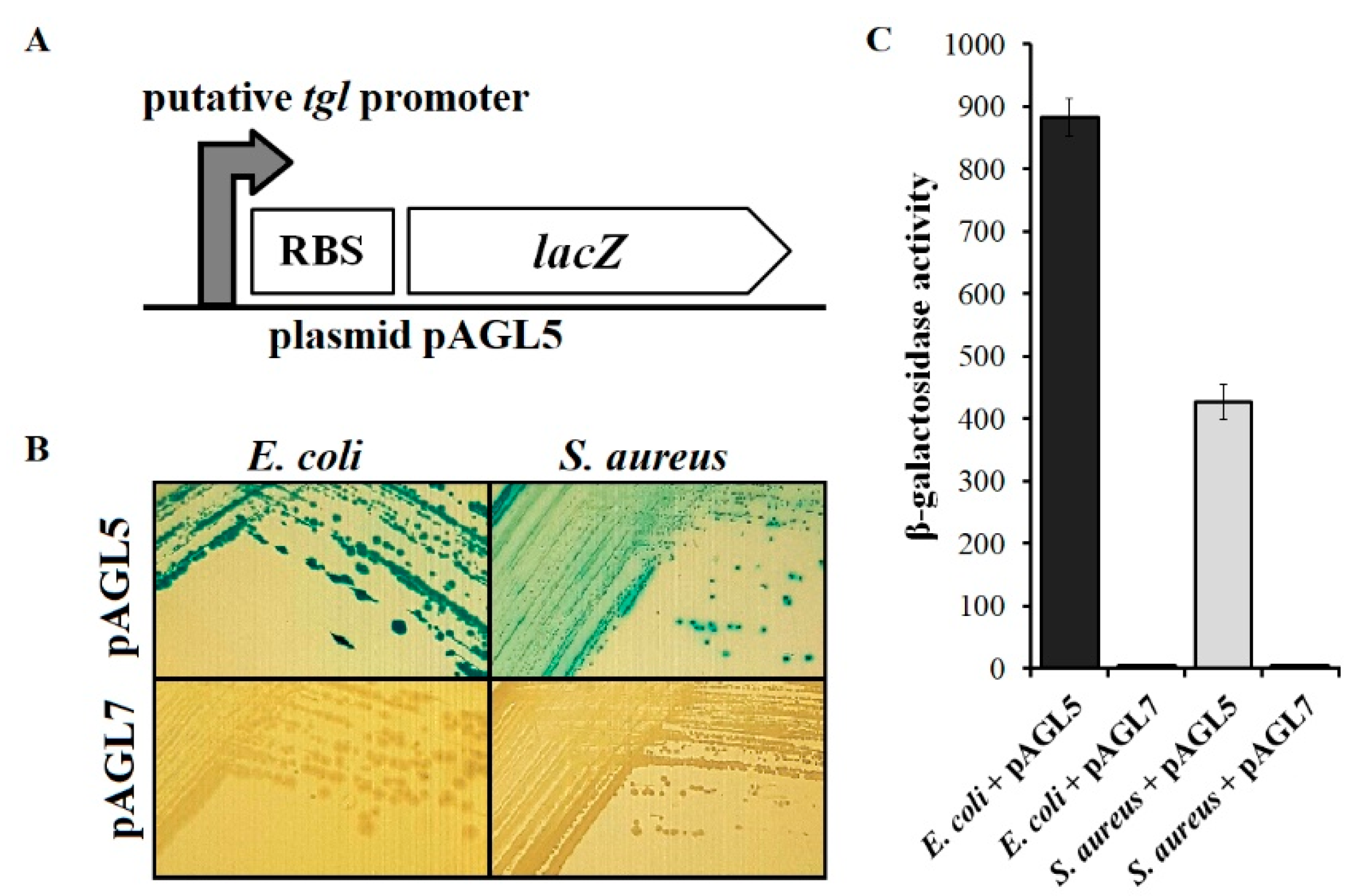
2.7. Assays of β-Galactosidase Activity
2.8. Preparation of S. aureus Cell Walls for Zymography
2.9. Overproduction of TglΔSP Protein and Zymography
2.10. Preparation of Nisin Stock Solution and Nisin Activity Assay
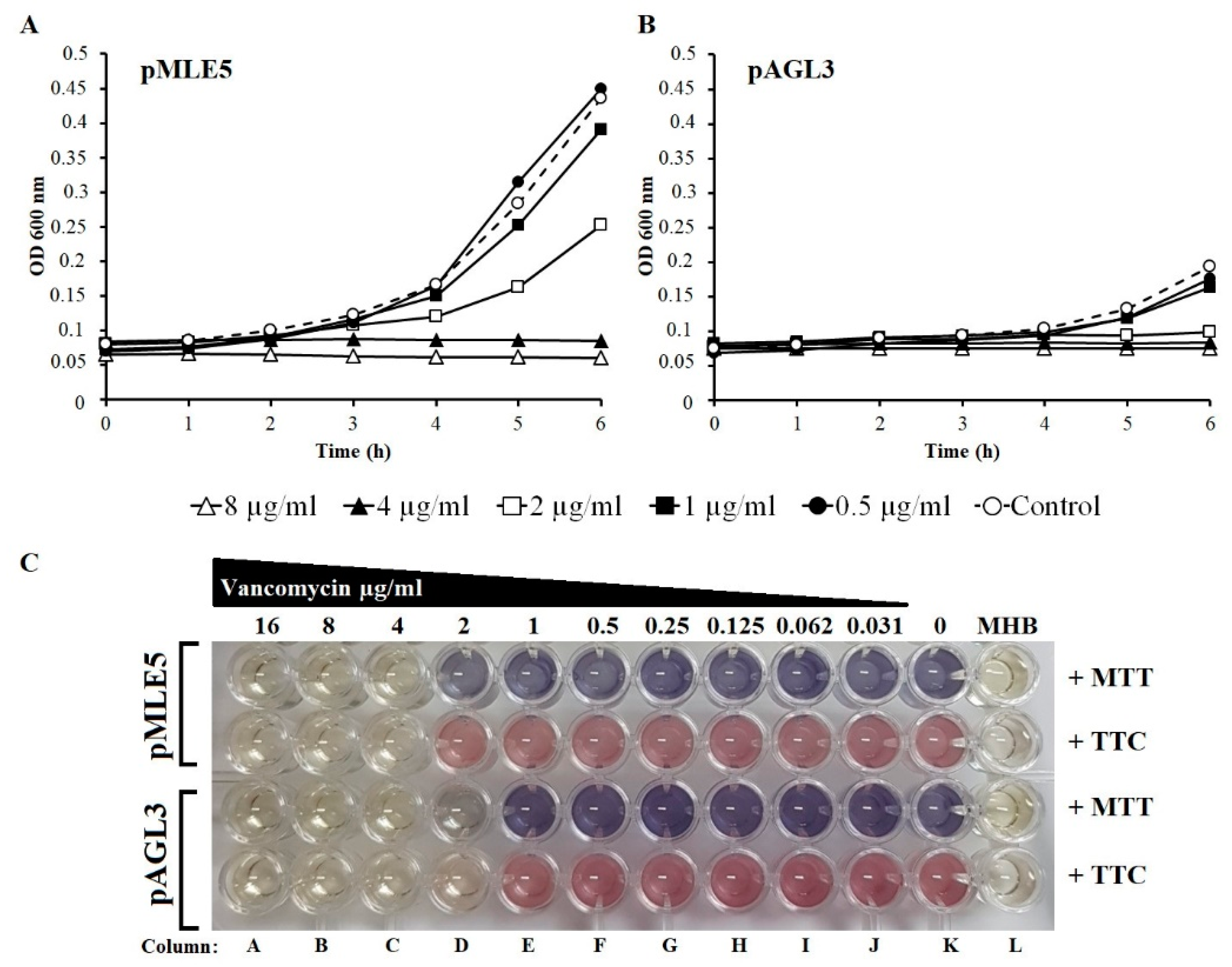
2.11. Testing the Influence of Tgl Protein Production on the Sensitivity of S. aureus to Vancomycin
3. Results
3.1. Analysis of the Amino Acid Sequence of Tgl Protein
3.2. Localization of Tgl Protein in S. aureus
3.3. Influence of Tgl on the Growth and Survivability of E. coli and S. aureus
3.4. Muralytic Activity of Tgl Protein against S. aureus Cell Walls
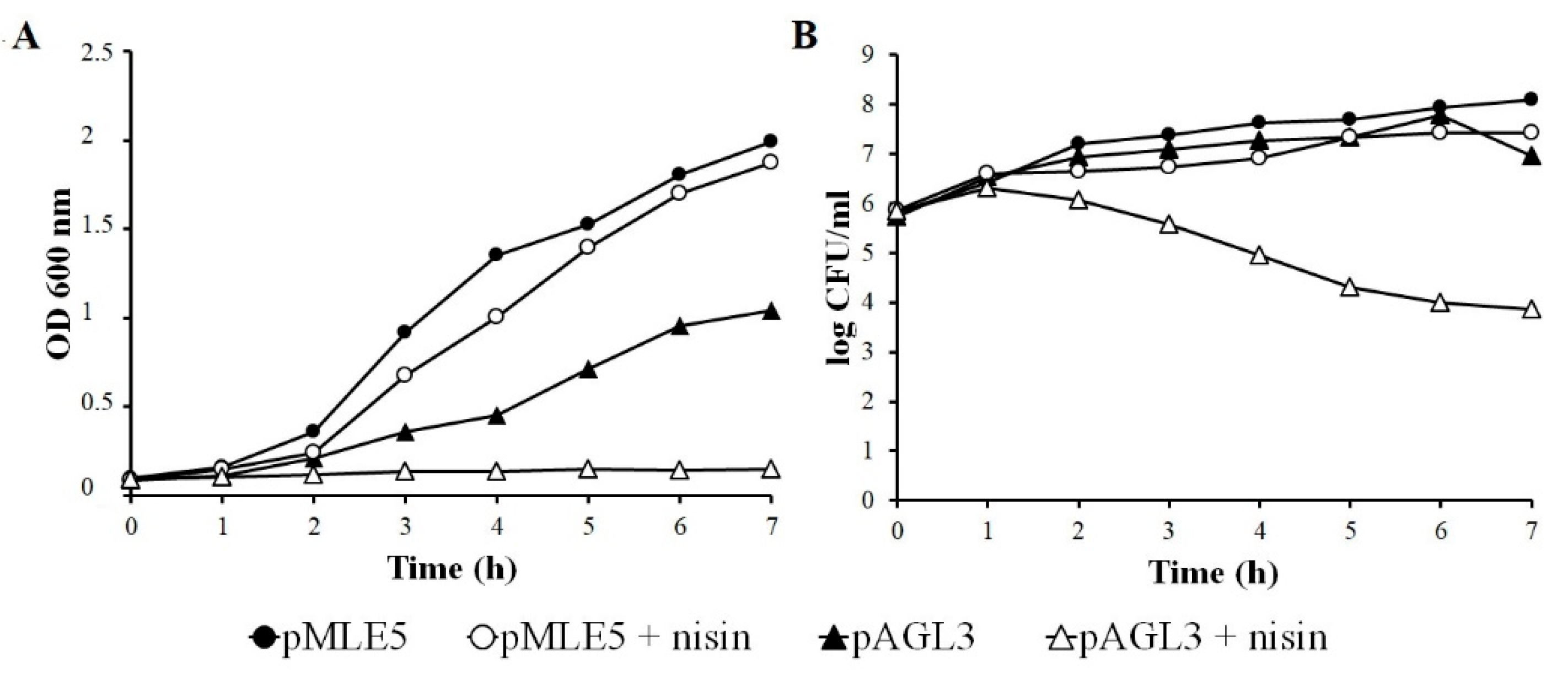
3.5. Influence of Nisin on Growth and Survivability of Tgl Producing S. aureus Cells
3.6. Analysis of Transcriptional Activity of Putative Early tgl Promoter
3.7. Susceptibility of Tgl-Producing S. aureus to Vancomycin
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lowy, F.D. Antimicrobial resistance: The example of Staphylococcus aureus. J. Clin. Investig. 2003, 111, 1265–1273. [Google Scholar] [CrossRef]
- Kaźmierczak, Z.; Górski, A.; Dąbrowska, K. Facing antibiotic resistance: Staphylococcus aureus phages as a medical tool. Viruses 2014, 6, 2551–2570. [Google Scholar] [CrossRef] [PubMed]
- Inal, J.M. Phage therapy: A reappraisal of bacteriophages as antibiotics. Arch. Immunol. Ther. Exp. 2003, 51, 237–244. [Google Scholar]
- Górski, A.; Międzybrodzki, R.; Borsowski, J.; Weber-Dąbrowska, B.; Łobocka, M.; Fortuna, W.; Letkiewicz, S.; Zimecki, M.; Filby, G. Bacteriophage therapy for the treatment of infections. Curr. Opin. Investig. Drugs 2009, 10, 766–774. [Google Scholar] [PubMed]
- Barylski, J.; Enault, F.; Dutilh, B.E.; Schuller, M.B.P.; Edwards, R.A.; Gillis, A.; Klumpp, J.; Knezevic, P.; Krupovic, M.; Kuhn, J.H.; et al. Analysis of Spounaviruses as a case study for the overdue reclassification of tailed phages. Syst. Biol. 2020, 69, 110–123. [Google Scholar] [CrossRef] [PubMed]
- Łobocka, M.B.; Hejnowicz, M.S.; Dąbrowski, K.; Gozdek, A.; Kosakowski, J.; Witkowska, M.; Ulatowska, M.; Weber-Dąbrowska, B.; Kwiatek, M.; Parasion, S.; et al. Genomics of staphylococcal Twort-like phages: Potential therapeutics of the post-antibiotic era. Adv. Virus Res. 2012, 83, 143–216. [Google Scholar] [CrossRef] [PubMed]
- Alves, D.R.; Gaudion, A.; Bean, J.E.; Perez Esteban, P.; Arnot, T.C.; Harper, D.R.; Kot, W.; Hansen, L.H.; Enright, M.C.; Jenkinsa, A.T.A. Combined use of bacteriophage K and a novel bacteriophage to reduce Staphylococcus aureus biofilm formation. Appl. Environ. Microbiol. 2014, 80, 6694–6703. [Google Scholar] [CrossRef]
- Fish, R.; Kutter, E.; Bryan, D.; Wheat, G.; Kuhl, S. Resolving digital staphylococcal osteomyelitis using bacteriophage- a case report. Antibiotics 2018, 7, 87. [Google Scholar] [CrossRef]
- Gill, J.J.; Pacan, J.C.; Carson, M.E.; Leslie, K.E.; Griffiths, M.W.; Sabour, P.M. Efficacy and pharmacokinetics of bacteriophage therapy in treatment of subclinical Staphylococcus aureus mastitis in lactating dairy cattle. Antimicrob. Agents Chemother. 2006, 50, 2912–2918. [Google Scholar] [CrossRef]
- Mendes, J.J.; Leandro, C.; Corte-Real, S.; Barbosa, R.; Cavaco-Silva, P.; Melo-Cristino, J.; Górski, A.; Garcia, M. Wound healing potential of topical bacteriophage therapy on diabetic cutaneous wounds. Wound Repair Regen. 2013, 21, 595–603. [Google Scholar] [CrossRef]
- Międzybrodzki, R.; Borysowski, J.; Weber-Dąbrowska, B.; Fortuna, W.; Letkiewicz, S.; Szufnarowski, K.; Pawełczyk, Z.; Rogóż, P.; Kłak, M.; Wojtasik, E.; et al. Clinical aspects of phage therapy. Adv. Virus Res. 2012, 83, 73–121. [Google Scholar] [CrossRef] [PubMed]
- Międzybrodzki, R.; Kłak, M.; Jończyk-Matysiak, E.; Bubak, B.; Wójcik, A.; Kaszowska, M.; Weber-Dąbrowska, B.; Łobocka, M.; Górski, A. Means to facilitate the overcoming of gastric juice barrier by a therapeutic staphylococcal bacteriophage A5/80. Front. Microbiol. 2017, 8, 467. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Cheng, M.; Zhai, S.; Xi, H.; Cai, R.; Wang, Z.; Zhang, H.; Wang, X.; Xue, Y.; Li, X.; et al. Preventive effect of the phage VB-SavM-JYL01 on rabbit necrotizing pneumonia caused by Staphylococcus aureus. Vet. Microbiol. 2019, 229, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Ooi, M.L.; Drilling, A.J.; Morales, S.; Fong, S.; Moraitis, S.; Macias-Valle, L.; Vreugde, S.; Psaltis, A.J.; Wormald, P.J. Safety and tolerability of bacteriophage therapy for chronic rhinosinusitis due to Staphylococcus aureus. JAMA Otolaryngol. Head Neck Surg. 2019. [Google Scholar] [CrossRef]
- Oliveira, H.; Sampaio, M.; Melo, L.D.R.; Dias, O.; Pope, W.H.; Hatfull, G.F.; Azeredo, J. Staphylococci phages display vast genomic diversity and evolutionary relationships. BMC Genom. 2019, 20, 357. [Google Scholar] [CrossRef]
- Pirnay, J.P.; Blasdel, B.G.; Bretaudeau, L.; Buckling, A.; Chanishvili, N.; Clark, J.R.; Corte-Real, S.; Debarbieux, L.; Dublanchet, A.; De Vos, D.; et al. Quality and safety requirements for sustainable phage therapy products. Pharm. Res. 2015, 32, 2173–2179. [Google Scholar] [CrossRef]
- Cui, Z.; Guo, X.; Dong, K.; Zhang, Y.; Li, Q.; Zhu, Y.; Zeng, L.; Tang, R.; Li, L. Safety assessment of Staphylococcus phages of the family Myoviridae based on complete genome sequences. Sci. Rep. 2017, 7, 41259. [Google Scholar] [CrossRef]
- Stapleton, M.R.; Horsburgh, M.J.; Hayhurst, E.J.; Wright, L.; Jonsson, I.M.; Tarkowski, A.; Kokai-Kun, J.F.; Mond, J.J.; Foster, S.J. Characterization of IsaA and SceD, two putative lytic transglycosylases of Staphylococcus aureus. J. Bacteriol. 2007, 189, 7316–7325. [Google Scholar] [CrossRef]
- Lorenz, U.; Ohlsen, K.; Karch, H.; Hecker, M.; Thiede, A.; Hacker, J. Human antibody response during sepsis against targets expressed by methicillin resistant Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 2000, 29, 145–153. [Google Scholar] [CrossRef]
- Sakata, N.; Terakubo, S.; Mukai, T. Subcellular location of the soluble lytic transglycosylase homologue in Staphylococcus aureus. Curr. Microbiol. 2005, 50, 47–51. [Google Scholar] [CrossRef]
- Lopes, A.A.; Yoshii, Y.; Yamada, S.; Nagakura, M.; Kinjo, Y.; Mizunoe, Y.; Okuda, K.I. Roles of lytic transglycosylases in biofilm formation and β-lactam resistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2019. [Google Scholar] [CrossRef] [PubMed]
- Islam, N.; Ross, J.M.; Marten, M.R. Proteome analyses of Staphylococcus aureus biofilm at elevated levels of NaCl. Clin. Microbiol. 2015, 4, 219. [Google Scholar] [PubMed]
- Resch, A.; Leicht, S.; Saric, M.; Pásztor, L.; Jakob, A.; Götz, F.; Nordheim, A. Comparative proteome analysis of Staphylococcus aureus biofilm and planktonic cells and correlation with transcriptome profiling. Proteomics 2006, 6, 1867–1877. [Google Scholar] [CrossRef]
- den Reijer, P.M.; Haisma, E.M.; Lemmens-den Toom, N.A.; Willemse, J.; Koning, R.I.; Demmers, J.A.; Dekkers, D.H.; Rijkers, E.; El Ghalbzouri, A.; Nibbering, P.H.; et al. Detection of alpha-toxin and other virulence factors in biofilms of Staphylococcus aureus on polystyrene and a human epidermal model. PLoS ONE 2016, 11, e0145722. [Google Scholar] [CrossRef]
- Gillaspy, A.F.; Worrell, V.; Orvis, J.; Roe, B.A.; Dyer, D.W.; Iandolo, J.J. Staphylococcus aureus NCTC8325 genome. In Gram-Positive Pathogens, 2nd ed.; Fischetti, V., Novick, R., Ferretti, J., Portnoy, D., Rood, J., Eds.; ASM Press: Washington, DC, USA, 2006; pp. 381–412. [Google Scholar]
- Dubrac, S.; Boneca, I.G.; Poupel, O.; Msadek, T. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J. Bacteriol. 2007, 189, 8257–8269. [Google Scholar] [CrossRef] [PubMed]
- Dubrac, S.; Msadek, T. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. J. Bacteriol. 2004, 186, 1175–1181. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.C.; Deck, J.; Edmondson, R.D.; Hart, M.E. Relative quantitative comparisons of the extracellular protein profiles of Staphylococcus aureus UAMS-1 and its sarA, agr, and sarA agr regulatory mutants using one-dimensional polyacrylamide gel electrophoresis and nanocapillary liquid chromatography coupled with tandem mass spectrometry. J. Bacteriol. 2008, 190, 5265–5278. [Google Scholar] [CrossRef]
- Ziebandt, A.K.; Weber, H.; Rudolph, J.; Schmid, R.; Höper, D.; Engelmann, S.; Hecker, M. Extracellular proteins of Staphylococcus aureus and the role of SarA and sigma B. Proteomics 2001, 1, 480–493. [Google Scholar] [CrossRef]
- Sakata, N.; Mukai, T. Production profile of the soluble lytic transglycosylase homologue in Staphylococcus aureus during bacterial proliferation. FEMS Immunol. Med. Microbiol. 2007, 49, 288–295. [Google Scholar] [CrossRef]
- van den Berg, S.; Koedijk, D.G.; Back, J.W.; Neef, J.; Dreisbach, A.; van Dijl, J.M.; Bakker-Woudenberg, I.A.; Buist, G. Active immunization with an octa-valent Staphylococcus aureus antigen mixture in models of S. aureus bacteremia and skin infection in mice. PLoS ONE 2015, 10, e0116847. [Google Scholar] [CrossRef]
- Ziebandt, A.K.; Kusch, H.; Degner, M.; Jaglitz, S.; Sibbald, M.J.; Arends, J.P.; Chlebowicz, M.A.; Albrecht, D.; Pantucek, R.; Doskar, J.; et al. Proteomics uncovers extreme heterogeneity in the Staphylococcus aureus exoproteome due to genomic plasticity and variant gene regulation. Proteomics 2010, 10, 1634–1644. [Google Scholar] [CrossRef]
- Dreisbach, A.; Hempel, K.; Buist, G.; Hecker, M.; Becher, D.; van Dijl, J.M. Profiling the surfacome of Staphylococcus aureus. Proteomics 2010, 10, 3082–3096. [Google Scholar] [CrossRef] [PubMed]
- Busche, T.; Hillion, M.; Van Loi, V.; Berg, D.; Walther, B.; Semmler, T.; Strommenger, B.; Witte, W.; Cuny, C.; Mellmann, A.; et al. Comparative secretome analyses of human and zoonotic Staphylococcus aureus isolates CC8, CC22, and CC398. Mol. Cell. Proteom. 2018, 17, 2412–2433. [Google Scholar] [CrossRef] [PubMed]
- Koedijk, D.G.A.M.; Pastrana, F.R.; Hoekstra, H.; Berg, S.V.D.; Back, J.W.; Kerstholt, C.; Prins, R.C.; Bakker-Woudenberg, I.A.J.M.; van Dijl, J.M.; Buist, G. Differential epitope recognition in the immunodominant staphylococcal antigen A of Staphylococcus aureus by mouse versus human IgG antibodies. Sci. Rep. 2017, 7, 8141. [Google Scholar] [CrossRef] [PubMed]
- van der Kooi-Pol, M.M.; de Vogel, C.P.; Westerhout-Pluister, G.N.; Veenstra-Kyuchukova, Y.K.; Duipmans, J.C.; Glasner, C.; Buist, G.; Elsinga, G.S.; Westra, H.; Bonarius, H.P.J.; et al. High anti-staphylococcal antibody titers in patients with epidermolysis bullosa relate to long-term colonization with alternating types of Staphylococcus aureus. J. Investig. Dermatol. 2013, 133, 847–850. [Google Scholar] [CrossRef] [PubMed]
- Romero Pastrana, F.; Neef, J.; Koedijk, D.G.A.M.; de Graaf, D.; Duipmans, J.; Jonkman, M.F.; Engelmann, S.; van Dijl, J.M.; Buist, G. Human antibody responses against non-covalently cell wall-bound Staphylococcus aureus proteins. Sci. Rep. 2018, 8, 3234. [Google Scholar] [CrossRef] [PubMed]
- Clarke, S.R.; Brummell, K.J.; Horsburgh, M.J.; McDowell, P.W.; Mohamad, S.A.; Stapleton, M.R.; Acevedo, J.; Read, R.C.; Day, N.P.; Peacock, S.J.; et al. Identification of in vivo-expressed antigens of Staphylococcus aureus and their use in vaccinations for protection against nasal carriage. J. Infect. Dis. 2006, 193, 1098–1108. [Google Scholar] [CrossRef]
- Ghasemzadeh-Moghaddam, H.; van Wamel, W.; van Belkum, A.; Hamat, R.A.; Tavakol, M.; Neela, V.K. Humoral immune consequences of Staphylococcus aureus ST239-associated bacteremia. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 255–263. [Google Scholar] [CrossRef]
- Lorenz, U.; Lorenz, B.; Schmitter, T.; Streker, K.; Erck, C.; Wehland, J.; Nickel, J.; Zimmermann, B.; Ohlsen, K. Functional antibodies targeting IsaA of Staphylococcus aureus augment host immune response and open new perspectives for antibacterial therapy. Antimicrob. Agents Chemother. 2011, 55, 165–173. [Google Scholar] [CrossRef]
- Oesterreich, B.; Lorenz, B.; Schmitter, T.; Kontermann, R.; Zenn, M.; Zimmermann, B.; Haake, M.; Lorenz, U.; Ohlsen, K. Characterization of the biological anti-staphylococcal functionality of hUK-66 IgG1, a humanized monoclonal antibody as substantial component for an immunotherapeutic approach. Hum. Vaccine Immunother. 2014, 10, 926–937. [Google Scholar] [CrossRef]
- van den Berg, S.; Bonarius, H.P.; van Kessel, K.P.; Elsinga, G.S.; Kooi, N.; Westra, H.; Bosma, T.; van der Kooi-Pol, M.M.; Koedijk, D.G.; Groen, H.; et al. A human monoclonal antibody targeting the conserved staphylococcal antigen IsaA protects mice against Staphylococcus aureus bacteremia. Int. J. Med. Microbiol. 2015, 305, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Burian, M.; Rautenberg, M.; Kohler, T.; Fritz, M.; Krismer, B.; Unger, C.; Hoffmann, W.H.; Peschel, A.; Wolz, C.; Goerke, C. Temporal expression of adhesion factors and activity of global regulators during establishment of Staphylococcus aureus nasal colonization. J. Infect. Dis. 2010, 201, 1414–1421. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, M.; Sekizuka, T.; Matsui, H.; Ohsuga, J.; Ohshima, T.; Hanaki, H. IS256-mediated overexpression of the WalKR two-component system regulon contributes to reduced vancomycin susceptibility in a Staphylococcus aureus clinical isolate. Front. Microbiol. 2019, 10, 1882. [Google Scholar] [CrossRef] [PubMed]
- Cafiso, V.; Bertuccio, T.; Spina, D.; Purrello, S.; Campanile, F.; Di Pietro, C.; Purrello, M.; Stefani, S. Modulating activity of vancomycin and daptomycin on the expression of autolysis cell-wall turnover and membrane charge genes in hVISA and VISA strains. PLoS ONE 2012, 7, e29573. [Google Scholar] [CrossRef] [PubMed]
- Pieper, R.; Gatlin-Bunai, C.L.; Mongodin, E.F.; Parmar, P.P.; Huang, S.T.; Clark, D.J.; Fleischmann, R.D.; Gill, S.R.; Peterson, S.N. Comparative proteomic analysis of Staphylococcus aureus strains with differences in resistance to the cell wall-targeting antibiotic vancomycin. Proteomics 2006, 6, 4246–4258. [Google Scholar] [CrossRef] [PubMed]
- Opoku-Temeng, C.; Onyedibe, K.I.; Aryal, U.K.; Sintim, H.O. Proteomic analysis of bacterial response to a 4-hydroxybenzylidene indolinone compound, which re-sensitizes bacteria to traditional antibiotics. J. Proteom. 2019, 202, 103368. [Google Scholar] [CrossRef]
- Drummelsmith, J.; Winstall, E.; Bergeron, M.G.; Poirier, G.G.; Ouellette, M. Comparative proteomics analyses reveal a potential biomarker for the detection of vancomycin-intermediate Staphylococcus aureus strains. J. Proteome Res. 2007, 6, 4690–4702. [Google Scholar] [CrossRef]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Selvarasu, S.; Ow, D.S.; Lee, S.Y.; Lee, M.M.; Oh, S.K.; Karimi, I.A.; Lee, D.Y. Characterizing Escherichia coli DH5alpha growth and metabolism in a complex medium using genome-scale flux analysis. Biotechnol. Bioeng. 2009, 102, 923–934. [Google Scholar] [CrossRef]
- Paliy, O.; Gunasekera, T.S. Growth of E. coli BL21 in minimal media with different gluconeogenic carbon sources and salt contents. Appl. Microbiol. Biotechnol. 2007, 73, 1169–1172. [Google Scholar] [CrossRef]
- Nair, D.; Memmi, G.; Hernandez, D.; Bard, J.; Beaume, M.; Gill, S.; Francois, P.; Cheung, A.L. Whole-genome sequencing of Staphylococcus aureus strain RN4220, a key laboratory strain used in virulence research, identifies mutations that affect not only virulence factors but also the fitness of the strain. J. Bacteriol. 2011, 193, 2332–2335. [Google Scholar] [CrossRef]
- Berscheid, A.; Sass, P.; Weber-Lassalle, K.; Cheung, A.L.; Bierbaum, G. Revisiting the genomes of the Staphylococcus aureus strains NCTC 8325 and RN4220. Int. J. Med. Microbiol. 2012, 302, 84–87. [Google Scholar] [CrossRef]
- Kwiatek, M.; Parasion, S.; Mizak, L.; Gryko, R.; Bartoszcze, M.; Kocik, J. Characterization of a bacteriophage, isolated from a cow with mastitis, that is lytic against Staphylococcus aureus strains. Arch. Virol. 2012, 157, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Grkovic, S.; Brown, M.H.; Hardie, K.M.; Firth, N.; Skurray, R.A. Stable low-copy-number Staphylococcus aureus shuttle vectors. Microbiology 2003, 149, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Schofield, D.A.; Westwater, C.; Hoel, B.D.; Werner, P.A.; Norris, J.S.; Schmidt, M.G. Development of a thermally regulated broad-spectrum promoter system for use in pathogenic gram-positive species. Appl. Environ. Microbiol. 2003, 69, 3385–3392. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning a Laboratory Manual, 2nd ed.; Cold Spring Harbor Lab Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Zhang, J.; Madden, T.L. PowerBLAST: A new network BLAST application for interactive or automated sequence analysis and annotation. Genome Res. 1997, 7, 649–656. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A Completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, H.; Melo, L.D.; Santos, S.B.; Nóbrega, F.L.; Ferreira, E.C.; Cerca, N.; Azeredo, J.; Kluskens, L.D. Molecular aspects and comparative genomics of bacteriophage endolysins. J. Virol. 2013, 87, 4558–4570. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.H. Experiments in Molecular Genetics; Cold Spring Harbor Lab Press: Cold Spring Harbor, NY, USA, 1972. [Google Scholar]
- Vaz, F.; Filipe, S.R. Preparation and analysis of crude autolytic enzyme extracts from Staphylococcus aureus. Bio-protocol 2015, 5, 1–12. [Google Scholar] [CrossRef]
- Schillingera, U.; Stilesb, M.E.; Holzapfela, W.H. Bacteriocin production by Carnobacterium piscicola LV 61. Int. J. Food Microbiol. 1993, 20, 131–147. [Google Scholar] [CrossRef]
- Franz, C.M.; Du Toit, M.; von Holy, A.; Schillinger, U.; Holzapfel, W.H. Production of nisin-like bacteriocins by Lactococcus lactis strains isolated from vegetables. J. Basic Microbiol. 1997, 37, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Tuncer, Y. Phenotypic and genotypic characterization of nisin-producing Lactococcus lactis subsp. lactis YB23 isolated from raw milk in Turkey. Biotechnol. Biotechnol. Equip. 2009, 23, 1504–1508. [Google Scholar] [CrossRef]
- Andrews, J.M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001, 48, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Thunnissen, A.M.; Dijkstra, A.J.; Kalk, K.H.; Rozeboom, H.J.; Engel, H.; Keck, W.; Dijkstra, B.W. Doughnut-shaped structure of a bacterial muramidase revealed by X-ray crystallography. Nature 1994, 367, 750–753. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Bockstael, K.; Nath, S.; Engelborghs, Y.; Anné, J.; Geukens, N. Enzymatic investigation of the Staphylococcus aureus type I signal peptidase SpsB- implications for the search for novel antibiotics. FEBS J. 2009, 276, 3222–3234. [Google Scholar] [CrossRef] [PubMed]
- Bockstael, K.; Geukens, N.; Rao, C.V.; Herdewijn, P.; Anné, J.; Van Aerschot, A. An easy and fast method for the evaluation of Staphylococcus epidermidis type I signal peptidase inhibitors. J. Microbiol. Methods 2009, 78, 231–237. [Google Scholar] [CrossRef]
- Arimori, T.; Kawamoto, N.; Shinya, S.; Okazaki, N.; Nakazawa, M.; Miyatake, K.; Fukamizo, T.; Ueda, M.; Tamada, T. Crystal structures of the catalytic domain of a novel glycohydrolase family 23 chitinase from Ralstonia sp. A-471 reveals a unique arrangement of the catalytic residues for inverting chitin hydrolysis. J. Biol. Chem. 2013, 288, 18696–18706. [Google Scholar] [CrossRef]
- Dik, D.A.; Marous, D.R.; Fisher, J.F.; Mobashery, S. Lytic transglycosylases: Concinnity in concision of the bacterial cell wall. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 503–542. [Google Scholar] [CrossRef]
- Vijayaraghavan, J.; Kumar, V.; Krishnan, N.P.; Kaufhold, R.T.; Zeng, X.; Lin, J.; van den Akker, F. Structural studies and molecular dynamics simulations suggest a processive mechanism of exolytic lytic transglycosylase from Campylobacter jejuni. PLoS ONE 2018, 13, e0197136. [Google Scholar] [CrossRef]
- Alcorlo, M.; Martínez-Caballero, S.; Molina, R.; Hermoso, J.A. Carbohydrate recognition and lysis by bacterial peptidoglycan hydrolases. Curr. Opin. Struct. Biol. 2017, 44. [Google Scholar] [CrossRef] [PubMed]
- Helland, R.; Larsen, R.L.; Finstad, S.; Kyomuhendo, P.; Larsen, A.N. Crystal structures of g-type lysozyme from Atlantic cod shed new light on substrate binding and the catalytic mechanism. Cell. Mol. Life Sci. 2009, 66, 2585–2598. [Google Scholar] [CrossRef] [PubMed]
- Thunnissen, A.M.; Isaacs, N.W.; Dijkstra, B.W. The catalytic domain of a bacterial lytic transglycosylase defines a novel class of lysozymes. Proteins 1995, 22, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Łobocka, M.; Kropinski, A.M.; Adriaenssens, E.M. Create One New Genus (Baoshanvirus) Including Two New Species in the Subfamily Twortvirinae, Family Herelleviridae. International Committee on Taxonomy of Viruses ICTV. Available online: https://talk.ictvonline.org/files/proposals/taxonomy_proposals_prokaryote1/m/bact04/8875 (accessed on 20 December 2019).
- Łobocka, M.; Kropinski, A.M.; Adriaenssens, E.M. Create One New Genus (Sciuriunavirus) Including One New Species in the Subfamily Twortvirinae, Family Herelleviridae. International Committee on Taxonomy of Viruses ICTV. Available online: https://talk.ictvonline.org/files/proposals/taxonomy_proposals_prokaryote1/m/bact04/8863 (accessed on 20 December 2019).
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Wiedemann, I.; Benz, R.; Sahl, H.G. Lipid II-mediated pore formation by the peptide antibiotic nisin: A black lipid membrane study. J. Bacteriol. 2004, 186, 3259–3261. [Google Scholar] [CrossRef]
- Vandersteegen, K.; Matthews, W.; Ceyssens, P.J.; Bilock, F.; De Vos, D.; Pirnay, J.P.; Noben, J.P.; Merabishvili, M.; Lipinska, U.; Hermans, K.; et al. Microbiological and molecular assesment of bacteriophage ISP for the control of Staphylococcus aureus. PLoS ONE 2011, 6, e24418. [Google Scholar] [CrossRef]
- Tse, H.; Chan, E.; Lam, C.W.; Leung, K.F.; Chow, P.; Lee, K.C.; Sze, K.H.; Cheung, S.K.; Tse, M.K.; Ho, P.L.; et al. Production of 2-aminophenoxazin-3-one by Staphylococcus aureus causes false-positive results in β-galactosidase assays. J. Clin. Microbiol. 2012, 50, 3780–3782. [Google Scholar] [CrossRef]
- McAleese, F.; Wu, S.W.; Sieradzki, K.; Dunman, P.; Murphy, E.; Projan, S.; Tomasz, A. Overexpression of genes of the cell wall stimulon in clinical isolates of Staphylococcus aureus exhibiting vancomycin-intermediate- S. aureus-type resistance to vancomycin. J. Bacteriol. 2006, 188, 1120–1133. [Google Scholar] [CrossRef]
- Chen, H.; Liu, Y.; Zhao, C.; Xiao, D.; Zhang, J.; Zhang, F.; Chen, M.; Wang, H. Comparative proteomics-based identification of genes associated with glycopeptide resistance in clinically derived heterogeneous vancomycin-intermediate Staphylococcus aureus strains. PLoS ONE 2013, 8, e66880. [Google Scholar] [CrossRef]
- Maor, Y.; Rahav, G.; Belausov, N.; Ben-David, D.; Smollan, G.; Keller, N. Prevalence and characteristics of heteroresistant vancomycin-intermediate Staphylococcus aureus bacteremia in a tertiary care center. J. Clin. Microbiol. 2007, 45, 1511–1514. [Google Scholar] [CrossRef]
- Smith, T.L.; Pearson, M.L.; Wilcox, K.R.; Cruz, C.; Lancaster, M.V.; Robinson-Dunn, B.; Tenover, F.C.; Zervos, M.J.; Band, J.D.; White, E.; et al. Emergence of vancomycin resistance in Staphylococcus aureus. Glycopeptide-intermediate Staphylococcus aureus working group. N. Engl. J. Med. 1999, 340, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Viertel, T.M.; Ritter, K.; Horz, H.P. Viruses versus bacteria-novel approaches to phage therapy as a tool against multidrug-resistant pathogens. J. Antimicrob. Chemother. 2014, 69, 2326–2336. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Myung, H. Complete genome of Staphylococcus aureus phage SA11. J. Virol. 2012, 86, 10232. [Google Scholar] [CrossRef] [PubMed]
- O'Flaherty, S.; Coffey, A.; Edwards, R.; Meaney, W.; Fitzgerald, G.F.; Ross, R.P. Genome of staphylococcal phage K: A new lineage of Myoviridae infecting Gram-positive bacteria with a low G+C content. J. Bacteriol. 2004, 186, 2862–2871. [Google Scholar] [CrossRef] [PubMed]
- Ravipaty, S.; Reilly, J.P. Comprehensive characterization of methicillin-resistant Staphylococcus aureus subsp. aureus COL secretome by two-dimensional liquid chromatography and mass spectrometry. Mol. Cell. Proteom. 2010, 9, 1898–1919. [Google Scholar] [CrossRef]
- O’Flaherty, S.; Coffey, A.; Meaney, W.; Fitzgerald, G.F.; Ross, R.P. The recombinant phage lysin LysK has a broad spectrum of lytic activity against clinically relevant staphylococci, including methicillin-resistant Staphylococcus aureus. J. Bacteriol. 2005, 187, 7161–7164. [Google Scholar] [CrossRef]
- Becker, S.C.; Dong, S.; Baker, J.R.; Foster-Frey, J.; Pritchard, D.G.; Donovan, D.M. LysK CHAP endopeptidase domain is required for lysis of live staphylococcal cells. FEMS Microbiol. Lett. 2009, 294, 52–60. [Google Scholar] [CrossRef]
- Taylor, A.; Das, B.C.; Van Heijenoort, J. Bacterial cell wall peptidoglycan fragments produced by phage λ or Vi II endolysin and containing 1,6-anhydro-N acetylmuramic acid. Eur. J. Biochem. 1975, 53, 47–54. [Google Scholar] [CrossRef]
- Miroshnikov, K.A.; Faizullina, N.M.; Sykilinda, N.N.; Mesyanzhinov, V.V. Properties of the endolytic transglycosylase encoded by gene 144 of Pseudomonas aeruginosa bacteriophage phiKZ. Biochemistry 2006, 71, 300–305. [Google Scholar] [CrossRef]
- Nelson, D.C.; Schmelcher, M.; Rodriguez-Rubio, L.; Klumpp, J.; Pritchard, D.G.; Dong, S.; Donovan, D.M. Endolysins as antimicrobials. Adv. Virus Res. 2012, 83, 299–365. [Google Scholar] [CrossRef]
- Becker, S.C.; Foster-Frey, J.; Donovan, D.M. The phage K lytic enzyme LysK and lysostaphin act synergistically to kill MRSA. FEMS Microbiol. Lett. 2008, 287, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Becker, S.C.; Roach, D.R.; Chauhan, V.S.; Shen, Y.; Foster-Frey, J.; Powell, A.M.; Bauchan, G.; Lease, R.A.; Mohammadi, H.; Harty, W.J.; et al. Triple-acting lytic enzyme treatment of drug-resistant and intracellular Staphylococcus aureus. Sci. Rep. 2016, 6, 25063. [Google Scholar] [CrossRef] [PubMed]
- Verheust, C.; Fornelos, N.; Mahillon, J. The Bacillus thuringiensis phage GIL01 encodes two enzymes with peptidoglycan hydrolase activity. FEMS Microbiol. Lett. 2004, 237, 289–295. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pohane, A.A.; Jain, V. Insights into the regulation of bacteriophage endolysin: Multiple means to the same end. Microbiology 2015, 161, 2269–2276. [Google Scholar] [CrossRef] [PubMed]
- São-José, C.; Parreira, R.; Vieira, G.; Santos, M.A. The N-terminal region of the Oenococcus oeni bacteriophage fOg44 lysin behaves as a bona fide signal peptide in Escherichia coli and as a cis-inhibitory element, preventing lytic activity on oenococcal cells. J. Bacteriol. 2000, 182, 5823–5831. [Google Scholar] [CrossRef]
- Nascimento, J.G.; Guerreiro-Pereira, M.C.; Costa, S.F.; São-José, C.; Santos, M.A. Nisin-triggered activity of Lys44, the secreted endolysin from Oenococcus oeni phage fOg44. J. Bacteriol. 2008, 190, 457–461. [Google Scholar] [CrossRef]
- Fernandes, S.; São-José, C. More than a hole: The holin lethal function may be required to fully sensitize bacteria to the lytic action of canonical endolysins. Mol. Microbiol. 2016, 102, 92–106. [Google Scholar] [CrossRef]
- Fernandes, S.; São-José, C. Probing the function of the two holin-like proteins of bacteriophage SPP1. Virology 2017, 500, 184–189. [Google Scholar] [CrossRef]
- Lapatsina, L.; Brand, J.; Poole, K.; Daumke, O.; Lewin, G.R. Stomatin-domain proteins. Eur. J. Cell Biol. 2012, 91, 240–245. [Google Scholar] [CrossRef]
- Rashid, R.; Kline, K.A. Wrecking Staph’s rafts: Staphylococcus aureus no longer unsinkable? Cell Chem. Biol. 2017, 24, 779–781. [Google Scholar] [CrossRef]
- Lopez, D.; Koch, G. Exploring functional membrane microdomains in bacteria: An overview. Curr. Opin. Microbiol. 2017, 36, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Lopez, D. Molecular composition of functional microdomains in bacterial membranes. Chem. Phys. Lipids 2015, 192, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Mielich-Süss, B.; Schneider, J.; Lopez, D. Overproduction of flotillin influences cell differentiation and shape in Bacillus subtilis. MBio 2013, 4, e00719-13. [Google Scholar] [CrossRef] [PubMed]
- Vandersteegen, K.; Kropinski, A.M.; Nash, J.H.; Noben, J.P.; Hermans, K.; Lavigne, R. Romulus and Remus, two phage isolates representing a distinct clade within the Twortlikevirus genus, display suitable properties for phage therapy applications. J. Virol. 2013, 87, 3237–3247. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, S.E.; Tseng, Y.H.; Lo, H.H.; Chen, S.T.; Wu, C.N. Genomic analysis of Staphylococcus phage Stau2 isolated from medical specimen. Virus Genes 2016, 52, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Jurczak-Kurek, A.; Gąsior, T.; Nejman-Faleńczyk, B.; Bloch, S.; Dydecka, A.; Topka, G.; Necel, A.; Jakubowska-Deredas, M.; Narajczyk, M.; Richert, M.; et al. Biodiversity of bacteriophages: Morphological and biological properties of a large group of phages isolated from urban sewage. Sci. Rep. 2016, 6, 34338. [Google Scholar] [CrossRef] [PubMed]
- Łoś, M.; Węgrzyn, G. Pseudolysogeny. Adv. Virus Res. 2012, 82, 339–349. [Google Scholar] [CrossRef]
- Latino, L.; Midoux, C.; Hauck, Y.; Vergnaud, G.; Pourcel, C. Pseudolysogeny and sequential mutations build multiresistance to virulent bacteriophages in Pseudomonas aeruginosa. Microbiology 2016, 162, 748–763. [Google Scholar] [CrossRef]
- Gilmer, D.B.; Schmitz, J.E.; Euler, C.W.; Fischetti, V.A. Novel bacteriophage lysin with broad lytic activity protects against mixed infection by Streptococcus pyogenes and methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2013, 57, 2743–2750. [Google Scholar] [CrossRef]
- Schuch, R.; Lee, H.M.; Schneider, B.C.; Sauve, K.L.; Law, C.; Khan, B.K.; Rotolo, J.A.; Horiuchi, Y.; Couto, D.E.; Raz, A.; et al. Combination therapy with lysin CF-301 and antibiotic is superior to antibiotic alone for treating methicillin-resistant Staphylococcus aureus-induced murine bacteremia. J. Infect. Dis. 2014, 209, 1469–1478. [Google Scholar] [CrossRef]
- Rashel, M.; Uchiyama, J.; Ujihara, T.; Uehara, Y.; Kuramoto, S.; Sugihara, S.; Yagyu, K.; Muraoka, A.; Sugai, M.; Hiramatsu, K.; et al. Efficient elimination of multidrug-resistant Staphylococcus aureus by cloned lysin derived from bacteriophage phi MR11. J. Infect. Dis. 2007, 196, 1237–1247. [Google Scholar] [CrossRef] [PubMed]
- Daniel, A.; Euler, C.; Collin, M.; Chahales, P.; Gorelick, K.J.; Fischetti, V.A. Synergism between a novel chimeric lysin and oxacillin protects against infection by methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2010, 54, 1603–1612. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Głowacka-Rutkowska, A.; Ulatowska, M.; Empel, J.; Kowalczyk, M.; Boreczek, J.; Łobocka, M. A Kayvirus Distant Homolog of Staphylococcal Virulence Determinants and VISA Biomarker Is a Phage Lytic Enzyme. Viruses 2020, 12, 292. https://doi.org/10.3390/v12030292
Głowacka-Rutkowska A, Ulatowska M, Empel J, Kowalczyk M, Boreczek J, Łobocka M. A Kayvirus Distant Homolog of Staphylococcal Virulence Determinants and VISA Biomarker Is a Phage Lytic Enzyme. Viruses. 2020; 12(3):292. https://doi.org/10.3390/v12030292
Chicago/Turabian StyleGłowacka-Rutkowska, Aleksandra, Magdalena Ulatowska, Joanna Empel, Magdalena Kowalczyk, Jakub Boreczek, and Małgorzata Łobocka. 2020. "A Kayvirus Distant Homolog of Staphylococcal Virulence Determinants and VISA Biomarker Is a Phage Lytic Enzyme" Viruses 12, no. 3: 292. https://doi.org/10.3390/v12030292
APA StyleGłowacka-Rutkowska, A., Ulatowska, M., Empel, J., Kowalczyk, M., Boreczek, J., & Łobocka, M. (2020). A Kayvirus Distant Homolog of Staphylococcal Virulence Determinants and VISA Biomarker Is a Phage Lytic Enzyme. Viruses, 12(3), 292. https://doi.org/10.3390/v12030292




