MiR-202-5p Inhibits RIG-I-Dependent Innate Immune Responses to RGNNV Infection by Targeting TRIM25 to Mediate RIG-I Ubiquitination
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Fish Strains, Cell Lines, and Reagents
2.3. Viral Challenge
2.4. Prediction of MiR-202-5p Target Genes
2.5. Plasmid Construction
2.6. RNA Isolation and qRT-PCR
2.7. Dual Luciferase Reporter Assay
2.8. Co-Immunoprecipitations (Co-IP) and Ubiquitination Assays
2.9. Statistics Analysis
3. Results
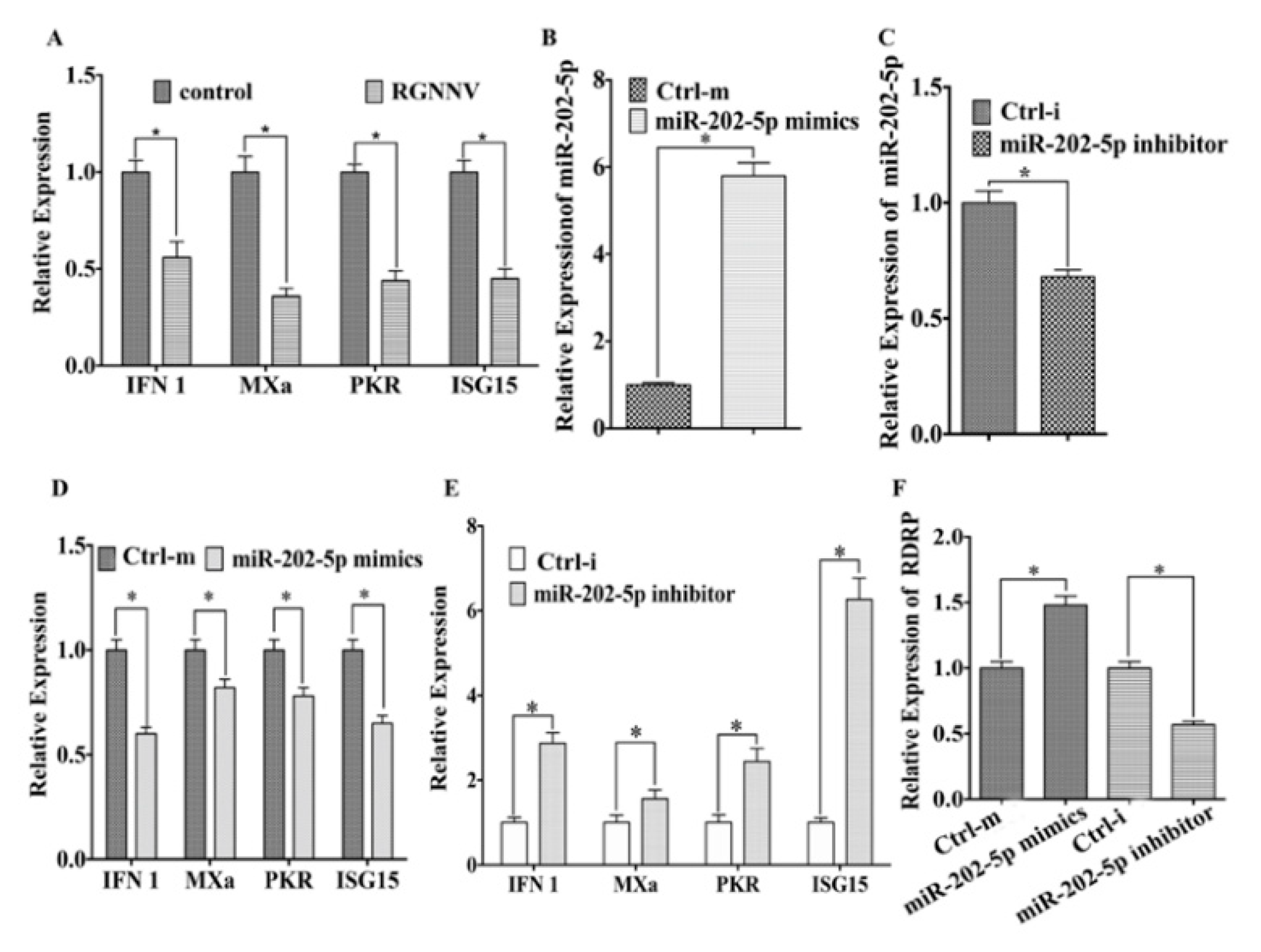
3.1. MiR-202-5p Expression is Up-Regulated Post RGNNV Infection In Vivo and In Vitro
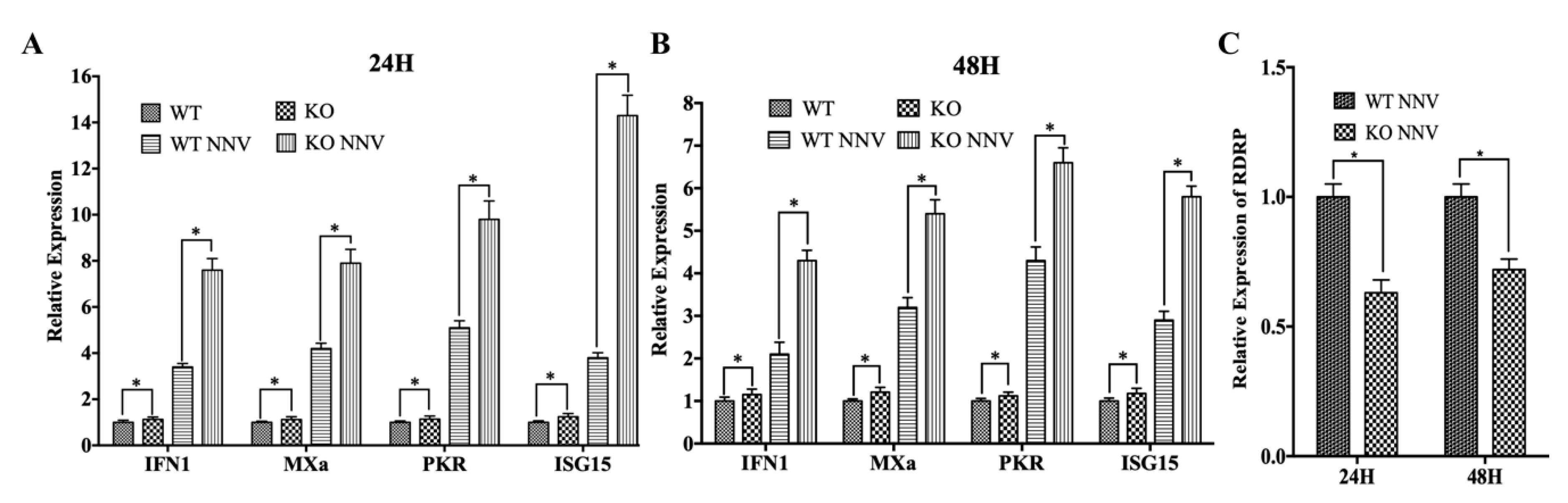
3.2. MiR-202-5p Suppresses the Expression of Antiviral Genes and Promotes RGNNV Replication
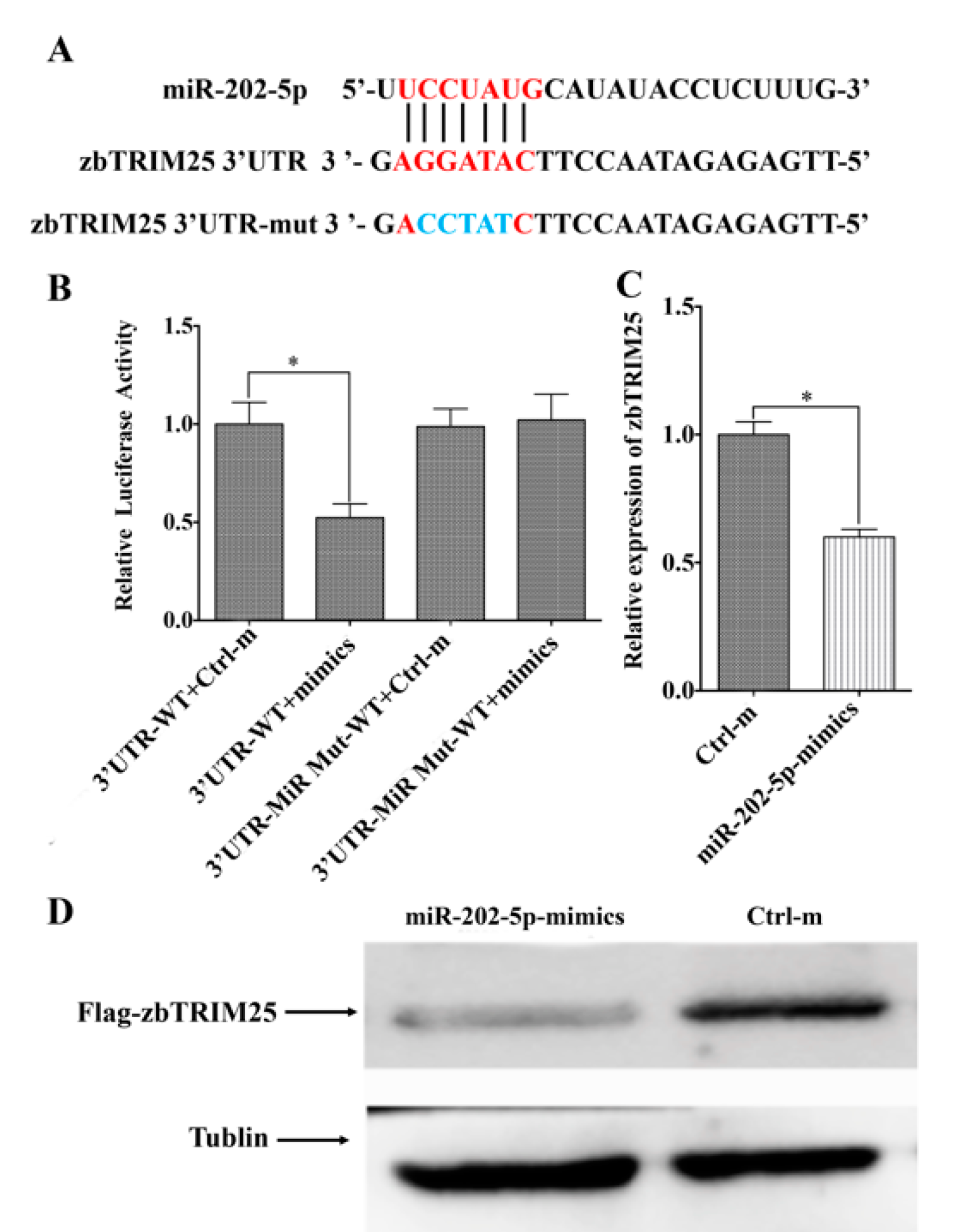
3.3. MiR-202-5p Targets zbTRIM25
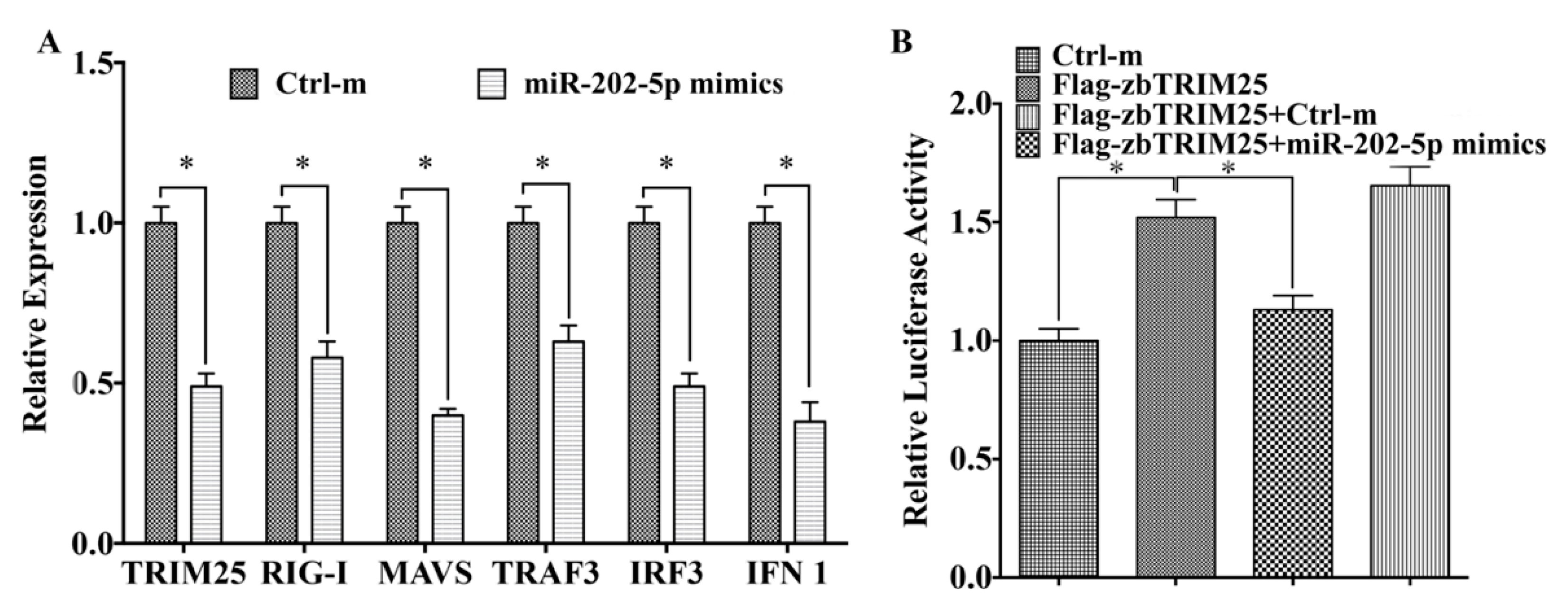
3.4. MiR-202-5p Negatively Regulates zbTRIM25-Mediated RLRs Signaling Pathway
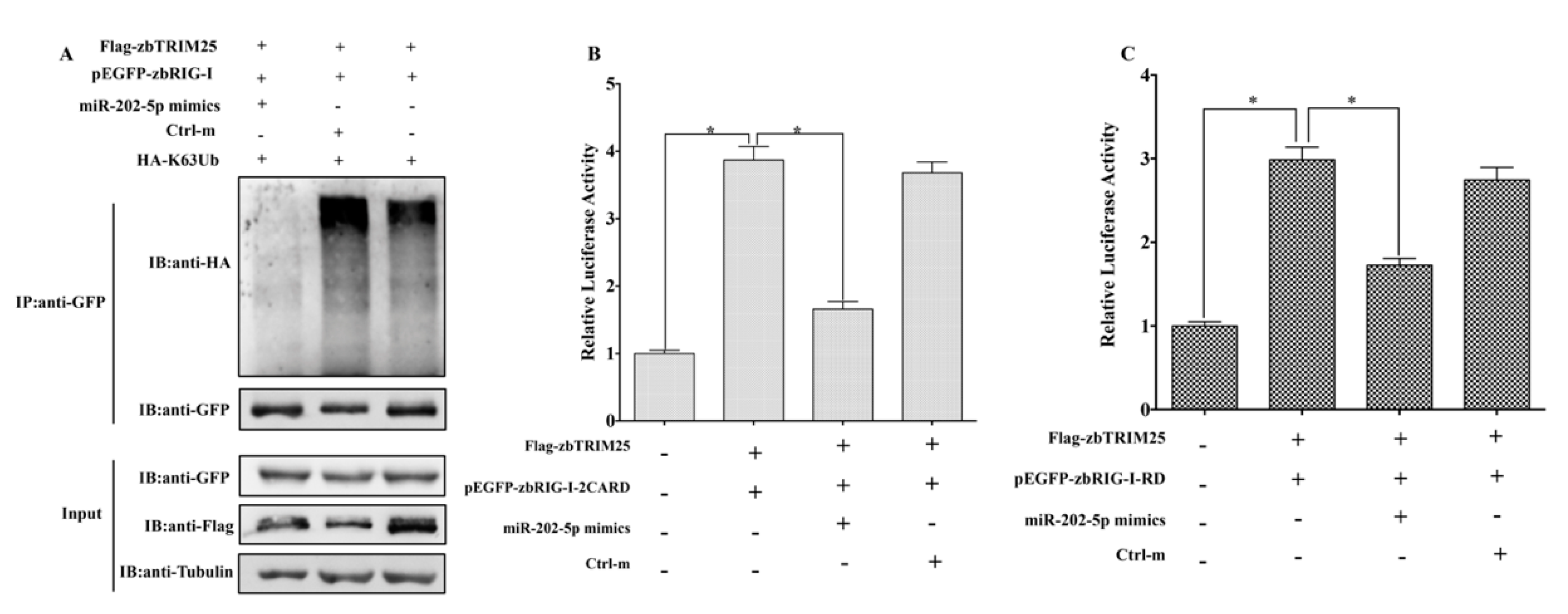
3.5. MiR-202-5p Suppresses zbTRIM25-Mediated zbRIG-I Activation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dolasia, K.; Bisht, M.K.; Pradhan, G.; Udgata, A.; Mukhopadhyay, S. TLRs/NLRs: Shaping the landscape of host immunity. Int. Rev. Immunol. 2018, 37, 3–19. [Google Scholar] [CrossRef]
- Eisenächer, K.; Krug, A. Regulation of RLR-Mediated innate immune signaling–It is all about keeping the balance. Eur. J. Cell Biol. 2012, 91, 36–47. [Google Scholar] [CrossRef]
- Zhang, H.; Ye, H.; Liu, S.; Deng, C.; Li, X.; Shi, P.; Zhang, B. West nile virus NS1 antagonizes interferon beta production by targeting RIG-I and MDA5. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Wang, W.; Jiang, M.; Liu, S.; Zhang, S.; Liu, W.; Ma, Y.; Zhang, L.; Zhang, J.; Cao, X. RNF122 suppresses antiviral type I interferon production by targeting RIG-I CARDs to mediate RIG-I degradation. Proc. Natl. Acad. Sci. USA 2016, 113, 9581–9586. [Google Scholar] [CrossRef]
- Costa, J.Z.; Thompson, K.D. Understanding the interaction between Betanodavirus and its host for the development of prophylactic measures for viral encephalopathy and retinopathy. Fish Shellfish Immunol. 2016, 53, 35–49. [Google Scholar] [CrossRef]
- Chen, H.; Liu, W.; Wu, S.; Chiou, P.P.; Li, Y.-H.; Chen, Y.; Lin, G.; Lu, M.; Wu, J. RIG-I specifically mediates group II type I IFN activation in nervous necrosis virus infected zebrafish cells. Fish Shellfish Immunol. 2015, 43, 427–435. [Google Scholar] [CrossRef]
- Jia, P.; Jia, K.; Chen, L.; Le, Y.; Jin, Y.; Zhang, J.; Zhu, L.; Zhang, L.; Yi, M. Identification and characterization of the melanoma differentiation-Associated gene 5 in sea perch, Lateolabrax japonicus. Dev. Comp. Immunol. 2016, 61, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Jia, P.; Zhang, J.; Jin, Y.; Zeng, L.; Jia, K.; Yi, M. Characterization and expression analysis of laboratory of genetics and physiology 2 gene in sea perch, Lateolabrax japonicus. Fish Shellfish Immunol. 2015, 47, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Deng, J.; Ye, Z.; Wang, J.; Gou, H.; Liu, W.; Zhao, M.; Liao, M.; Yi, L.; Chen, J. Absence of autophagy promotes apoptosis by modulating the ROS-Dependent RLR signaling pathway in classical swine fever virus-infected cells. Autophagy 2016, 12, 1738–1758. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Kai, Y.; Chi, S. Persistently betanodavirus-Infected barramundi (Lates calcarifer) exhibit resistances to red sea bream iridovirus infection. Dev. Comp. Immunol. 2013, 41, 666–674. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.-W.; Chao, Y.-M.; Guo, T.-C.; Santi, N.; Evensen, O.; Kasani, S.K.; Hong, J.-R.; Wu, J.-L. The interferon response is involved in nervous necrosis virus acute and persistent infection in zebrafish infection model. Mol. Immunol. 2008, 45, 1146–1152. [Google Scholar] [CrossRef] [PubMed]
- Dai, R.; Ahmed, S.A. MicroRNA, a new paradigm for understanding immunoregulation, inflammation, and autoimmune diseases. Transl. Res. 2011, 157, 163–179. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Pan, Y.; Fu, H.; Zhang, J. microRNA-205 and microRNA-338-3p reduces cell apoptosis in prostate carcinoma tissue and lncap prostate carcinoma cells by directly targeting the B-Cell lymphoma 2 (Bcl-2) gene. Med. Sci. Monit. 2019, 25, 1122–1132. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Huang, Y.; Guo, Y.; Xiong, Y.; Zhu, S.; Xu, L.; Lu, J.; Li, X.; Wan, J.; Lu, Y.; et al. microRNA-690 regulates induced pluripotent stem cells (iPSCs) differentiation into insulin-producing cells by targeting Sox9. Stem. Cell Res. Ther. 2019, 10, 59. [Google Scholar] [CrossRef]
- Hou, J.; Wang, P.; Lin, L.; Liu, X.; Ma, F.; An, H.; Wang, Z.; Cao, X. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J. Immunol. 2009, 183, 2150–2158. [Google Scholar] [CrossRef]
- Xu, T.; Chu, Q.; Cui, J.; Bi, D. Inducible microRNA-3570 feedback inhibits the RIG-I-Dependent innate immune response to rhabdovirus in teleost fish by targeting MAVS/IPS-1. J. Virol. 2017, 92, e01594-17. [Google Scholar] [CrossRef]
- Jin, Y.L.; Liu, W.; Xiang, Y.X.; Zhang, W.W.; Zhang, H.; Jia, K.T.; Yi, M.S. Maternal miR-202-5p is required for zebrafish primordial germ cell migration by protecting small GTPase Cdc42. J. Mol. Cell Biol. 2019. [Google Scholar] [CrossRef]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef]
- Jin, Y.L.; Chen, L.M.; Le, Y.; Li, Y.L.; Hong, Y.H.; Jia, K.T.; Yi, M.S. Establishment of a cell line with high transfection efficiency from zebrafish Danio rerio embryos and its susceptibility to fish viruses: Characterization of a d. rerio cell line. J. Fish Biol. 2017, 91, 1018–1031. [Google Scholar] [CrossRef]
- Jin, Y.L.; Jia, K.T.; Zhang, W.W.; Xiang, Y.X.; Jia, P.; Liu, W.; Yi, M.S. Zebrafish TRIM25 promotes innate immune response to RGNNV infection by targeting 2CARD and RD regions of RIG-I for K63-Linked ubiquitination. Front. Immunol. 2019, 10, 12. [Google Scholar] [CrossRef]
- Zhang, W.; Li, Z.; Jia, P.; Liu, W.; Yi, M.; Jia, K. Interferon regulatory factor 3 from sea perch (Lateolabrax japonicus) exerts antiviral function against nervous necrosis virus infection. Dev. Comp. Immunol. 2018, 88, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, W.; Jin, Y.; Jia, P.; Jia, K.; Yi, M. MiR-202-5p is a novel germ plasm-Specific microRNA in zebrafish. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Jia, K.T.; Wu, Y.Y.; Liu, Z.Y.; Mi, S.; Zheng, Y.W.; He, J.; Weng, S.P.; Li, S.C.; He, J.G.; Guo, C.J. Mandarin fish caveolin 1 interaction with major capsid protein of infectious spleen and kidney necrosis virus and its role in early stages of infection. J. Virol. 2013, 87, 3027–3038. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Ketkar, H.; Geng, T.; Lo, E.; Wang, L.; Xi, J.; Sun, Q.; Zhu, Z.; Cui, Y.; Yang, L.; et al. Zika Virus non-Structural protein 4A blocks the RLR-MAVS signaling. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Ashraf, U.; Ye, J.; Duan, X.; Zohaib, A.; Wang, W.; Chen, Z.; Zhu, B.; Li, Y.; Chen, H.; et al. MicroRNA-22 negatively regulates poly(I:C)-triggered type I interferon and inflammatory cytokine production via targeting mitochondrial antiviral signaling protein (MAVS). Oncotarget 2016, 7, 76667–76683. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Yi, L.; Feng, S.; Zhao, L.; Li, J.; Zhou, M.; Liang, R.; Gu, N.; Wu, Z.; Tu, J.; et al. Characterization of microRNAs in orange-Spotted grouper (Epinephelus coioides) fin cells upon red-Spotted grouper nervous necrosis virus infection. Fish Shellfish Immunol. 2017, 63, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Jia, K.; Zhang, J.; Jia, P.; Zeng, L.; Jin, Y.; Yuan, Y.; Chen, J.; Hong, Y.; Yi, M. Identification of microRNAs in zebrafish spermatozoa. Zebrafish 2015, 12, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Wainwright, E.N.; Jorgensen, J.S.; Kim, Y.; Truong, V.; Bagheri-Fam, S.; Davidson, T.; Svingen, T.; Fernandez-Valverde, S.L.; McClelland, K.S.; Taft, R.J.; et al. SOX9 regulates microRNA miR-202-5p/3p expression during mouse testis differentiation. Biol. Reprod. 2013, 89, 34. [Google Scholar] [CrossRef]
- Gack, M.U.; Shin, Y.C.; Joo, C.-H.; Urano, T.; Liang, C.; Sun, L.; Takeuchi, O.; Akira, S.; Chen, Z.; Inoue, S.; et al. TRIM25 RING-Finger E3 ubiquitin ligase is essential for RIG-I-Mediated antiviral activity. Nature 2007, 446, 916–920. [Google Scholar] [CrossRef]
- Lian, H.; Zang, R.; Wei, J.; Ye, W.; Hu, M.-M.; Chen, Y.; Zhang, X.; Guo, Y.; Lei, C.; Yang, Q.; et al. The zinc-Finger protein ZCCHC3 binds RNA and facilitates viral RNA sensing and activation of the RIG-I-Like receptors. Immunity 2018, 49, 438–448.e5. [Google Scholar] [CrossRef]
- Pauli, E.-K.; Chan, Y.K.; Davis, M.E.; Gableske, S.; Wang, M.K.; Feister, K.F.; Gack, M.U. The ubiquitin-Specific protease USP15 promotes RIG-I-mediated antiviral signaling by deubiquitylating TRIM25. Sci. Signal 2014, 7, ra3. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Choi, J.; Yoon, I.; Lee, J.K.; Inn, K. Positive regulatory role of c-Src-mediated TRIM25 tyrosine phosphorylation on RIG-I ubiquitination and RIG-I-mediated antiviral signaling pathway. Cell. Immunol. 2018, 332, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Gack, M.U.; Albrecht, R.A.; Urano, T.; Inn, K.-S.; Huang, I.-C.; Carnero, E.; Farzan, M.; Inoue, S.; Jung, J.U.; García-Sastre, A. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 2009, 5, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.; Gack, M.U. Post-Translational control of intracellular pathogen sensing pathways. Trends Immunol. 2017, 38, 39–52. [Google Scholar] [CrossRef]
- Xu, C.; He, X.; Zheng, Z.; Zhang, Z.; Wei, C.; Guan, K.; Hou, L.; Zhang, B.; Zhu, L.; Cao, Y.; et al. Downregulation of microRNA miR-526a by enterovirus inhibits RIG-I-Dependent innate immune response. J. Virol. 2014, 88, 11356–11368. [Google Scholar] [CrossRef]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Jin, Y.; Zhang, W.; Xiang, Y.; Jia, P.; Yi, M.; Jia, K. MiR-202-5p Inhibits RIG-I-Dependent Innate Immune Responses to RGNNV Infection by Targeting TRIM25 to Mediate RIG-I Ubiquitination. Viruses 2020, 12, 261. https://doi.org/10.3390/v12030261
Liu W, Jin Y, Zhang W, Xiang Y, Jia P, Yi M, Jia K. MiR-202-5p Inhibits RIG-I-Dependent Innate Immune Responses to RGNNV Infection by Targeting TRIM25 to Mediate RIG-I Ubiquitination. Viruses. 2020; 12(3):261. https://doi.org/10.3390/v12030261
Chicago/Turabian StyleLiu, Wei, Yilin Jin, Wanwan Zhang, Yangxi Xiang, Peng Jia, Meisheng Yi, and Kuntong Jia. 2020. "MiR-202-5p Inhibits RIG-I-Dependent Innate Immune Responses to RGNNV Infection by Targeting TRIM25 to Mediate RIG-I Ubiquitination" Viruses 12, no. 3: 261. https://doi.org/10.3390/v12030261
APA StyleLiu, W., Jin, Y., Zhang, W., Xiang, Y., Jia, P., Yi, M., & Jia, K. (2020). MiR-202-5p Inhibits RIG-I-Dependent Innate Immune Responses to RGNNV Infection by Targeting TRIM25 to Mediate RIG-I Ubiquitination. Viruses, 12(3), 261. https://doi.org/10.3390/v12030261





