Plant Molecular Responses to Potato Virus Y: A Continuum of Outcomes from Sensitivity and Tolerance to Resistance
Abstract
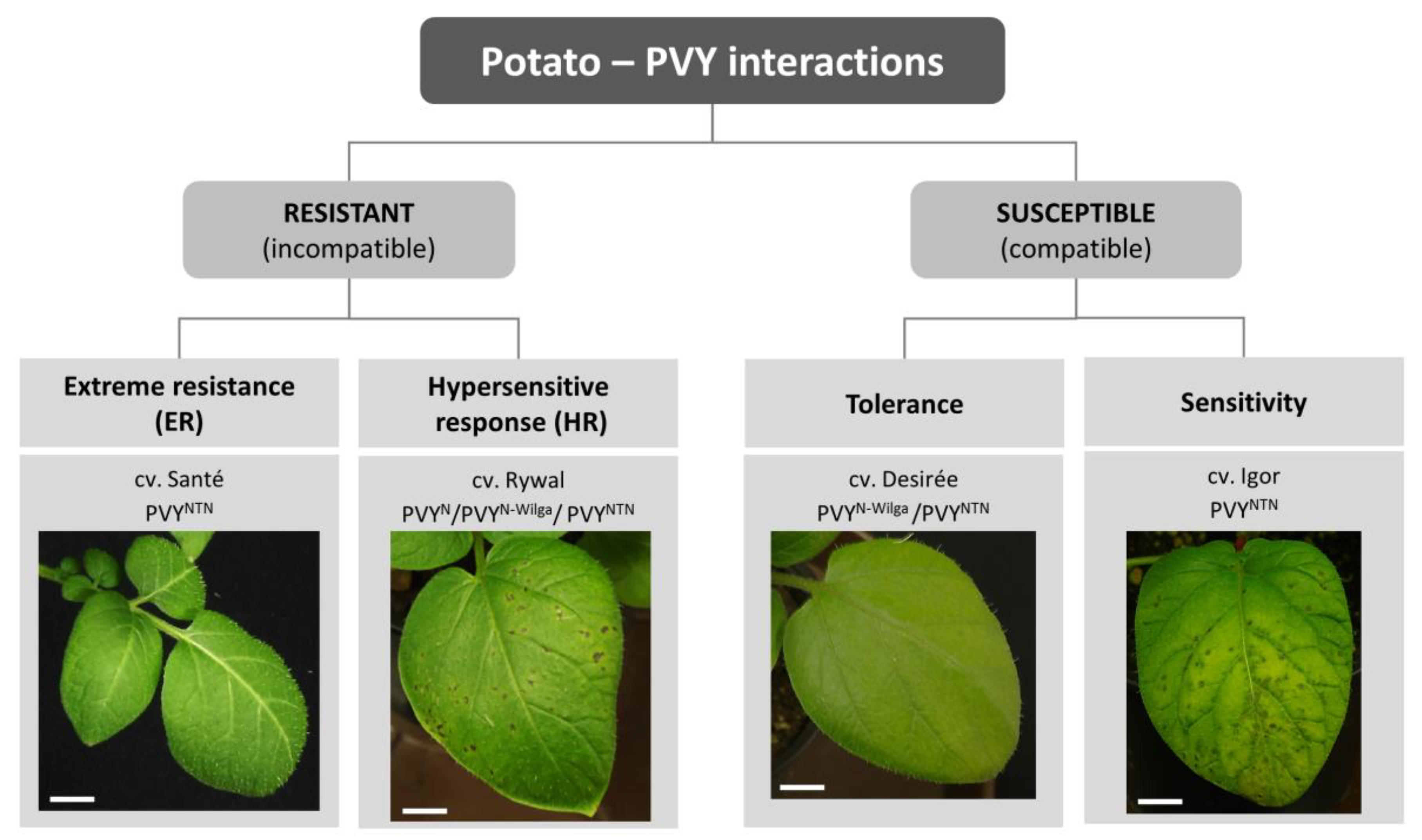
:1. Introduction
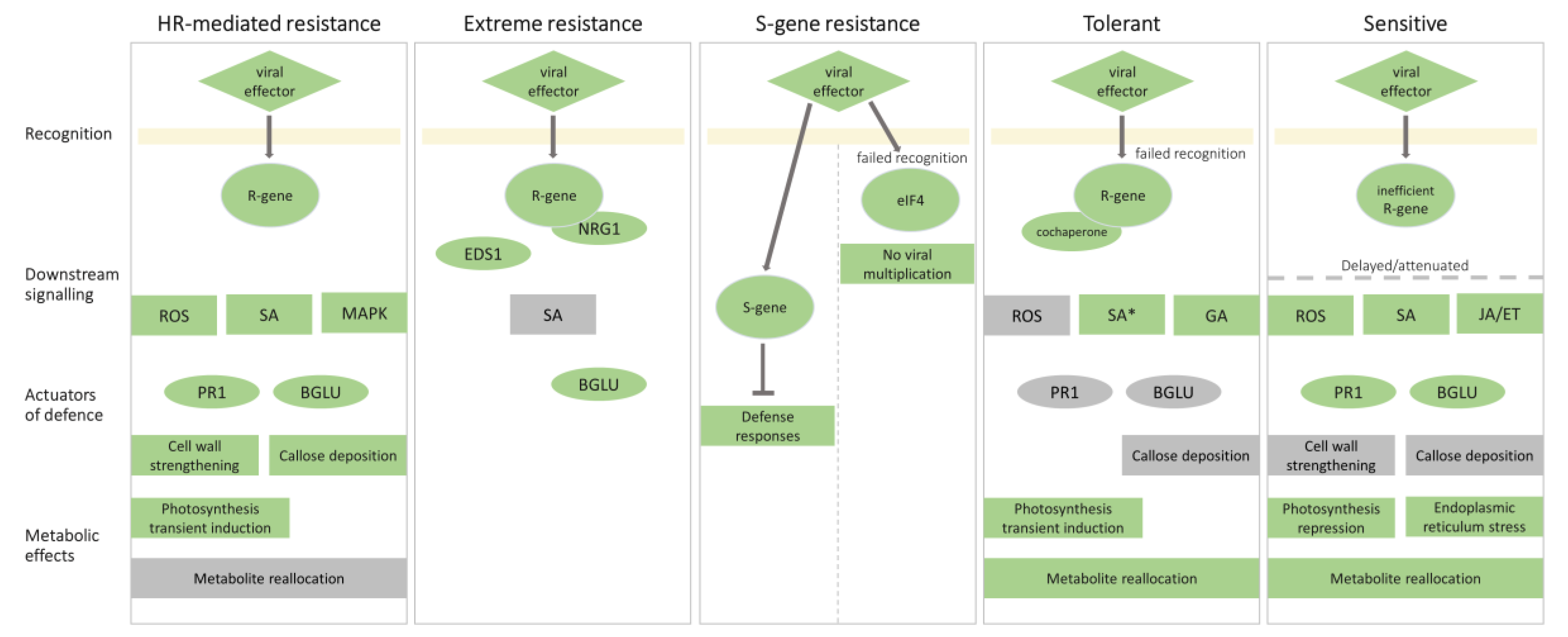
2. Molecular Mechanisms Underlying Resistance to PVY
2.1. Hypersensitive Response-Mediated Resistance: Response at the Right Place at the Right Time
2.2. Extreme Resistance Response: No Signs of Battle
2.3. S-Gene Conferred Resistance: Can’t Live without You
3. Susceptible Response to PVY
3.1. Sensitive Response: Too Late or too Weak
3.2. Tolerant Response: Balance between Plant Defense and Virus Counterdefense
4. Future Outlook
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kreuze, J.F.; Jeevalatha, A.; Figueira, A.R. Viral Diseases in Potato. In The Potato Crop; Campos, H., Ortiz, O., Eds.; Springer: Cham, Switzerland, 2020; pp. 389–430. [Google Scholar]
- Bokx, J.A.; van der Want, J.P.H. Viruses of Potatoes and Seed-Potato Production. Potato Res. 1988, 31, 405–406. [Google Scholar]
- Valkonen, J.P.T. Viruses: Economical Losses and Biotechnological Potential. In Potato Biology and Biotechnology: Advances and Perspectives; Vreugdenhil, D., Bradshaw, J., Gebhardt, C., Govers, F., Mackerron, D.K.L., Taylor, M.A., Ross, H.A., Eds.; Elsevier Science BV: Oxford, UK; San Diego, CA, USA, 2007; pp. 619–641. [Google Scholar]
- Lacomme, C.; Glais, L.; Bellstedt, D.U.; Dupuis, B.; Karasev, A.V.; Jacquot, E. Transmission and Epidemiology of Potato virus Y. In Potato Virus Y: Biodiversity, Pathogenicity, Epidemiology and Management; Lacomme, C., Glais, L., Bellstedt, D.U., Dupuis, B., Karasev, A.V., Jacquot, E., Eds.; Springer: Cham, Switzerland, 2017; pp. 141–176. [Google Scholar]
- Chung, B.Y.-W.; Miller, W.A.; Atkins, J.F.; Firth, A.E. An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. USA 2008, 105, 5897–5902. [Google Scholar] [CrossRef] [Green Version]
- Kežar, A.; Kavčič, L.; Polák, M.; Nováček, J.; Gutiérrez-Aguirre, I.; Žnidarič, M.T.; Coll, A.; Stare, K.; Gruden, K.; Ravnikar, M.; et al. Structural basis for the multitasking nature of the potato virus Y coat protein. Sci. Adv. 2019, 5, eaaw3808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofius, D.; Maier, A.T.; Dietrich, C.; Jungkunz, I.; Börnke, F.; Maiss, E.; Sonnewald, U. Capsid protein-mediated recruitment of host DnaJ-like proteins is required for Potato virus Y infection in tobacco plants. J. Virol. 2007, 81, 11870–11880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hafrén, A.; Hofius, D.; Rönnholm, G.; Sonnewald, U.; Mäkinen, K. HSP70 and its cochaperone CPIP promote potyvirus infection in Nicotiana benthamiana by regulating viral coat protein functions. Plant Cell 2010, 22, 523–535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lõhmus, A.; Hafrén, A.; Mäkinen, K. Coat protein regulation by CK2, CPIP, HSP70 and CHIP is required for Potato virus A replication and coat protein accumulation. J. Virol. 2017, 91, e01316-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valli, A.; Gallo, A.; Rodamilans, B.; Lopez-Moya, J.; Garcia, J.A. The HCPro from the Potyviridae family: An enviable multitasking Helper Component that every virus would like to have. Mol. Plant Pathol. 2018, 19, 744–763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coutinho de Oliveira, L.; Volpon, L.; Rahardjo, A.K.; Osborne, M.J.; Culjkovic-Kraljacic, B.; Trahan, C.; Oeffinger, M.; Kwok, B.H.; Borden, K.L.B. Structural studies of the eIF4E—VPg complex reveal a direct competition for capped RNA: Implications for translation. Proc. Natl. Acad. Sci. USA 2019, 116, 24056–24065. [Google Scholar] [CrossRef]
- Cooper, J.I.; Jones, A.T. The Use of Terms for Responses of Plants to Viruses: A Reply to Recent Proposals. Phytopathology 1984, 73, 127–128. [Google Scholar] [CrossRef] [Green Version]
- Valkonen, J.P.T. Elucidation of virus-host interactions to enhance resistance breeding for control of virus diseases in potato. Breed. Sci. 2015, 65, 69–76. [Google Scholar] [CrossRef] [Green Version]
- Michel, V.; Julio, E.; Candresse, T.; Cotucheau, J.; Decorps, C.; Volpatti, R.; Moury, B.; Glais, L.; Dorlhac de Borne, F.; Decroocq, V.; et al. NtTPN1: A RPP8-like R gene required for Potato virus Y-induced veinal necrosis in tobacco. Plant J. 2018, 95, 700–714. [Google Scholar] [CrossRef] [PubMed]
- Karasev, A.V.; Gray, S.M. Continuous and Emerging Challenges of Potato virus Y in Potato. Annu. Rev. Phytopathol. 2013, 51, 571–586. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, A.J.; Ohshima, K.; Yasaka, R.; Mohammadi, M.; Gibbs, M.J.; Jones, R.A.C. The phylogenetics of the global population of potato virus Y and its necrogenic recombinants. Virus Evol. 2017, 3, vex002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nigam, D.; LaTourrette, K.; Souza, P.F.N.; Garcia-Ruiz, H. Genome-Wide Variation in Potyviruses. Front. Plant Sci. 2019, 10, 1439. [Google Scholar] [CrossRef] [Green Version]
- Kogovšek, P.; Pompe-Novak, M.; Baebler, Š.; Rotter, A.; Gow, L.; Gruden, K.; Foster, G.D.; Boonham, N.; Ravnikar, M. Aggressive and mild Potato virus Y isolates trigger different specific responses in susceptible potato plants. Plant Pathol. 2010, 59, 1121–1132. [Google Scholar] [CrossRef] [Green Version]
- Yin, Z.; Xie, F.; Michalak, K.; Pawełkowicz, M.; Zhang, B.; Murawska, Z.; Lebecka, R.; Zimnoch-Guzowska, E. Potato cultivar Etola exhibits hypersensitive resistance to PVY NTN and partial resistance to PVY Z-NTN and PVY N-Wi strains and strain-specific alterations of certain host miRNAs might correlate with symptom severity. Plant Pathol. 2017, 66, 539–550. [Google Scholar] [CrossRef]
- Lukan, T.; Pompe-Novak, M.; Baebler, Š.; Tušek Žnidarič, M.; Kladnik, A.; Blejec, A.; Zagorščak, M.; Stare, K.; Dušak, B.; Coll, A.; et al. Spatial accumulation of salicylic acid is in effector-triggered immunity of potato against viruses regulated by RBOHD. bioRxiv 2020. [Google Scholar] [CrossRef]
- Makarova, S.; Makhotenko, A.; Spechenkova, N.; Love, A.J.; Kalinina, N.O.; Taliansky, M. Interactive Responses of Potato (Solanum tuberosum L.) Plants to Heat Stress and Infection With Potato Virus Y. Front. Microbiol. 2018, 9, 2582. [Google Scholar] [CrossRef] [Green Version]
- García-Marcos, A.; Pacheco, R.; Martiáñez, J.; González-Jara, P.; Díaz-Ruíz, J.R.; Tenllado, F. Transcriptional changes and oxidative stress associated with the synergistic interaction between Potato virus X and Potato virus Y and their relationship with symptom expression. Mol. Plant Microbe Interact. 2009, 22, 1431–1444. [Google Scholar] [CrossRef] [Green Version]
- Pacheco, R.; García-Marcos, A.; Barajas, D.; Martiáñez, J.; Tenllado, F. PVX-potyvirus synergistic infections differentially alter microRNA accumulation in Nicotiana benthamiana. Virus Res. 2012, 165, 231–235. [Google Scholar] [CrossRef]
- Liang, Z.; Dickison, V.; Singh, M.; Xiong, X.; Nie, X. Studies of tomato plants in response to infections with PVX and different PVY isolates reveal a remarkable PVX-PVYNTN synergism and diverse expression profiles of genes involved in different pathways. Eur. J. Plant Pathol. 2016, 144, 55–71. [Google Scholar] [CrossRef]
- Moreno Gonçalves, A.B.; Lopez-Moya, J.J. When viruses play team sports: Mixed infections in plants. Phytopathology 2020, 110, 29–48. [Google Scholar] [CrossRef] [PubMed]
- McCormack, M.E.; Lopez, J.A.; Crocker, T.H.; Mukhtar, M.S. Making the right connections: Network biology and plant immune system dynamics. Curr. Plant Biol. 2016, 5, 2–12. [Google Scholar] [CrossRef] [Green Version]
- Van Bel, M.; Diels, T.; Vancaester, E.; Kreft, L.; Botzki, A.; Van De Peer, Y.; Coppens, F.; Vandepoele, K. PLAZA 4.0: An integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 2018, 46, D1190–D1196. [Google Scholar] [CrossRef] [PubMed]
- Ramšak, Ž.; Coll, A.; Stare, T.; Tzfadia, O.; Baebler, Š.; Van de Peer, Y.; Gruden, K. Network modelling unravels mechanisms of crosstalk between ethylene and salicylate signalling in potato. Plant Physiol. 2018, 178, 488–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gantner, J.; Ordon, J.; Kretschmer, C.; Guerois, R.; Stuttmann, J. An EDS1-SAG101 complex is essential for tTNL-mediated immunity in Nicotiana benthamiana. Plant Cell 2019, 31, 2456–2474. [Google Scholar] [CrossRef] [Green Version]
- The Potato Genome Sequencing Consortium. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–195. [Google Scholar] [CrossRef]
- Petek, M.; Zagorščak, M.; Ramšak, Ž.; Sanders, S.; Tseng, E.; Zouine, M.; Coll, A.; Gruden, K. Cultivar-specic transcriptome and pan-transcriptome reconstruction of tetraploid potato. bioRxiv 2019, 1–12. [Google Scholar] [CrossRef]
- Wu, X.; Valli, A.; García, J.A.; Zhou, X.; Cheng, X. The Tug-of-War between plants and viruses: Great progress and many remaining questions. Viruses 2019, 11, 203. [Google Scholar] [CrossRef] [Green Version]
- Calil, I.P.; Fontes, E.P.B. Plant immunity against viruses: Antiviral immune receptors in focus. Ann. Bot. 2017, 119, 711–723. [Google Scholar] [CrossRef] [Green Version]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Gou, X.; He, K.; Xi, D.; Du, J.; Lin, H.; Li, J. BAK1 and BKK1 in Arabidopsis thaliana confer reduced susceptibility to turnip crinkle virus. Eur. J. Plant Pathol. 2010, 127, 149–156. [Google Scholar] [CrossRef]
- Kørner, C.J.; Klauser, D.; Niehl, A.; Domínguez-Ferreras, A.; Chinchilla, D.; Boller, T.; Heinlein, M.; Hann, D.R. The immunity regulator BAK1 contributes to resistance against diverse RNA viruses. Mol. Plant Microbe Interact. 2013, 26, 1271–1280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niehl, A.; Wyrsch, I.; Boller, T.; Heinlein, M. Double-stranded RNAs induce a pattern-triggered immune signaling pathway in plants. New Phytol. 2016, 211, 1008–1019. [Google Scholar] [CrossRef] [Green Version]
- Niehl, A.; Heinlein, M. Perception of double-stranded RNA in plant antiviral immunity. Mol. Plant Pathol. 2019, 20, 1203–1210. [Google Scholar] [CrossRef]
- Dangl, J.L.; Jones, J.D.G. A pentangular plant inflammasome. Science 2019, 364, 31–32. [Google Scholar] [CrossRef]
- Valkonen, J.P.T.; Gebhardt, C.; Zimnoch-Guzowska, E.; Watanabe, K.; Lacomme, C.; Glais, L.; Bellstedt, D.U.; Dupuis, B.; Karasev, A.V.; Jacquot, E. Resistance to Potato virus Y. In Potato Virus Y: Biodiversity, Pathogenicity, Epidemiology and Management; Lacomme, C., Glais, L., Bellstedt, D.U., Dupuis, B., Karasev, A.V., Jacquot, E., Eds.; Springer: Cham, Switzerland, 2017; pp. 207–241. [Google Scholar]
- Gaguancela, O.A.; Źuñiga, L.P.; Arias, A.V.; Halterman, D.; Flores, F.J.; Johansen, I.E.; Wang, A.; Yamaji, Y.; Verchot, J. The IRE1/bZIP60 pathway and bax inhibitor 1 suppress systemic accumulation of potyviruses and potexviruses in arabidopsis and nicotiana benthamiana plants. Mol. Plant Microbe Interact. 2016, 29, 750–766. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Hu, M.; Wang, J.; Qi, J.; Han, Z.; Wang, G.; Qi, Y.; Wang, H.W.; Zhou, J.M.; Chai, J. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 2019, 364, eaav5870. [Google Scholar] [CrossRef]
- Wang, J.; Wang, J.; Hu, M.; Wu, S.; Qi, J.; Wang, G.; Han, Z.; Qi, Y.; Gao, N.; Wang, H.W.; et al. Ligand-triggered allosteric ADP release primes a plant NLR complex. Science 2019, 364, eaav5868. [Google Scholar] [CrossRef]
- Gilroy, E.M.; Taylor, R.M.; Hein, I.; Boevink, P.; Sadanandom, A.; Birch, P.R.J. CMPG1-dependent cell death follows perception of diverse pathogen elicitors at the host plasma membrane and is suppressed by Phytophthora infestans RXLR effector AVR3a. New Phytol. 2011, 190, 653–666. [Google Scholar] [CrossRef]
- Zavaliev, R.; Levy, A.; Gera, A.; Epel, B.L. Subcellular Dynamics and Role of Arabidopsis β-1,3-Glucanases in Cell-to-Cell Movement of Tobamoviruses. Mol. Plant Microbe Interact. 2013, 26, 1016–1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zorzatto, C.; MacHado, J.P.B.; Lopes, K.V.G.; Nascimento, K.J.T.; Pereira, W.A.; Brustolini, O.J.B.; Reis, P.A.B.; Calil, I.P.; Deguchi, M.; Sachetto-Martins, G.; et al. NIK1-mediated translation suppression functions as a plant antiviral immunity mechanism. Nature 2015, 520, 679–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Zhang, C.; Li, Y.; Wu, G.; Hou, X.; Zhou, X.; Wang, A. Beclin1 restricts RNA virus infection in plants through suppression and degradation of the viral polymerase. Nat. Commun. 2018, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Liu, H.; Gong, Y.; Tao, Y.; Jiang, L.; Zuo, W.; Yang, Q.; Ye, J.; Lai, J.; Wu, J.; et al. An Atypical Thioredoxin Imparts Early Resistance to Sugarcane Mosaic Virus in Maize. Mol. Plant 2017, 10, 483–497. [Google Scholar] [CrossRef] [Green Version]
- Ishibashi, K.; Saruta, M.; Shimizu, T.; Shu, M.; Anai, T.; Komatsu, K.; Yamada, N.; Katayose, Y.; Ishikawa, M.; Ishimoto, M.; et al. Soybean antiviral immunity conferred by dsRNase targets the viral replication complex. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, M.; Neriya, Y.; Yamaji, Y.; Namba, S. Recessive resistance to plant viruses: Potential resistance genes beyond translation initiation factors. Front. Microbiol. 2016, 7, 1695. [Google Scholar] [CrossRef] [Green Version]
- Szajko, K.; Chrzanowska, M.; Witek, K.; Strzelczyk-Zyta, D.; Zagórska, H.; Gebhardt, C.; Hennig, J.; Marczewski, W. The novel gene Ny-1 on potato chromosome IX confers hypersensitive resistance to Potato virus Y and is an alternative to Ry genes in potato breeding for PVY resistance. Theor. Appl. Genet. 2008, 116, 297–303. [Google Scholar] [CrossRef] [Green Version]
- Szajko, K.; Strzelczyk-Zyta, D.; Marczewski, W. Ny-1 and Ny-2 genes conferring hypersensitive response to potato virus Y (PVY) in cultivated potatoes: Mapping and marker-assisted selection validation for PVY resistance in potato breeding. Mol. Breed. 2014, 34, 267–271. [Google Scholar] [CrossRef] [Green Version]
- Tomczyńska, I.; Jupe, F.; Hein, I.; Marczewski, W.; Sliwka, J. Hypersensitive response to Potato virus Y in potato cultivar Sárpo Mira is conferred by the Ny-Smira gene located on the long arm of chromosome IX. Mol. Breed. 2014, 34, 471–480. [Google Scholar] [CrossRef] [Green Version]
- Künstler, A.; Bacsó, R.; Gullner, G.; Hafez, Y.M.; Király, L. Staying alive—Is cell death dispensable for plant disease resistance during the hypersensitive response? Physiol. Mol. Plant Pathol. 2016, 93, 75–84. [Google Scholar] [CrossRef]
- Lukan, T.; Baebler, Š.; Pompe-Novak, M.; Guček, K.; Zagorščak, M.; Coll, A.; Gruden, K. Cell Death Is Not Sufficient for the Restriction of Potato Virus Y Spread in Hypersensitive Response-Conferred Resistance in Potato. Front. Plant Sci. 2018, 9, 168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szajko, K.; Strzelczyk-Żyta, D.; Marczewski, W. Comparison of leaf proteomes of potato (Solanum tuberosum L.) genotypes with ER- and HR-mediated resistance to PVY infection. Eur. J. Plant Pathol. 2018, 150, 375–385. [Google Scholar] [CrossRef]
- Balint-Kurti, P. The plant hypersensitive response: Concepts, control and consequences. Mol. Plant Pathol. 2019, 20, 1163–1178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baebler, Š.; Witek, K.; Petek, M.; Stare, K.; Tušek-Znidaric, M.; Pompe-Novak, M.; Renaut, J.; Szajko, K.; Strzelczyk-Zyta, D.; Marczewski, W.; et al. Salicylic acid is an indispensable component of the Ny-1 resistance-gene-mediated response against Potato virus Y infection in potato. J. Exp. Bot. 2014, 65, 1095–1109. [Google Scholar] [CrossRef] [PubMed]
- Otulak-kozieł, K.; Kozieł, E. The Respiratory Burst Oxidase Homolog D (RbohD) Cell and Tissue Distribution in Potato—Potato Virus Y (PVY NTN) Hypersensitive and Susceptible Reactions. Int. J. Mol. Sci. 2019, 20, 2741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bigeard, J.; Hirt, H. Nuclear signaling of plant MAPKs. Front. Plant Sci. 2018, 9, 469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazar, A.; Coll, A.; Dobnik, D.; Baebler, Š.; Bedina-Zavec, A.; Žel, J.; Gruden, K. Involvement of Potato (Solanum tuberosum L.) MKK6 in Response to Potato virus Y. PLoS ONE 2014, 9, e104553. [Google Scholar] [CrossRef]
- Dobnik, D.; Lazar, A.; Stare, T.; Gruden, K.; Vleeshouwers, V.G.; Žel, J. Solanum venturii, a suitable model system for virus-induced gene silencing studies in potato reveals StMKK6 as an important player in plant immunity. Plant Methods 2016, 12, 29. [Google Scholar] [CrossRef] [Green Version]
- Pieterse, C.M.J.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C.M. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef] [Green Version]
- Mur, L.A.J.; Kenton, P.; Lloyd, A.J.; Ougham, H.; Prats, E. The hypersensitive response; the centenary is upon us but how much do we know? J. Exp. Bot. 2008, 59, 501–520. [Google Scholar] [CrossRef] [Green Version]
- Goyer, A.; Hamlin, L.; Crosslin, J.M.; Buchanan, A.; Chang, J.H. RNA-Seq analysis of resistant and susceptible potato varieties during the early stages of potato virus Y infection. BMC Genom. 2015, 16, 472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iglesias, V.A.; Meins, F. Movement of plant viruses is delayed in a β-1,3-glucanase-deficient mutant showing a reduced plasmodesmatal size exclusion limit and enhanced callose deposition. Plant J. 2000, 21, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Otulak-Kozieł, K.; Kozieł, E.; Bujarski, J. Spatiotemporal Changes in Xylan-1/Xyloglucan and Xyloglucan Xyloglucosyl Transferase (XTH-Xet5) as a Step-In of Ultrastructural Cell Wall Remodelling in Potato–Potato Virus Y (PVYNTN) Hypersensitive and Susceptible Reaction. Int. J. Mol. Sci. 2018, 19, 2287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otulak-Kozieł, K.; Kozieł, E.; Lockhart, B. Plant Cell Wall Dynamics in Compatible and Incompatible Potato Response to Infection Caused by Potato Virus Y (PVYNTN). Int. J. Mol. Sci. 2018, 19, 862. [Google Scholar] [CrossRef] [Green Version]
- Dobnik, D.; Baebler, Š.; Kogovšek, P.; Pompe-Novak, M.; Štebih, D.; Panter, G.; Janez, N.; Morisset, D.; Žel, J.; Gruden, K. β-1,3-glucanase class III promotes spread of PVY(NTN) and improves in planta protein production. Plant Biotechnol. Rep. 2013, 7, 547–555. [Google Scholar] [CrossRef] [Green Version]
- Chowdury, R.; Lasky, D.; Karki, H.; Zhang, Z.; Goyer, A.; Halterman, D.; Rakotondrafara, A.M. HCPro suppression of callose deposition contributes to strain specific resistance against potato virus Y. Phytopathology 2019. [Google Scholar] [CrossRef]
- Tian, Y.P.; Valkonen, J.P.T. Genetic determinants of Potato virus Y required to overcome or trigger hypersensitive resistance to PVY strain group o controlled by the gene Ny in Potato. Mol. Plant Microbe Interact. 2013, 26, 297–305. [Google Scholar] [CrossRef] [Green Version]
- Valkonen, J.P.T.; Palohuhta, J.P. Resistance to potato virus A and potato virus Y in potato cultivars grown in Finland. Agric. Food Sci. Finland 1996, 5, 57–62. [Google Scholar] [CrossRef]
- Seo, J.-K.; Kwon, S.-J.; Cho, W.K.; Choi, H.-S.; Kim, K.-H. Type 2C Protein Phosphatase Is a Key Regulator of Antiviral Extreme Resistance Limiting Virus Spread. Sci. Rep. 2014, 4, 5905. [Google Scholar] [CrossRef] [Green Version]
- Cockerham, G. Potato breeding for virus resistance. Ann. Appl. Biol. 1943, 30, 105–108. [Google Scholar] [CrossRef]
- Flis, B.; Hennig, J.; Strzelczyk-Zyta, D.; Gebhardt, C.; Marczewski, W. The Ry-fsto gene from Solanum stoloniferum for extreme resistant to Potato virus Y maps to potato chromosome XII and is diagnosed by PCR marker GP122 718 in PVY resistant potato cultivars. Mol. Breed. 2005, 15, 95–101. [Google Scholar] [CrossRef] [Green Version]
- Hosaka, K.; Hosaka, Y.; Mori, M.; Maida, T.; Matsunaga, H. Detection of a simplex RAPD marker linked to resistance to potato virus Y in a tetraploid potato. Am. J. Potato Res. 2001, 78, 191–196. [Google Scholar] [CrossRef]
- Hämäläinen, J.H.; Watanabe, K.N.; Valkonen, J.P.T.; Arihara, A.; Plaisted, R.L.; Pehu, E.; Miller, L.; Slack, S.A. Mapping and marker-assisted selection for a gene for extreme resistance to potato virus Y. Theor. Appl. Genet. 1997, 94, 192–197. [Google Scholar] [CrossRef]
- Cockerham, G. Genetical studies on resistance to potato viruses X and Y. Heredity (Edinburgh) 1970, 25, 309–348. [Google Scholar] [CrossRef]
- Szajko, K.; Yin, Z.; Marczewski, W. Accumulation of miRNA and mRNA Targets in Potato Leaves Displaying Temperature-Dependent Responses to Potato Virus Y. Potato Res. 2019, 62, 379–392. [Google Scholar] [CrossRef] [Green Version]
- Torrance, L.; Cowan, G.H.; McLean, K.; MacFarlane, S.; Al-Abedy, A.N.; Armstrong, M.; Lim, T.-Y.; Hein, I.; Bryan, G.J. Natural resistance to Potato virus Y in Solanum tuberosum Group Phureja. Theor. Appl. Genet. 2020. [Google Scholar] [CrossRef] [Green Version]
- Thiele, G.; Thiesen, K.; Bonierbale, M.; Walker, T. Targeting the poor and hungry with potato science. Potato J. 2010, 37, 75–86. [Google Scholar]
- Grech-Baran, M.; Witek, K.; Szajko, K.; Witek, A.I.; Morgiewicz, K.; Wasilewicz-Flis, I.; Jakuczun, H.; Marczewski, W.; Jones, J.D.; Hennig, J. Extreme resistance to Potato Virus Y in potato carrying the Rysto gene is mediated by a TIR-NLR immune receptor. Plant Biotechnol. J. 2019, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Mestre, P.; Brigneti, G.; Baulcombe, D.C. An Ry-mediated resistance response in potato requires the intact active site of the Nla proteinase from potato virus Y. Plant J. 2000, 23, 653–661. [Google Scholar] [CrossRef] [Green Version]
- Mestre, P.; Brigneti, G.; Durrant, M.C.; Baulcombe, D.C. Potato virus Y NIa protease activity is not sufficient for elicitation of Ry -mediated disease resistance in potato. Plant J. 2003, 36, 755–761. [Google Scholar] [CrossRef]
- Kim, S.B.; Kang, W.H.; Huy, H.N.; Yeom, S.I.; An, J.T.; Kim, S.; Kang, M.Y.; Kim, H.J.; Jo, Y.D.; Ha, Y.; et al. Divergent evolution of multiple virus-resistance genes from a progenitor in Capsicum spp. New Phytol. 2017, 213, 886–899. [Google Scholar] [CrossRef] [PubMed]
- Dogimont, C.; Palloix, A.; Daubze, A.M.; Marchoux, G.; Selassie, K.G.; Pochard, E. Genetic analysis of broad spectrum resistance to potyviruses using doubled haploid lines ofpepper (Capsicum annuum L.). Euphytica 1996, 88, 231–239. [Google Scholar] [CrossRef]
- Kim, S.-B.; Lee, H.-Y.; Seo, S.; Lee, J.H.; Choi, D. RNA-Dependent RNA Polymerase (NIb) of the Potyviruses Is an Avirulence Factor for the Broad-Spectrum Resistance Gene Pvr4 in Capsicum annuum cv. CM334. PLoS ONE 2015, 10, e0119639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, J.E.; Holub, E.B.; Frost, L.N.; Falk, A.; Gunn, N.D.; Daniels, M.J. Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 1996, 8, 2033–2046. [Google Scholar] [PubMed] [Green Version]
- Aarts, N.; Metz, M.; Holub, E.; Staskawicz, B.J.; Daniels, M.J.; Parker, J.E. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 1998, 95, 10306–10311. [Google Scholar] [CrossRef] [Green Version]
- Peart, J.R.; Mestre, P.; Lu, R.; Malcuit, I.; Baulcombe, D.C. NRG1, a CC-NB-LRR protein, together with N, a TIR-NB-LRR protein, mediates resistance against tobacco mosaic virus. Curr. Biol. 2005, 15, 968–973. [Google Scholar] [CrossRef] [Green Version]
- Castel, B.; Ngou, P.M.; Cevik, V.; Redkar, A.; Kim, D.S.; Yang, Y.; Ding, P.; Jones, J.D.G. Diverse NLR immune receptors activate defence via the RPW8-NLR NRG1. New Phytol. 2019, 222, 966–980. [Google Scholar] [CrossRef]
- Baebler, Š.; Krecic-Stres, H.; Rotter, A.; Kogovsek, P.; Cankar, K.; Kok, E.J.; Gruden, K.; Kovac, M.; Zel, J.; Pompe-Novak, M.; et al. PVY NTN elicits a diverse gene expression response in different potato genotypes in the first 12 h after inoculation. Mol. Plant Pathol. 2009, 10, 263–275. [Google Scholar] [CrossRef]
- Stare, T.; Stare, K.; Weckwerth, W.; Wienkoop, S.; Gruden, K. Comparison between Proteome and Transcriptome Response in Potato (Solanum tuberosum L.) Leaves. Proteomes 2017, 5, 14. [Google Scholar] [CrossRef] [Green Version]
- Baebler, Š.; Stare, K.; Kovac, M.; Blejec, A.; Prezelj, N.; Stare, T.; Kogovšek, P.; Pompe-Novak, M.; Rosahl, S.; Ravnikar, M.; et al. Dynamics of responses in compatible potato-Potato virus Y interaction are modulated by salicylic acid. PLoS ONE 2011, 6, e29009. [Google Scholar] [CrossRef] [Green Version]
- Mäkinen, K. Plant susceptibility genes as a source for potyvirus resistance. Ann. Appl. Biol. 2019, 1–8. [Google Scholar] [CrossRef]
- Michel, V.; Julio, E.; Candresse, T.; Cotucheau, J.; Decorps, C.; Volpatti, R.; Moury, B.; Glais, L.; Jacquot, E.; de Borne, F.D.; et al. A complex eIF4E locus impacts the durability of va resistance to Potato virus Y in tobacco. Mol. Plant Pathol. 2019, 20, 1051–1066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takakura, Y.; Udagawa, H.; Shinjo, A.; Koga, K. Mutation of a Nicotiana tabacum L. eukaryotic translation-initiation factor gene reduces susceptibility to a resistance-breaking strain of Potato virus Y. Mol. Plant Pathol. 2018, 19, 2124–2133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavatorta, J.; Perez, K.W.; Gray, S.M.; Van Eck, J.; Yeam, I.; Jahn, M. Engineering virus resistance using a modified potato gene. Plant Biotechnol. J. 2011, 9, 1014–1021. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez Sanchez, P.A.; Babujee, L.; Jaramillo Mesa, H.; Arcibal, E.; Gannon, M.; Halterman, D.; Jahn, M.; Jiang, J.; Rakotondrafara, A.M. Overexpression of a modified eIF4E regulates potato virus y resistance at the transcriptional level in potato. BMC Genom. 2020, 21, 18. [Google Scholar] [CrossRef]
- Guo, Y.; Jia, M.; Yang, Y.; Zhan, L.; Cheng, X.; Cai, J.; Zhang, J.; Yang, J.; Liu, T.; Fu, Q.; et al. Integrated analysis of tobacco miRNA and mRNA expression profiles under PVY infection provids insight into tobacco-PVY interactions. Sci. Rep. 2017, 7, 4895. [Google Scholar] [CrossRef]
- Ivanov, K.I.; Eskelin, K.; Lõhmus, A.; Mäkinen, K. Molecular and cellular mechanisms underlying potyvirus infection. J. Gen. Virol. 2014, 95, 1415–1429. [Google Scholar] [CrossRef] [Green Version]
- Vuorinen, A.L.; Kelloniemi, J.; Valkonen, J.P.T. Why do viruses need phloem for systemic invasion of plants? Plant Sci. 2011, 181, 355–363. [Google Scholar] [CrossRef]
- Mandadi, K.K.K.; Scholthof, K.-B. Plant immune responses against viruses: How does a virus cause disease? Plant Cell 2013, 25, 1489–1505. [Google Scholar] [CrossRef] [Green Version]
- Llave, C. Dynamic cross-talk between host primary metabolism and viruses during infections in plants. Curr. Opin. Virol. 2016, 19, 50–55. [Google Scholar] [CrossRef] [Green Version]
- Sedlar, A.; Gerič Stare, B.; Mavrič Pleško, I.; Dolničar, P.; Maras, M.; Šuštar-Vozlič, J.; Baebler; Gruden, K.; Meglič, V. Expression and regulation of programmed cell death-associated genes in systemic necrosis of PVYNTN susceptible potato tubers. Plant Pathol. 2018, 67, 1238–1252. [Google Scholar] [CrossRef]
- Kogovšek, P.; Pompe-Novak, M.; Petek, M.; Fragner, L.; Weckwerth, W.; Gruden, K. Primary metabolism, phenylpropanoids and antioxidant pathways are regulated in potato as a response to potato virus y infection. PLoS ONE 2016, 11, e0146135. [Google Scholar] [CrossRef] [PubMed]
- Stare, T.; Ramšak, Ž.; Blejec, A.; Stare, K.; Turnšek, N.; Weckwerth, W.; Wienkoop, S.; Vodnik, D.; Gruden, K. Bimodal dynamics of primary metabolism-related responses in tolerant potato-Potato virus Y interaction. BMC Genom. 2015, 16, 716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paudel, D.B.; Sanfaçon, H. Exploring the diversity of mechanisms associated with plant tolerance to virus infection. Front. Plant Sci. 2018, 9, 1575. [Google Scholar] [CrossRef]
- Lamm, C.E.; Kraner, M.E.; Hofmann, J.; Börnke, F.; Mock, H.P.; Sonnewald, U. Hop/sti1—A two-faced cochaperone involved in pattern recognition receptor maturation and viral infection. Front. Plant Sci. 2017, 8, 1754. [Google Scholar] [CrossRef] [Green Version]
- Križnik, M.; Petek, M.; Dobnik, D.; Ramšak, Ž.; Baebler, Š.; Pollmann, S.; Kreuze, J.F.; Žel, J.; Gruden, K. Salicylic Acid Perturbs sRNA-Gibberellin Regulatory Network in Immune Response of Potato to Potato virus Y Infection. Front. Plant Sci. 2017, 8, 2192. [Google Scholar] [CrossRef] [Green Version]
- Antolín-Llovera, M.; Petutsching, E.K.; Ried, M.K.; Lipka, V.; Nürnberger, T.; Robatzek, S.; Parniske, M. Knowing your friends and foes—Plant receptor-like kinases as initiators of symbiosis or defence. New Phytol. 2014, 204, 791–802. [Google Scholar] [CrossRef]
- Prigigallo, M.I.; Križnik, M.; De Paola, D.; Catalano, D.; Gruden, K.; Finetti-Sialer, M.M.; Cillo, F. Potato Virus Y Infection Alters Small RNA Metabolism and Immune Response in Tomato. Viruses 2019, 11, 1100. [Google Scholar] [CrossRef] [Green Version]
- Stare, T.; Ramšak, Ž.; Križnik, M.; Gruden, K. Multiomics analysis of tolerant interaction of potato with potato virus Y. Sci. Data 2019, 6, 250. [Google Scholar] [CrossRef]
- Bukovinszki, Á.; Götz, R.; Johansen, E.; Maiss, E.; Balázs, E. The role of the coat protein region in symptom formation on Physalis floridana varies between PVY strains. Virus Res. 2007, 127, 122–125. [Google Scholar] [CrossRef]
- Rupar, M.; Florence, F.; Michel, T.; Ion, G.-A.; Agnès, D.; Laurent, G.; Maja, K.; David, D.; Kristina, G.; Emmanuel, J.; et al. Fluorescently Tagged Potato virus Y: A Versatile Tool for Functional Analysis of Plant-Virus Interactions. Mol. Plant Microbe Interact. 2015, 28, 739–750. [Google Scholar] [CrossRef] [PubMed]
- Cordero, T.; Mohamed, M.A.; López-Moya, J.-J.; Daròs, J.-A. A Recombinant Potato virus Y Infectious Clone Tagged with the Rosea1 Visual Marker (PVY-Ros1) Facilitates the Analysis of Viral Infectivity and Allows the Production of Large Amounts of Anthocyanins in Plants. Front. Microbiol. 2017, 8, 611. [Google Scholar] [CrossRef] [PubMed]
- Hilleary, R.; Choi, W.G.; Kim, S.H.; Lim, S.D.; Gilroy, S. Sense and sensibility: The use of fluorescent protein-based genetically encoded biosensors in plants. Curr. Opin. Plant Biol. 2018, 46, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Yin, K.; Gao, C.; Qiu, J.-L. Progress and prospects in plant genome editing. Nat. Plants 2017, 3, 17107. [Google Scholar] [CrossRef]
- Perraki, A.; Gronnier, J.; Gouguet, P.; Boudsocq, M.; Deroubaix, A.F.; Simon, V.; German-Retana, S.; Legrand, A.; Habenstein, B.; Zipfel, C.; et al. REM1.3′s phospho-status defines its plasma membrane nanodomain organization and activity in restricting PVX cell-to-cell movement. PLoS Pathog. 2018, 14, e1007378. [Google Scholar] [CrossRef]
- Da Silva, W.L.; Ingram, J.; Hackett, C.A.; Coombs, J.J.; Douches, D.; Bryan, G.J.; De Jong, W.; Gray, S. Mapping loci that control tuber and foliar symptoms caused by PVY in autotetraploid potato (Solanum tuberosum L.). G3 Genes Genomes Genet. 2017, 7, 3587–3595. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baebler, Š.; Coll, A.; Gruden, K. Plant Molecular Responses to Potato Virus Y: A Continuum of Outcomes from Sensitivity and Tolerance to Resistance. Viruses 2020, 12, 217. https://doi.org/10.3390/v12020217
Baebler Š, Coll A, Gruden K. Plant Molecular Responses to Potato Virus Y: A Continuum of Outcomes from Sensitivity and Tolerance to Resistance. Viruses. 2020; 12(2):217. https://doi.org/10.3390/v12020217
Chicago/Turabian StyleBaebler, Špela, Anna Coll, and Kristina Gruden. 2020. "Plant Molecular Responses to Potato Virus Y: A Continuum of Outcomes from Sensitivity and Tolerance to Resistance" Viruses 12, no. 2: 217. https://doi.org/10.3390/v12020217
APA StyleBaebler, Š., Coll, A., & Gruden, K. (2020). Plant Molecular Responses to Potato Virus Y: A Continuum of Outcomes from Sensitivity and Tolerance to Resistance. Viruses, 12(2), 217. https://doi.org/10.3390/v12020217





