Insights into the Functions of eIF4E-Binding Motif of VPg in Potato Virus A Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Plants and Agrobacterium
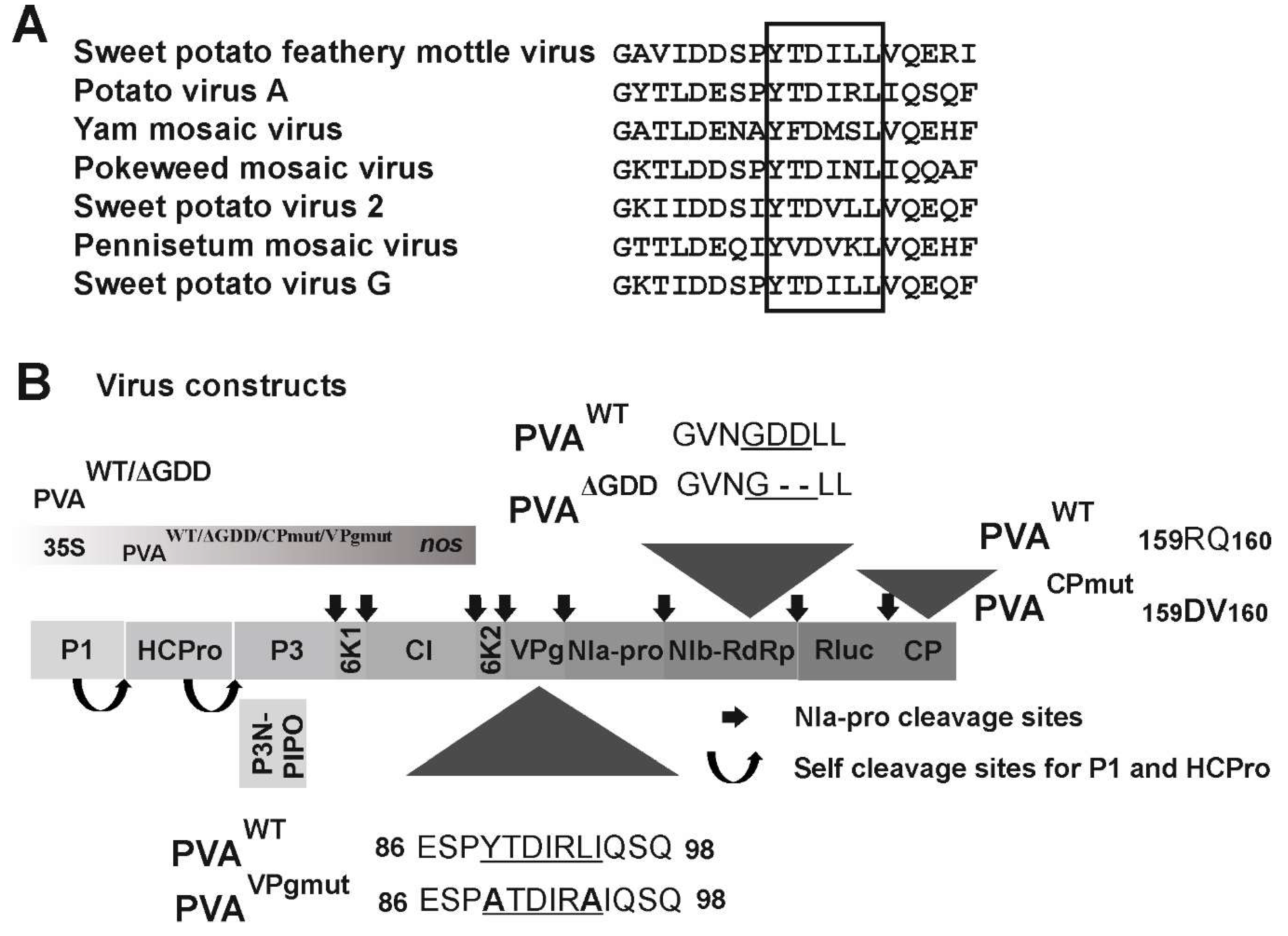
2.2. Viral and Protein Expression Constructs
2.3. Agrobacterium Infiltration and Sample Collection
2.4. Quantification of Viral Gene Expression by 3’RLUC Assay
2.5. Reverse Transcription-Quantitative PCR
2.6. Confocal Microscopy
2.7. Quantification of PGs by Epifluorescence Microscopy
2.8. Electron Microscopy (EM)
3. Results
3.1. The Central Region of PVA VPg Carries a Consensus Sequence for eIF(iso)4E Binding
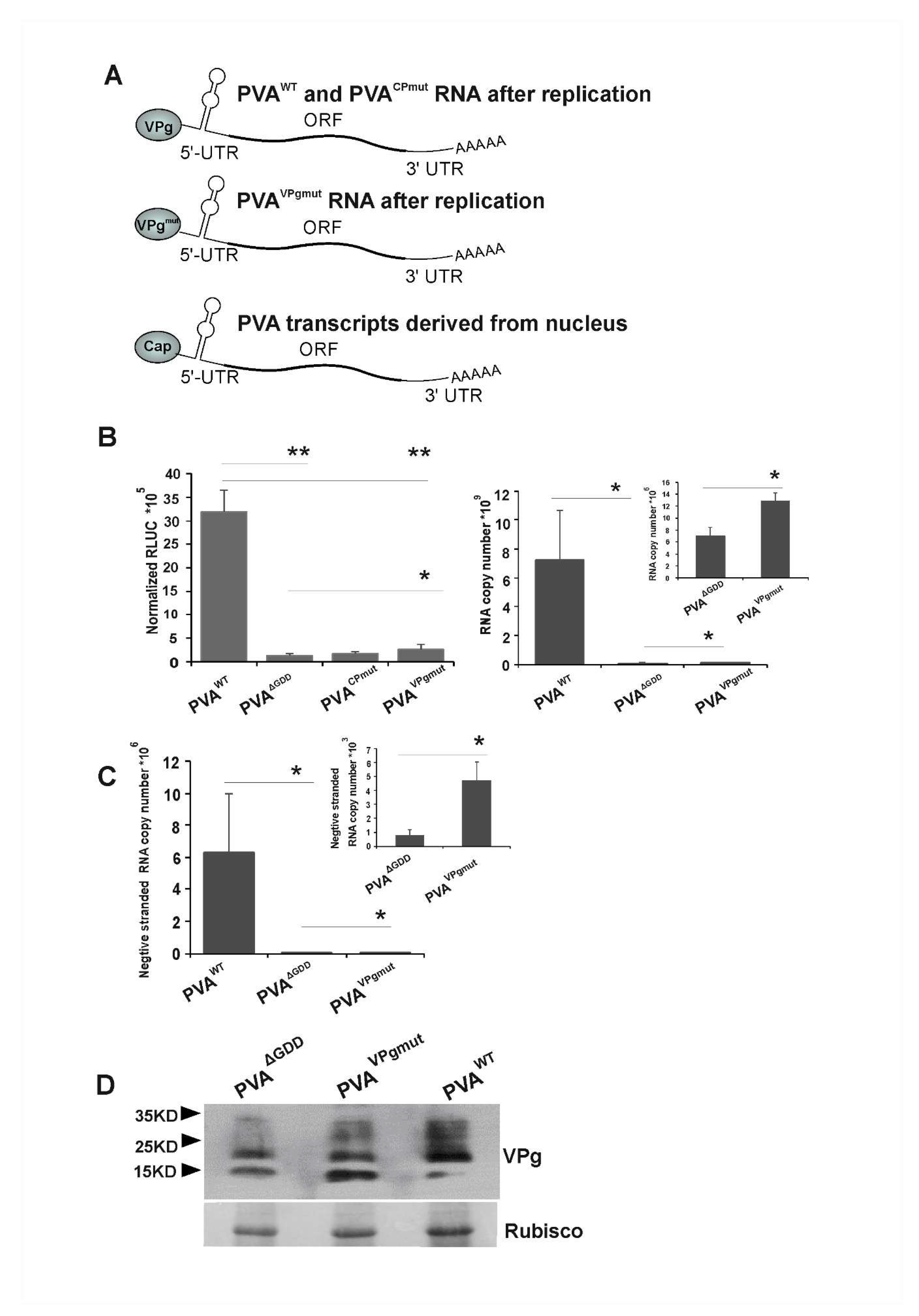
3.2. Low Level of Viral Replication Occurs in N. benthamiana Leaves Infiltrated with PVAVPgmut
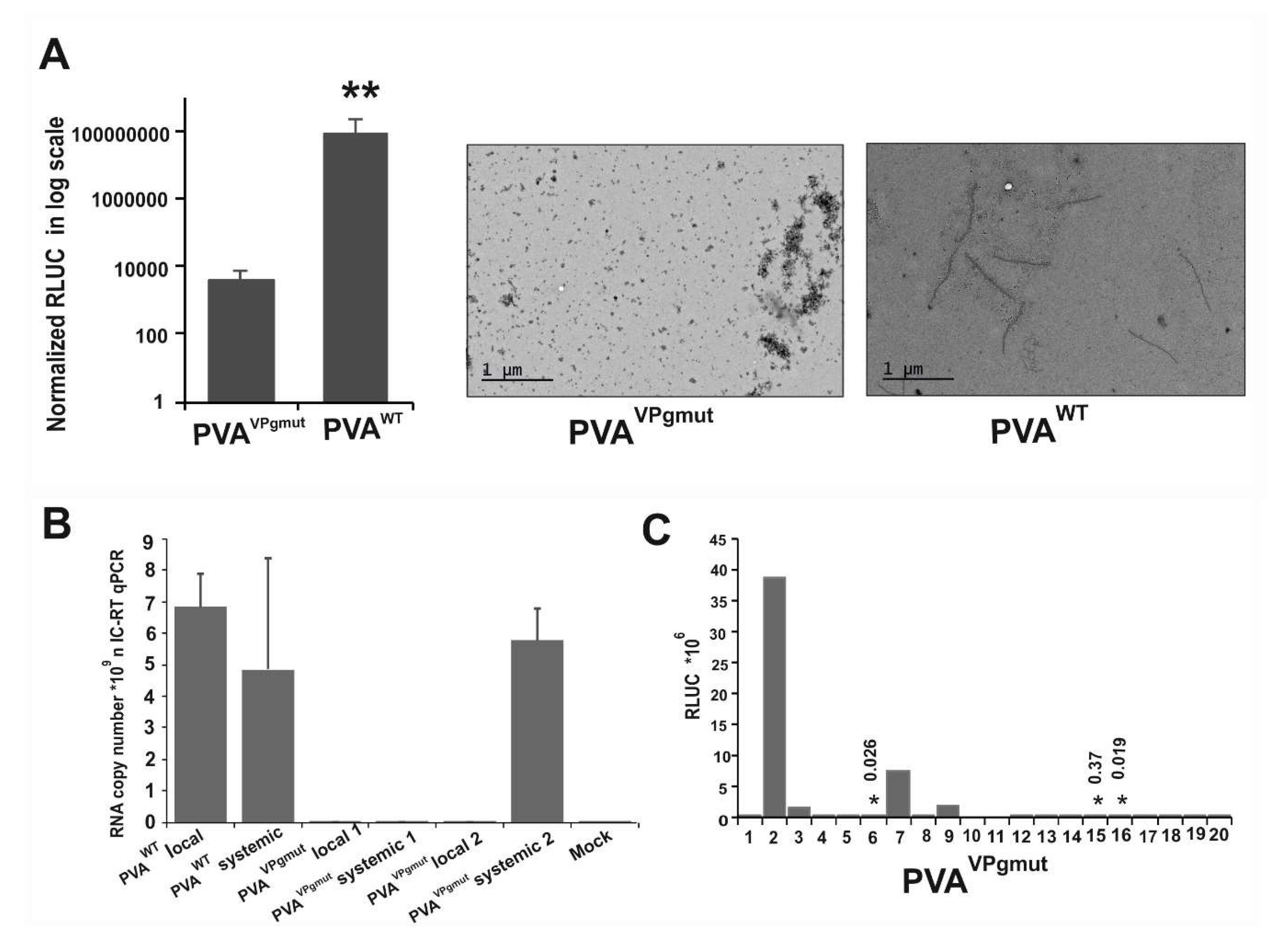
3.3. PVAVPgmut Fails to Move Systemically but Reverts Readily to PVAWT
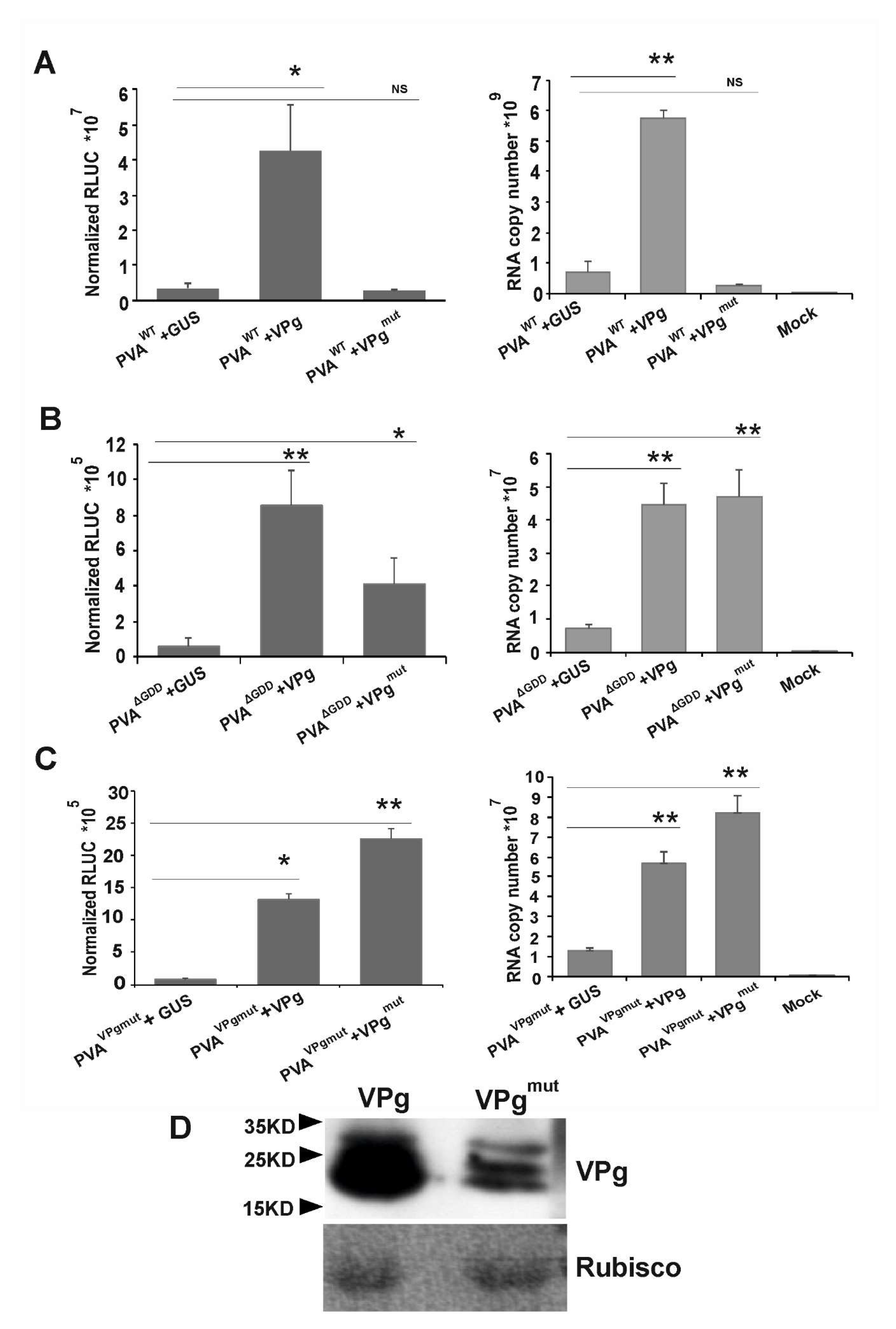
3.4. VPgmut is Not Able to Enhance PVAWT RNA Stability and 3’RLUC Expression
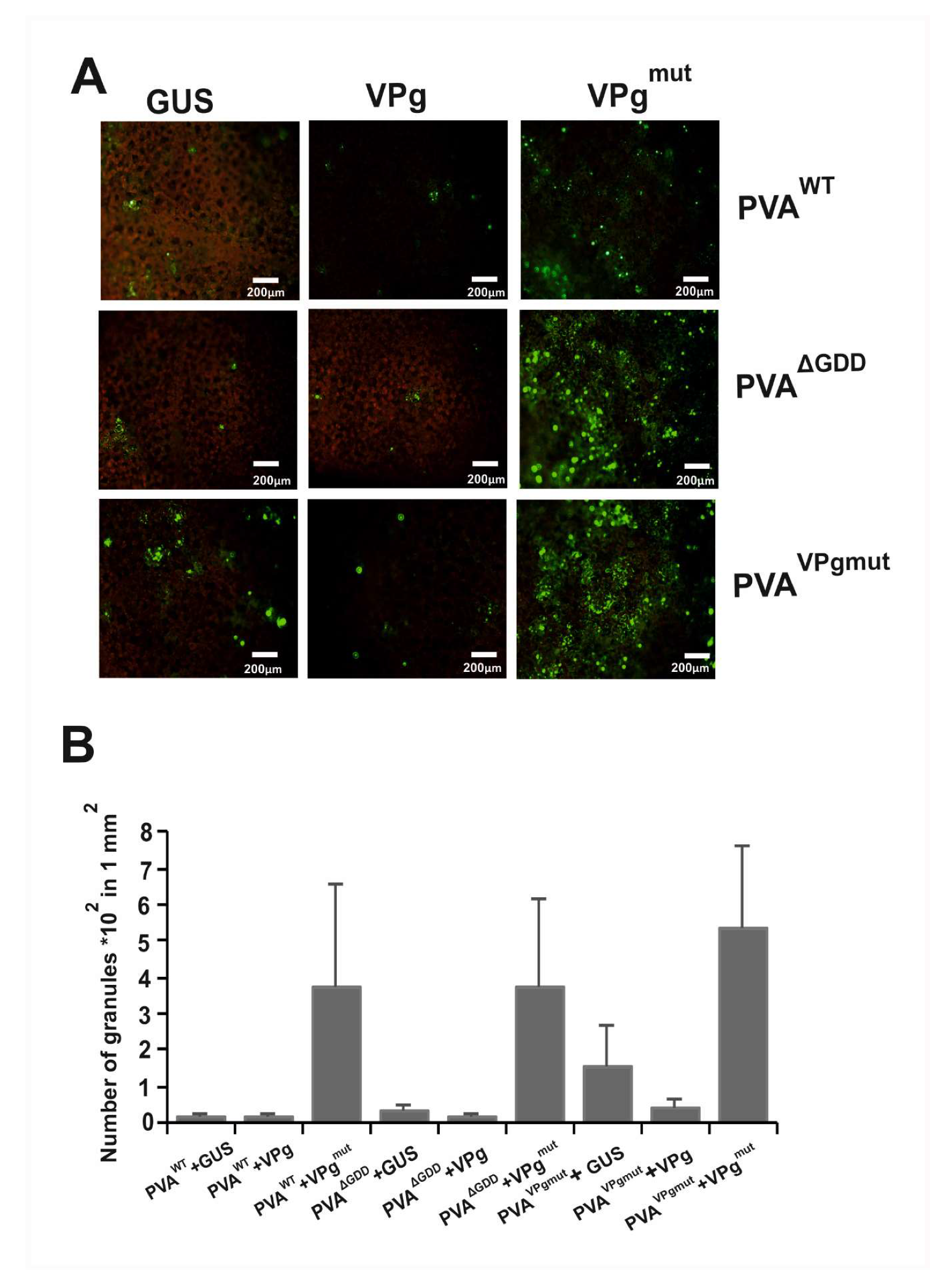
3.5. The Amount of PVA-Induced Granules Increases in the Presence of VPgmut
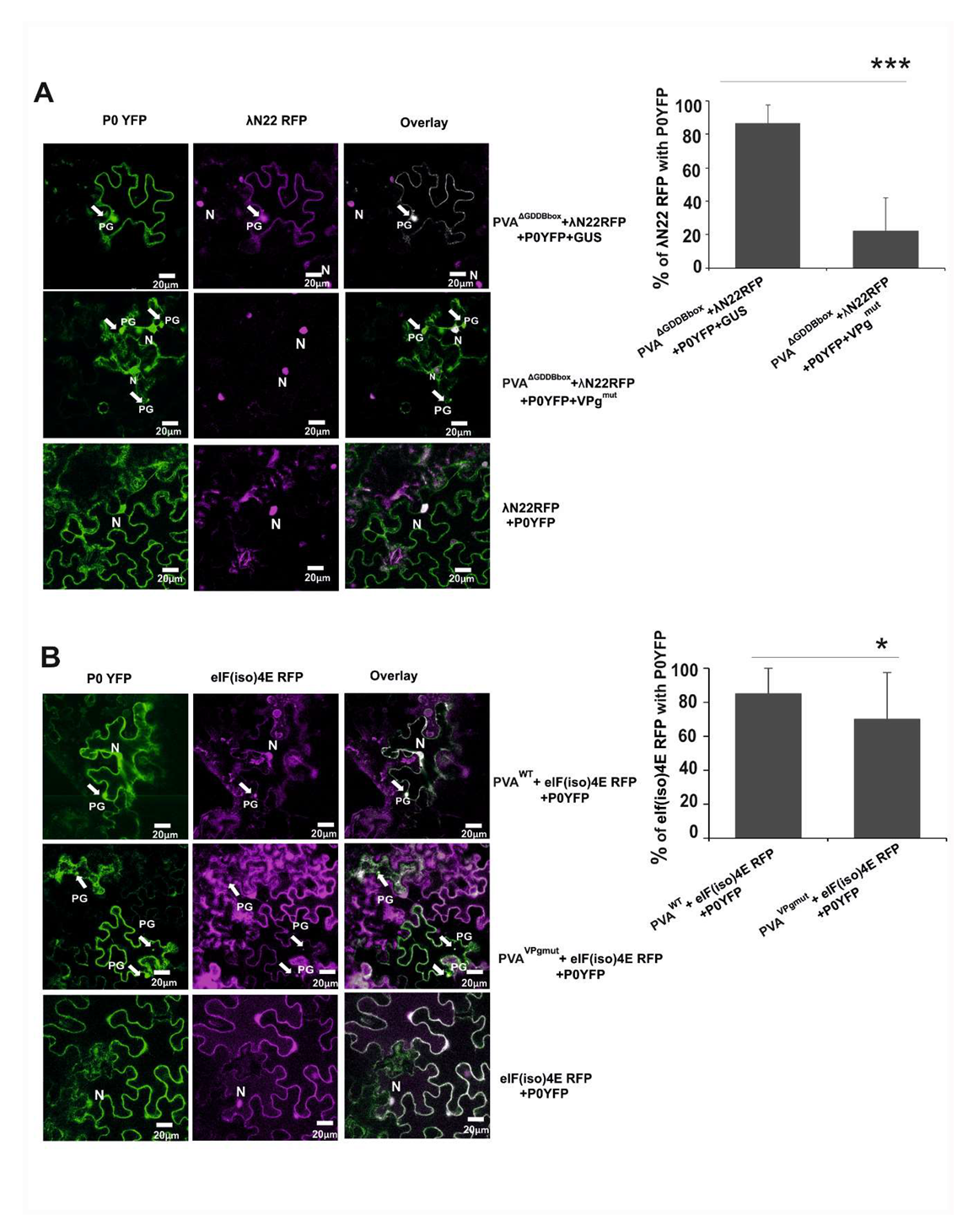
3.6. The Amount of Viral RNA is Reduced in P0-Contining Foci in the Presence of VPgmut
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Scholthof, K.B.; Adkins, S.; Czosnek, H.; Palukaitis, P.; Jacquot, E.; Hohn, T.; Hohn, B.; Saunders, K.; Candresse, T.; Ahlquist, P.; et al. Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 2011, 12, 938–954. [Google Scholar] [CrossRef] [PubMed]
- Topisirovic, I.; Sonenberg, N. Translational control by the eukaryotic ribosome. Cell 2011, 145, 333–334. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Browning, K.S. Plant translation initiation factors: It is not easy to be green. Biochem. Soc. Trans. 2004, 32, 589–591. [Google Scholar] [CrossRef]
- Gingras, A.C.; Raught, B.; Sonenberg, N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999, 68, 913–963. [Google Scholar] [CrossRef] [PubMed]
- Rajamäki, M.L.; Valkonen, J.P. The 6K2 protein and the VPg of potato virus A are determinants of systemic infection in Nicandra physaloides. Mol. Plant Microbe Interact. 1999, 12, 1074–1081. [Google Scholar] [CrossRef] [PubMed]
- Rajamäki, M.L.; Valkonen, J.P. Viral genome-linked protein (VPg) controls accumulation and phloem-loading of a potyvirus in inoculated potato leaves. Mol. Plant Microbe Interact. 2002, 15, 138–149. [Google Scholar] [CrossRef]
- Léonard, S.; Plante, D.; Wittmann, S.; Daigneault, N.; Fortin, M.G.; Laliberté, J.F. Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J. Virol. 2000, 74, 7730–7737. [Google Scholar] [CrossRef]
- Borgstrøm, B.; Johansen, I.E. Mutations in pea seedborne mosaic virus genome-linked protein VPg after pathotype-specific virulence in Pisum sativum. Mol. Plant Microbe Interact. 2001, 14, 707–714. [Google Scholar] [CrossRef]
- Moury, B.; Morel, C.; Johansen, E.; Guilbaud, L.; Souche, S.; Ayme, V.; Caranta, C.; Palloix, A.; Jacquemond, M. Mutations in potato virus Y genome-linked protein determine virulence toward recessive resistances in Capsicum annuum and Lycopersicon hirsutum. Mol. Plant Microbe Interact. 2004, 17, 322–329. [Google Scholar] [CrossRef]
- Gallois, J.L.; Charron, C.; Sánchez, F.; Pagny, G.; Houvenaghel, M.C.; Moretti, A.; Ponz, F.; Revers, F.; Caranta, C.; German-Retana, S. Single amino acid changes in the turnip mosaic virus viral genome-linked protein (VPg) confer virulence towards Arabidopsis thaliana mutants knocked out for eukaryotic initiation factors eIF(iso)4E and eIF(iso)4G. J. Gen. Virol. 2010, 91, 288–289. [Google Scholar] [CrossRef]
- Perez, K.; Yeam, I.; Kang, B.C.; Ripoll, D.R.; Kim, J.; Murphy, J.F.; Jahn, M.M. Tobacco etch virus infectivity in Capsicum spp. is determined by a maximum of three amino acids in the viral virulence determinant VPg. Mol. Plant Microbe Interact. 2012, 25, 1562–1573. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Krishnaswamy, S. Eukaryotic translation initiation factor 4E-mediated recessive resistance to plant viruses and its utility in crop improvement. Mol. Plant Pathol. 2012, 13, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Robaglia, C.; Caranta, C. Translation initiation factors: A weak link in plant RNA virus infection. Trends Plant Sci. 2006, 11, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Truniger, V.; Aranda, M.A. Recessive resistance to plant viruses. Adv. Virus Res. 2009, 75, 119–159. [Google Scholar]
- Okade, H.; Fujita, Y.; Miyamoto, S.; Tomoo, K.; Muto, S.; Miyoshi, H.; Natsuaki, T.; Rhoads, R.E.; Ishida, T. Turnip mosaic virus genome-linked protein VPg binds C-terminal region of cap-bound initiation factor 4E orthologue without exhibiting host cellular specificity. J. Biochem. 2009, 145, 299–307. [Google Scholar] [CrossRef]
- Sato, M.; Nakahara, K.; Yoshii, M.; Ishikawa, M.; Uyeda, I. Selective involvement of members of the eukaryotic initiation factor 4E family in the infection of Arabidopsis thaliana by potyviruses. FEBS Lett. 2005, 579, 1167–1171. [Google Scholar] [CrossRef]
- Nicaise, V.; Gallois, J.L.; Chafiai, F.; Allen, L.M.; Schurdi-Levraud, V.; Browning, K.S.; Candresse, T.; Caranta, C.; Le Gall, O.; German-Retana, S. Coordinated and selective recruitment of eIF4E and eIF4G factors for potyvirus infection in Arabidopsis thaliana. FEBS Lett. 2007, 581, 1041–1046. [Google Scholar] [CrossRef]
- Duprat, A.; Caranta, C.; Revers, F.; Menand, B.; Browning, K.S.; Robaglia, C. The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J. 2002, 32, 927–934. [Google Scholar] [CrossRef]
- Nicaise, V.; German-Retana, S.; Sanjuán, R.; Dubrana, M.P.; Mazier, M.; Maisonneuve, B.; Candresse, T.; Caranta, C.; Le Gall, O. The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the Potyvirus Lettuce mosaic virus. Plant Physiol. 2003, 132, 1272–1282. [Google Scholar] [CrossRef]
- Jenner, C.E.; Nellist, C.F.; Barker, G.C.; Walsh, J.A. Turnip mosaic virus (TuMV) is able to use alleles of both eIF4E and eIF(iso)4E from multiple loci of the diploid Brassica rapa. Mol. Plant Microbe Interact. 2010, 23, 1498–1505. [Google Scholar] [CrossRef]
- Sanfaçon, H. Plant Translation Factors and Virus Resistance. Viruses 2015, 7, 3392–3419. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, M.; Neriya, Y.; Yamaji, Y.; Namba, S. Recessive Resistance to Plant Viruses: Potential Resistance Genes Beyond Translation Initiation Factors. Front. Microbiol. 2016, 7, 1695. [Google Scholar] [CrossRef] [PubMed]
- Shopan, J.; Mou, H.; Zhang, L.; Zhang, C.; Ma, W.; Walsh, J.A.; Hu, Z.; Yang., J.; Zhang, M. Eukaryotic translation initiation factor 2B-beta (eIF2Bβ), a new class of plant virus resistance gene. Plant J. 2017, 90, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Charron, C.; Nicolaï, M.; Gallois, J.L.; Robaglia, C.; Moury, B.; Palloix, A.; Caranta, C. Natural variation and functional analyses provide evidence for co-evolution between plant eIF4E and potyviral VPg. Plant J. 2008, 54, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Nicaise, V. Crop immunity against viruses: outcomes and future challenges. Front. Plant Sci. 2014, 5, 660. [Google Scholar] [CrossRef] [PubMed]
- Moury, B.; Charron, C.; Janzac, B.; Simon, V.; Gallois, J.L.; Palloix, A.; Caranta, C. Evolution of plant eukaryotic initiation factor 4E (eIF4E) and potyvirus genome-linked protein (VPg): a game of mirrors impacting resistance spectrum and durability. Infect. Genet. Evol. 2014, 27, 472–480. [Google Scholar] [CrossRef]
- Monzingo, A.F.; Dhaliwal, S.; Dutt-Chaudhuri, A.; Lyon, A.; Sadow, J.H.; Hoffman, D.W.; Robertus, J.D.; Browning, K.S. The structure of eukaryotic translation initiation factor-4E from wheat reveals a novel disulfide bond. Plant. Physiol. 2007, 143, 1504–1518. [Google Scholar] [CrossRef]
- Walter, J.; Charon, J.; Hu, Y.; Lachat, J.; Leger, T.; Lafforgue, G.; Barra, A.; Michon, T. Comparative analysis of mutational robustness of the intrinsically disordered viral protein VPg and of its interactor eIF4E. PLoS ONE 2019, 14, e0211725. [Google Scholar] [CrossRef]
- Bruun-Rasmussen, M.; Møller, I.S.; Tulinius, G.; Hansen, J.K.; Lund, O.S.; Johansen, I.E. The same allele of translation initiation factor 4E mediates resistance against two Potyvirus spp. in Pisum sativum. Mol. Plant Microbe Interact. 2007, 20, 1075–1082. [Google Scholar] [CrossRef]
- Coutinho de Oliveira, L.; Volpon, L.; Rahardjo, A.K.; Osborne, M.J.; Culjkovic-Kraljacic, B.; Trahan, C.; Oeffinger, M.; Kwok, B.H.; Borden, K.L.B. Structural studies of the eIF4E-VPg complex reveal a direct competition for capped RNA: Implications for translation. Proc. Natl. Acad. Sci. USA 2019, 116, 24056–24065. [Google Scholar] [CrossRef]
- Puurand, U.; Valkonen, J.P.; Mäkinen, K.; Rabenstein, F.; Saarma, M. Infectious in vitro transcripts from cloned cDNA of the potato A potyvirus. Virus Res. 1996, 40, 135–140. [Google Scholar] [CrossRef]
- Michon, T.; Estevez, Y.; Walter, J.; German-Retana, S.; Le Gall, O. The potyviral virus genome-linked protein VPg forms a ternary complex with the eukaryotic initiation factors eIF4E and eIF4G and reduces eIF4E affinity for a mRNA cap analogue. FEBS J. 2006, 273, 1312–1322. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Miyoshi, H.; Gallie, D.R.; Goss, D.J. Potyvirus genome-linked protein, VPg, directly affects wheat germ in vitro translation: interactions with translation initiation factors eIF4F and eIFiso4F. J. Biol. Chem. 2008, 283, 1340–1349. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, H.; Okade, H.; Muto, S.; Suehiro, N.; Nakashima, H.; Tomoo, K.; Natsuaki, T. Turnip mosaic virus VPg interacts with Arabidopsis thaliana eIF(iso)4E and inhibits in vitro translation. Biochimie 2008, 90, 1427–1434. [Google Scholar] [CrossRef]
- Eskelin, K.; Hafrén, A.; Rantalainen, K.I.; Mäkinen, K. Potyviral VPg enhances viral RNA Translation and inhibits reporter mRNA translation in planta. J. Virol. 2011, 85, 9210–9221. [Google Scholar] [CrossRef] [PubMed]
- Eskelin, K.; Suntio, T.; Hyvärinen, S.; Hafren, A.; Mäkinen, K. Renilla luciferase-based quantitation of Potato virus A infection initiated with Agrobacterium infiltration of N. benthamiana leaves. J. Virol. Methods 2010, 164, 101–110. [Google Scholar] [CrossRef]
- Hafrén, A.; Lõhmus, A.; Mäkinen, K. Formation of Potato Virus A-Induced RNA Granules and Viral Translation Are Interrelated Processes Required for Optimal Virus Accumulation. PLoS Pathog. 2015, 11, e1005314. [Google Scholar] [CrossRef]
- Hafrén, A.; Hofius, D. NBR1-mediated antiviral xenophagy in plant immunity. Autophagy 2017, 13, 2000–2001. [Google Scholar] [CrossRef]
- Tavert-Roudet, G.; Anne, A.; Barra, A.; Chovin, A.; Demaille, C.; Michon, T. The Potyvirus Particle Recruits the Plant Translation Initiation Factor eIF4E by Means of the VPg covalently Linked to the Viral RNA. Mol. Plant Microbe Interact. 2017, 30, 754–762. [Google Scholar] [CrossRef]
- Lellis, A.D.; Kasschau, K.D.; Whitham, S.A.; Carrington, J.C. Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr. Biol. 2002, 12, 1046–1051. [Google Scholar] [CrossRef]
- Contreras-Paredes, C.A.; Silva-Rosales, L.; Daròs, J.A.; Alejandri-Ramírez, N.D.; Dinkova, T.D. The absence of eukaryotic initiation factor eIF(iso)4E affects the systemic spread of a Tobacco etch virus isolate in Arabidopsis thaliana. Mol. Plant Microbe Interact. 2013, 26, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Marcotrigiano, J.; Gingras, A.C.; Sonenberg, N.; Burley, S.K. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell 1999, 3, 707–716. [Google Scholar] [CrossRef]
- Ala-Poikela, M. Coordinated functions of the HCpro and VPg proteins of Potato virus A (PVA) with the translation initiation factors eIF4E and eIF(iso)4E. PhD Thesis, University of Helsinki, Helsinki, Finland, 17 June 2014. [Google Scholar]
- Ala-Poikela, M.; Goytia, E.; Haikonen, T.; Rajamäki, M.L.; Valkonen, J.P.T. Helper component proteinase of the genus Potyvirus is an interaction partner of translation initiation factors eIF(iso)4E and eIF4E and contains a 4E binding motif. J. Virol. 2011, 85, 6784–6794. [Google Scholar] [CrossRef] [PubMed]
- Ala-Poikela, M.; Rajamäki, M.L.; Valkonen, J.P.T. A Novel Interaction Network Used by Potyviruses in Virus-Host Interactions at the Protein Level. Viruses 2019, 11, 1158. [Google Scholar] [CrossRef]
- Puurand, U.; Mäkinen, K.; Paulin, L.; Saarma, M. The nucleotide sequence of potato virus A genomic RNA and its sequence similarities with other potyviruses. J. Gen. Virol. 1994, 75, 457–461. [Google Scholar] [CrossRef]
- Hafrén, A.; Eskelin, K.; Mäkinen, K. Ribosomal protein P0 promotes Potato virus A infection and functions in viral translation together with VPg and eIF(iso)4E. J. Virol. 2013, 87, 4302–4312. [Google Scholar] [CrossRef]
- Liu, D.; Shi, L.; Han, C.; Yu, J.; Li, D.; Zhang, Y. Validation of reference genes for gene expression studies in virus infected Nicotiana benthamiana using quantitative real-time PCR. PLoS ONE 2012, 7, e46451. [Google Scholar] [CrossRef]
- Rhoads, R.E. eIF4E: New family members, new binding partners, new roles. J. Biol. Chem. 2009, 284, 16711–16715. [Google Scholar] [CrossRef]
- Puustinen, P.; Mäkinen, K. Uridylylation of the potyvirus VPg by viral replicase NIb correlates with the nucleotide binding capacity of VPg. J. Biol. Chem. 2004, 279, 38103–38110. [Google Scholar] [CrossRef]
- Oruetxebarria, I.; Guo, D.; Merits, A.; Mäkinen, K.; Saarma, M.; Valkonen, J.P.T. Identification of the genome-linked protein in virions of Potato virus A, with comparison to other members in genus Potyvirus. Virus Res. 2001, 73, 103–112. [Google Scholar] [CrossRef]
- Valli, A.; Gallo, A.; Calvo, M.; de Jesús Pérez, J.; García, J.A. A novel role of the potyviral helper component proteinase contributes to enhance the yield of viral particles. J. Virol. 2014, 17, 9808–9818. [Google Scholar] [CrossRef]
- Gallo, A.; Valli, A.; Calvo, M.; García, J.A. A functional link between RNA replication and virion assembly in the Potyvirus. J. Virol. 2018, 92, e02179-17. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Hafren, A.; Mäkinen, K. Dynamics of Protein Accumulation from the 3’ End of Viral RNA are different from those in the rest of the genome in Potato Virus A infection. J. Virol. 2019, 12, 00721-19. [Google Scholar] [CrossRef] [PubMed]
- Schönberger, J.; Hammes, U.Z.; Dresselhaus, T. In vivo visualization of RNA in plants cells using the λN₂₂ system and a GATEWAY-compatible vector series for candidate RNAs. Plant J. 2012, 71, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Anindya, R.; Chittori, S.; Savithri, H.S. Tyrosine 66 of Pepper vein banding virus genome-linked protein is uridylylated by RNA-dependent RNA polymerase. Virology 2005, 336, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.V.; Wimmer, E. Initiation of protein-primed picornavirus RNA synthesis. Virus Res. 2015, 206, 12–26. [Google Scholar] [CrossRef] [PubMed]
- Pumplin, N.; Voinnet, O. RNA silencing suppression by plant pathogens: defense, counter-defense and counter-counter-defense. Nat. Rev. Microbiol. 2013, 11, 745–760. [Google Scholar] [CrossRef]
- Ivanov, K.I.; Eskelin, K.; Bašić, M.; De, S.; Lõhmus, A.; Varjosalo, M.; Mäkinen, K. Molecular insights into the function of the viral RNA silencing suppressor HC-pro. Plant J. 2016, 85, 30–45. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Roudet-Tavert, G.; Michon, T.; Walter, J.; Delaunay, T.; Redondo, E.; Le Gall, O. Central domain of a potyvirus VPg is involved in the interaction with the host translation initiation factor eIF4E and the viral protein HcPro. J. Gen. Virol. 2007, 88, 1029–1033. [Google Scholar] [CrossRef]
- Tavert-Roudet, G.; Abdul-Razzak, A.; Doublet, B.; Walter, J.; Delaunay, T.; German-Retana, S.; Michon, T.; Le Gall, O.; Candresse, T. The C terminus of lettuce mosaic potyvirus cylindrical inclusion helicase interacts with the viral VPg and with lettuce translation eukaryotic initiation factor 4E. J. Gen. Virol. 2012, 93, 184–193. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saha, S.; Mäkinen, K. Insights into the Functions of eIF4E-Binding Motif of VPg in Potato Virus A Infection. Viruses 2020, 12, 197. https://doi.org/10.3390/v12020197
Saha S, Mäkinen K. Insights into the Functions of eIF4E-Binding Motif of VPg in Potato Virus A Infection. Viruses. 2020; 12(2):197. https://doi.org/10.3390/v12020197
Chicago/Turabian StyleSaha, Shreya, and Kristiina Mäkinen. 2020. "Insights into the Functions of eIF4E-Binding Motif of VPg in Potato Virus A Infection" Viruses 12, no. 2: 197. https://doi.org/10.3390/v12020197
APA StyleSaha, S., & Mäkinen, K. (2020). Insights into the Functions of eIF4E-Binding Motif of VPg in Potato Virus A Infection. Viruses, 12(2), 197. https://doi.org/10.3390/v12020197




