Abstract
DEAD-box helicases are a large family of conserved RNA-binding proteins that belong to the broader group of cellular DExD/H helicases. Members of the DEAD-box helicase family have roles throughout cellular RNA metabolism from biogenesis to decay. Moreover, there is emerging evidence that cellular RNA helicases, including DEAD-box helicases, play roles in the recognition of foreign nucleic acids and the modulation of viral infection. As intracellular parasites, viruses must evade detection by innate immune sensing mechanisms and degradation by cellular machinery while also manipulating host cell processes to facilitate replication. The ability of DEAD-box helicases to recognize RNA in a sequence-independent manner, as well as the breadth of cellular functions carried out by members of this family, lead them to influence innate recognition and viral infections in multiple ways. Indeed, DEAD-box helicases have been shown to contribute to intracellular immune sensing, act as antiviral effectors, and even to be coopted by viruses to promote their replication. However, our understanding of the mechanisms underlying these interactions, as well as the cellular roles of DEAD-box helicases themselves, is limited in many cases. We will discuss the diverse roles that members of the DEAD-box helicase family play during viral infections.
1. The DEAD-box Helicase Family of RNA-Binding Proteins
The broad family of DExD/H helicases, including DEAD-box helicases and the related DEAH-box helicases, belongs to superfamily 2 of the RNA helicase family. This family is characterized by a series of amino acid motifs that form the RNA and ATP binding sites of the helicase core [1,2]; DEAD-box helicases contain the amino acid sequence DEAD in motif II as well as an additional upstream Q motif [3]. DEAD-box helicases are conserved from bacteria to mammals [4]. Structurally, the DEAD-box helicase core contains two domains resembling bacterial RecA, which cooperatively bind RNA and ATP, interacting with RNA along the sugar-phosphate backbone [4]. Because of the lack of interactions with the RNA nucleotide bases, the binding of DEAD-box helicases to RNA is thought to be generally sequence-independent but structure-dependent [5,6].
In vitro studies of DEAD-box helicase unwinding activity have revealed ATP-dependent RNA helicase function, but these proteins tend to have only weakly processive helicase activity, and instead are thought to promote local RNA:RNA rearrangements [7]. Beyond their activity on RNA duplexes, studies have also revealed that some members of this family have other activities regulating nucleic acid binding, including the modulation of RNA:DNA interactions, DNA:DNA interactions, and RNA-protein (RNP) complex remodeling [8,9].
Although DEAD-box helicases share a conserved core domain, variable N- and C-terminal regions allow members of this protein family to act on diverse targets. These flanking domains target the helicases to particular RNAs or proteins; the diversity of these regions accounts for the breadth of function observed within this protein family [7]. As a result of their diversity of binding and function, DEAD-box RNA helicases are involved in all aspects of cellular RNA metabolism, including transcription, splicing, microRNA biogenesis, ribosomal RNA processing, RNA export, translation, RNP granule formation, and decay [10]. Moreover, they are also involved in cellular stress responses, contributing to double-strand break (DSB) repair [11], stress granule formation, and antimicrobial responses [12].
2. DEAD-Box Helicases in Canonical Innate Immune Sensing
Innate immune defenses depend upon the recognition of invading pathogens by the host, and viral infections, including RNA viruses, are largely sensed through nucleic acid recognition. RNA viruses present a host cell with various foreign RNA features, such as unique ends, structured RNA elements, or dsRNA replication intermediates, which can be recognized as hallmarks of infection. RNA binding proteins thus play important roles in the sensing of viral infection, and DExD/H helicases are well-suited to recognize structural features characteristic of viral RNAs.
The canonical cytoplasmic sensors of viral RNA, the RIG-I-like receptors (RLRs) RIG-I and MDA-5, contain DExD/H-box helicase domains [13]. RIG-I recognizes uncapped viral RNAsand short dsRNAs containing a 5′ triphosphate, while MDA-5 binds longer dsRNA replication intermediates [13,14,15]. In addition to the helicase core domain, these proteins contain caspase activation and recruitment domains (CARDs).RNA recognition by these RLRs leads to oligomerization of CARDs and activation of antiviral signaling complexes [16,17]. RIG-I and MDA5 both signal through the CARD-domain-containing adapter protein MAVS, causing the polymerization and activation of MAVS on the mitochondrial membrane [14]. This leads to the activation of IRF3 and IRF7, promoting the transcription of IFN-β [14]. Type I interferons subsequently induce the transcription of hundreds of interferon-stimulated genes (ISGs) that control infection, including a number of RNA helicases (Figure 1).
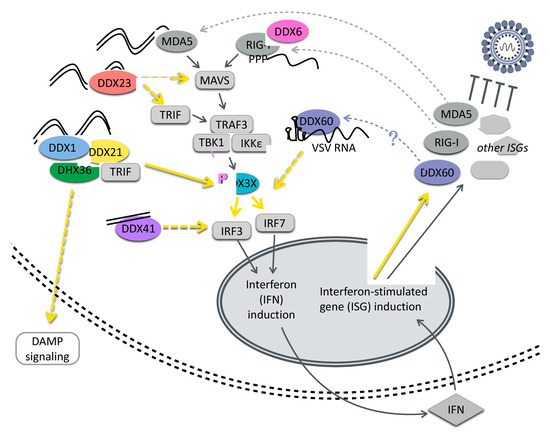
Figure 1.
DEAD-box helicases contribute to innate immune signaling. DEAD-box helicases have been demonstrated to serve as sensors for nucleic acids, including dsRNA, cytoplasmic DNA, and viral RNAs, leading to the induction of interferon and interferon-stimulated genes. DEAD-box helicases also contribute to innate immune signaling downstream of nucleic acid sensing by mediating protein–protein interactions and promoting DAMP signaling. Moreover, some DEAD-box helicases are ISGs, suggesting positive feedback on their innate immune roles. RIG-I and MDA-5, while considered members of the RIG-I-like receptors (RLR) family, contain related DExD/H-box RNA binding domains. DEAD-box helicases are shown in color and other cellular components are shown in gray. Dotted yellow lines are used where there may be intermediate steps in the indicated relationship. (DAMP, damage-associated molecular pattern; IFN, interferon; ISG, interferon-stimulated gene; TLR, toll-like receptor; VSV, vesicular stomatitis virus).
DEAD-box helicases have been shown to contribute to cytosolic viral RNA recognition and to feed into these canonical immune signaling pathways, sometimes acting in concert with DEAH-box helicases. Several DEAD-box helicases promote interferon induction or other inflammatory signaling. DDX1 contributes to IFN-β activation during transmissible gastroenteritis virus infection via its interaction with the nonstructural protein nsP14 [18] and interacts with RelA potentiating NF-κB-dependent transcription, a function which requires intact helicase activity [19]. DDX21 is antiviral against dengue virus, promoting IFN-β induction during infection; in turn, the dengue virus ns2B/3 protein complex directs the proteasomal degradation of DDX21 to counteract this signaling [20]. A complex of the helicases DDX1, DDX21, and DHX36 interacts with the adaptor protein TRIF and cytosolic dsRNA, leading to a TLR3-independent activation of IRF3/7 and interferon induction, thus suggesting an additional role for TRIF beyond its known functions in toll-like receptor (TLR) signaling and endosomal sensing [21]. DDX21 has been further implicated in inflammatory signaling via the production of endogenous damage-associated molecular patterns (DAMPs). During influenza virus infection, DDX21 and TRIF are required for the secretion of the TLR DAMP ligand S100A9, which serves as a DAMP signal and results in the rapid production of inflammatory cytokines, including TNF and IL-6 [22].
Cytosolic DNA recognition represents another important surveillance mechanism for stimulating innate immune pathways in response to invasion by DNA viruses and bacteria, as the cytoplasm has a dearth of DNA under normal conditions. Much of this signaling is now known to occur via the cGAS-STING pathway, where DNA recognition by cGAS leads to the production of the cyclic dinucleotide cGAMP, which signals through STING, leading to phosphorylation of IRF3 by TBK1 [23,24]. However, DEAD-box helicases can also contribute to dsDNA sensing: DDX41 senses dsDNA from diverse intracellular pathogens, including bacteria and DNA viruses, and upregulates interferon and cytokine responses [25]. DDX41 has been shown to bind DNA via the walker A and B motifs of its helicase domain, to interact with STING, and to be required for the activation of TBK1 [25].
While the innate immune roles of some DEAD-box proteins involve nucleic acid sensing, other innate immune functions involve protein–protein interactions with DEAD-box-helicases and are nucleic acid independent. DDX3X may serve as a scaffold for protein–protein interactions to promote the transduction of innate immune signaling cascades. The binding and phosphorylation of DDX3X by TBK1 facilitates IFN-β induction in response to diverse stimuli including poly(I:C), poly(dA:dT), and Listeria monocytogenes infection [26]. DDX3X also interacts with IKKε to mediate the phosphorylation of IRF3, and with IKKα to support its activation downstream of TLR7 [27,28].
Some DEAD-box helicases are themselves ISGs, further reinforcing their antiviral activity via positive feedback. RIG-I, MDA5, DDX60, and DDX60L are canonical ISGs [29,30,31]. DDX60 and DDX60L share approximately 70% amino acid identity and both have antiviral functions; however, their roles are distinct [29,31]. DDX60 binds vesicular stomatitis virus (VSV) ssRNA and dsRNA in vitro and promotes RLR activation [31]. DDX60 also contributes to RIG-I-independent degradation of HCV RNA, although its role in this process is unclear [32]. Activation of the EGF growth factor receptor attenuates DDX60 activity, so viruses known to activate EGF including influenza A virus, HCV, and VSV may be able to counteract DDX60 functions [32]. A study contrasting the responses of hepatocyte cell lines to IFN-γ uncovered the importance of DDX60L expression for controlling HCV infection, and ectopic expression of DDX60L was also found to restrict HCV infection independent of interferon signaling, suggesting an additional direct effector mechanism [29].
2.1. Negative Regulation of Innate Immune Responses by DEAD-Box Helicases and Viral Antagonism
Some DEAD-box helicase family members are negative regulators of interferon responses. This can occur via competition for RNA substrates or protein–protein interactions. For example, DDX24 binds dsRNA and ssRNA and sequesters it away from RLRs, thus interfering with IRF7 activation [33].
In the virus–host arms race, viruses have evolved diverse countermeasures to evade detection by the host and evade host immune responses. The existence of such countermeasures targeting DEAD-box helicase proteins further underscores these proteins’ importance as antiviral factors. A variety of viruses hijack or target DDX3X to promote infection. The binding of DDX3X by the vaccinia virus K7 protein interferes with DDX3X-dependent TBK1/IKKε activation [34]. Moreover, DDX3X binding to hepatitis B virus (HBV) polymerase also interferes with DDX3X-dependent TBK1/IKKε activation early in infection, while at later stages the helicase function of DDX3X is required to limit HBV transcription [35,36]. Hepatitis C virus (HCV) takes advantage of the immune modulatory function of DDX3X to support its own replication: the binding of DDX3X to the HCV 3′UTR activates IKKα-dependent noncanonical NF-κB transcription, promoting lipogenesis, which aids in viral replication [37]. However, DDX3X negatively regulates type I interferon production during arenavirus infection, and interactions between DDX3X and arenavirus nucleoprotein promote viral replication [38].
The relocalization of nuclear DDX21 to the cytoplasm during dengue virus infection contributes to interferon responses; this is consistent with previous reports of DDX21 involvement in innate immune sensing [20,21,22]. In turn, DENV NS2B/3 protease counteracts DDX21, leading to its proteasome-dependent degradation [20].
2.2. Conserved Interferon-Independent Antiviral Functions of DEAD-Box Helicases
DEAD-box helicases can also contribute to antiviral effector functions independent of interferon signaling (Figure 2) [39]. Studies in invertebrates and plants have allowed the identification of several antiviral helicases which are necessarily interferon-independent, as these taxa lack an interferon system. Moreover, many of these antiviral functions are conserved in mammals. The screening of DEAD-box helicases in a Drosophila cell culture model of arthropod-borne RNA virus infection has revealed antiviral functions for several DEAD-box helicases that also have human homologs, including DDX6, DDX17, DDX24, and DDX56 [40,41].
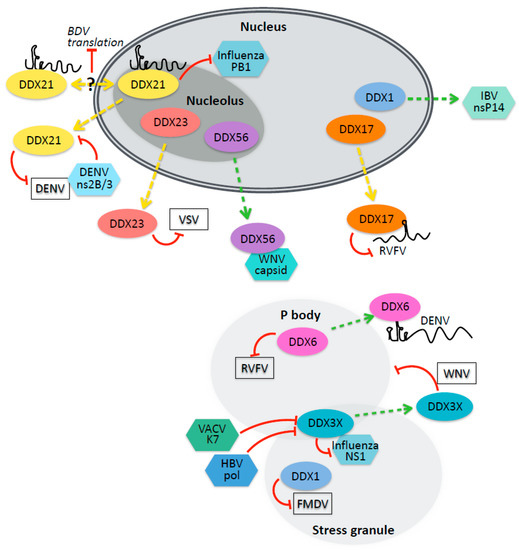
Figure 2.
DEAD-box helicases with diverse cellular roles and localization interact with viral components and influence infection. DEAD-box helicases residing in nuclear, p-body, and stress granule compartments, have been implicated in viral infection. Interactions involve viral proteins, viral RNAs, or remain undefined. DEAD-box helicases can have positive or negative effects on infection. In some cases, viruses have been shown to counteract antiviral helicases, to alter the localization of helicases, or to disrupt RNA-protein (RNP) complexes. The red lines indicate negative interactions; the green arrows indicate positive interactions; and the dotted lines indicate protein relocalization. (VSV, vesicular stomatitis virus; DENV, dengue virus; RVFV, Rift Valley fever virus; WNV, West Nile virus; BDV, Borna disease virus; IBV, infections bronchitis virus, FMDV, foot and mouth disease virus; HBV, hepatitis B virus; VACV, vaccinia virus).
DDX17 and DDX5 are nuclear resident paralogs that have diverse functions in transcription, splicing, miRNA biogenesis, mRNA export, and ribosome biogenesis [42]. DDX17 but not DDX5 was found to be antiviral against Rift Valley Fever Virus (RVFV). Moreover, DDX17 but not DDX5 relocalized to the cytoplasm during RVFV infection [41]. DDX17 activity against RVFV depends on the binding of a stem-loop structure onto the small segment of the tripartite RNA genome, and the addition of that stem-loop region to an unrelated virus rendered the chimeric virus sensitive to DDX17′s antiviral effect [41]. DDX17 may also have an antiviral function as a cofactor for the zinc-finger antiviral protein (ZAP), unwinding ZAP-bound viral mRNA to promote viral RNA decay by the RNA exosome [43,44]. The Arabidopsis homolog of DDX17, RH30, has also been shown to be antiviral against tombus viruses such as tomato bushy stunt virus and cucumber necrosis virus, both in plant hosts and a yeast surrogate host model [45]. RH30 relocalizes from the nucleus to the sites of viral replication and interacts with both viral proteins and viral RNA, particularly with structured cis-acting elements, thus interfering with template recruitment to the replicase complex [45]. In contrast to the antiviral roles of DDX17, knockdown of its paralog DDX5 attenuates replication of Japanese encephalitis virus (JEV), and helicase activity of DDX5 is required for this function [46]. DDX5 is recruited to the replication sites of JEV in the cytoplasm, and binds to the viral 3′ UTR to promote infection [46]. DDX3 is also recruited to JEV replication foci and directly promotes JEV replication by binding the untranslated regions [47].
DDX23 is a nuclear resident helicase with known roles in splicing. Studies in the invertebrate chordate amphioxus identified DDX23 as a dsRNA binding protein, and found that dsRNA binding was conserved in human cells [48]. How DDX23 controls viral infection in amphioxus is unclear, and whether DDX23 binding directly to viral RNA also impacts infection has not been explored. In humans, DDX23 binding to dsRNA potentiates antiviral signaling downstream of TRIF and MAVS [48]. Moreover, after dsRNA treatment or VSV infection, DDX23 translocates from the nucleus to the cytoplasm, suggesting that the relocalization of DDXs may be a common strategy to repurpose these RNA binding proteins for antiviral defense [48].
3. Cell Biological Interfaces of DEAD-Box Helicase Activity and Viral Infections
3.1. Nucleolar Helicases and Viral Infection
The nucleolus is the site of ribosomal RNA (rRNA) transcription, rRNA biogenesis, and assembly of ribosomal subunits. Nucleoli form as sites of concentrated processing factors around clusters of rDNA genes, and liquid–liquid phase separation drives the organization of subcompartments in which rRNA is cleaved and modified [49,50]. Many DEAD-box helicases localize to the nucleolus and participate in ribosome biogenesis; their precise functions are generally poorly understood, but they are thought to be involved in the remodeling of pre-ribosomal RNP complexes to promote rRNA biogenesis and in the release of snoRNAs after rRNA modification [51].
There is a growing appreciation for the role of the nucleolus in cellular stress responses, including responses to viral infections [49,52]. Known roles for nucleolar proteins in viral infection can involve interactions with RNA or proteins of nuclear-replicating viruses, the relocalization of nucleolar proteins to other cellular compartments, or targeting by viral proteins that localize to the nucleolus. For example, the nucleolar helicase DDX56, which normally functions in rRNA biogenesis [53], promotes West Nile virus infection [54,55,56]. The West Nile virus capsid biochemically interacts with DDX56 during infection, both in nucleoli and in the cytoplasm, and DDX56 helicase activity is required to promote the packaging of viral RNA into particles [54,55,56]. In the case of HIV-1, a DDX56–Gag interaction facilitates HIV-1 particle assembly, suggesting a broader role for DDX56 in viral assembly [57].
DDX21 is another nucleolar helicase with cellular functions in ribosomal RNA production [58] that also has a variety of moonlighting functions. DDX21 has been shown to be antiviral against both nuclear- and cytoplasmic-replicating RNA viruses through a variety of mechanisms. In the nucleus, DDX21 interacts with the Borna disease virus (BDV) RNA, binding the 5′ UTR of the BDV X/P mRNA and decreasing its translation [59]. In vitro RNA folding assays suggested that DDX21 binding causes structural alterations in the 5′UTR of the viral mRNA, thus interfering with the reinitiation of translation by ribosomes on this polycistronic message [59]. Influenza also replicates in the nucleus, but the activity of DDX21 against influenza appears not to require nucleic acid binding [60]; instead, a protein–protein interaction between DDX21 and the influenza PB1 polymerase inhibits the assembly of the viral replicase complex until being disrupted later in infection by increasing levels of the influenza NS1 protein [60]. During infection with DENV, DDX21 relocates to the cytoplasm and promotes interferon responses, as described above [20].
Taken together, these examples paint a picture where nucleolar proteins are influenced by, and exert effects on, cytoplasmic processes.
3.2. P-Body and Stress Granule Helicases in Viral Infection
P bodies are nonmembranous organelles in the cytosol that contribute to the regulation of cellular RNAs—resident RNAs can be stored, translationally repressed, and/or degraded [61]. Many viruses interface with P bodies in diverse ways (Reviewed in [62]). DDX6 resides in P bodies and facilitates P body assembly, as well as promoting the decapping and turnover of cellular mRNAs [63]. DDX6 also has cellular roles in the regulation of gene expression via translational repression [64].
DDX6 has been shown to have pro- or anti-viral roles in a variety of viral infections, including several arthropod-borne viruses. DDX6 enhances RIG-I signaling, interacting with both RIG-I and influenza viral RNA in infected cells [65]. However, DDX6 was also identified in a screen for suppressors of aberrant ISG activity; the deletion of DDX6 activates innate immune signaling pathways and primes cells to be more responsive to IFN, likely via the disruption of cytoplasmic RNA turnover and the accumulation of self RNA in P bodies [66]. Bunyaviruses snatch mRNA caps from host transcripts in P bodies, and thus cellular factors such as DDX6, which promote the decapping of host mRNAs and thereby deplete the pool of capped RNAs in P bodies, attenuate bunyaviral infection including La Crosse virus (LACV) and RVFV [40]. In mosquitoes, DDX6 is antiviral against the flaviviruses West Nile virus and Zika virus and is counteracted by the noncoding sfRNA derived from the flavivirus 3′UTR, which binds to DDX6 and sequesters it [67]. Human DDX6 is similarly antiviral against ZIKV and is sequestered by sfRNA [68].
Viruses can also hijack P-body components to aid in replication. DDX6 and other P body components have been found by mass spectrometry to bind the dengue virus 3′ UTR RNA, but in constrast to other flaviviruses, DENV infection is attenuated by DDX6 knockdown [69]. West Nile virus also subverts P body function by disrupting P bodies and recruiting component proteins including LSM1, GW182, DDX3, and XRN1 to viral replication sites where they positively contribute to replication by an unknown mechanism [70].
Stress granules are another type of nonmembranous organelle which form when translation is impaired [71]. The stress granule resident helicase DDX3X has diverse roles in interferon signaling (described in other sections of this review) as well as in other aspects of cell biology. DDX3X is antiviral against influenza A virus infection via its role in nucleating stress granules: the C-terminal domain interacts with influenza NS1 to sequester the viral protein in stress granules and limit the amount available to carry out viral replication [72]. It is also possible that many of the other roles of DDX3X in interferon signaling and viral replication are taking place in stress granules; however, this has not been explored. In plants and yeast, the helicases RH20/Ded1p (DDX3) are coopted to form part of the tombus virus replicase complex, and their presence helps to maintain full-length genome integrity and prevent recombination [73]. DDX3 was also shown to contribute to export of unspliced HIV mRNA, supporting HIV replication [74].
DDX1 is another stress granule resident protein [12] and has been found to inhibit or facilitate diverse viral infections including HIV, foot and mouth disease virus (FMDV), and transmissible gastroenteritis virus [18,71,75,76]. DDX1 was identified in a two-hybrid screen for HIV-Rev interactors and was shown to modulate the localization of Rev and the splicing of HIV mRNA. [77]. DDX1 acts as a chaperone to remodel the structure of the Rev-responsive element (RRE) RNA sequence, promoting increased Rev binding [75,78] and acting as a cofactor for Rev oligomerization [79]. The RRE can adopt several distinct structural conformations with differing affinities for Rev and functional consequences for HIV replication [80]. Another yeast two-hybrid screen led to the discovery that DDX1 supports the replication of the coronavirus infectious bronchitis virus (IBV), relocalizing to viral replication centers and interacting with the viral nonstructural protein nsP14; the ability of nsP14 to interact with DDX1 is conserved in severe acute respiratory syndrome coronavirus (SARS-CoV) [81]. Moreover, in a yeast model of Tombus virus infection, Ded1 and Dbp2 enhance viral replication by unwinding 3′ secondary structure to promote plus-strand RNA synthesis [82]. DDX1 inhibits foot and mouth disease virus and increases IFNβ production in infected cells [76]. DDX1 acts as a coactivator of NF-κB-mediated transcription via interaction with RelA, a function which requires intact helicase activity [19].
These interactions highlight the ways that both viruses and hosts can use RNP interactions to promote or antagonize one another’s function (Figure 2). The ability of some viruses to use DEAD-box helicase remodeling functions on their own RNA suggest that there may be other settings in which DEAD-box-helicase remodeling of viral RNA structure is detrimental to replication.
4. Summary and Discussion
The anti-viral and pro-viral DEAD-box helicase relationships that have been described so far in the literature likely represent only the tip of the iceberg in terms of the multitude of roles that DEAD-box helicases play in innate immunity and the modulation of viral infection (Table 1). These relationships were discovered through diverse approaches and have revealed interactions of viral infection with many aspects of RNA biology. Several examples have been identified through mass spectrometry approaches, arising either from studies investigating binding of viral proteins to host factors [35,60,83,84] or to nucleic acids [21,69,85]. Interactome studies have provided additional evidence for the importance of DEAD-box helicases in HIV and other viral infections [77,79,84,85,86,87]. Biochemical studies of HIV-Rev, alone or Rev in combination with RRE RNA, have also identified positive roles for DDX3X, DDX5, DDX17, and DDX21 in HIV infection, although some of these interactions could be indirect [84]. Other DEAD-box helicases have been found genetically by screening to identify genes that affect infection [41,88]. The genetic screening of DEAD-box helicases in poxvirus infection revealed both positive and negative regulation of infection by DEAD-box helicases [88]. And still other studies have focused on changes in gene expression that indicate either cellular innate immune responses or a viral manipulation of the host cell environment [31,89,90]. DDX3 was found to be upregulated in response to HIV-Tat expression, and contributes to the export of unspliced HIV mRNA, supporting HIV replication [74]. Microarray studies of HIV latency and reactivation found that DDX18 and DDX39 are upregulated during HIV latency and early reactivation and that the expression of additional DEAD-box helicases DDX10, DDX21, DDX23, and DDX52 is induced immediately following reactivation [89]. The biochemical characterization of these interactions provides crucial mechanistic insight into how these proteins impact infection. Confirming and defining protein–protein interactions with methods such as coimmunoprecipitation and defining regions of RNA binding by CLIP-seq have elucidated the mechanism of action of some of these proteins during viral infection.

Table 1.
DEAD-box helicase relationships with viral infections. Interactions are categorized as pro-viral if they support viral replication, and they are categorized as anti-viral if they counteract viral infection either directly or by activating innate immune signaling. Viral countermeasures are listed if a viral component disrupts an otherwise antiviral interaction. Other categories of interactions including gene expression changes are listed in the otherwise implicated column.
Although much remains to be defined about the antiviral roles of the DEAD-box helicase family, some broad themes have emerged. First, the functions of DEAD-box helicases in viral infection appear to often, but not always, rely on direct interactions between the host DEAD-box protein and viral RNAs. Some DEAD/DEAH-box helicases, such as the RLRs RIG-I and MDA-5, are specialized innate immune sensors, and others, such as DDX3X, have cellular roles but also contribute to innate immune signaling pathways. Still others bind viral RNA as antiviral effectors, whether induced by interferon signaling in the case of DDX60L or independently of known innate immune pathways, as is the case for DDX17 and DDX56. Continuing to biochemically define the RNA features recognized by antiviral helicases and the ways those RNA structures may be altered by helicase activity could help us identify functionally-important RNA structures in viral replication. Additionally, in recent years, the application of RNA probing technologies such as SHAPE to viral RNAs is beginning to enable detailed analysis of secondary and tertiary structures that may be functionally relevant [30,91,92]. A comparison of SHAPE data to DEAD-box helicase binding sites may enhance our understanding of structural motifs that stimulate innate immunity.
Second, DEAD-box helicase/virus interactions are highly context-dependent: different viruses are affected in different ways by the same DEAD-box helicases, and although RNA binding is important in many cases, there also exist examples where helicase function is dispensable for the antiviral activities that have been characterized. Detailed mechanistic studies naturally lag far behind the high-throughput genomic and proteomic identification of virus-host interactions, but a deeper investigation of the virus and host determinants underlying antiviral DEAD-box helicase function may illuminate key characteristics governing the outcome of a virus-helicase interaction.
Third, our understanding of the antiviral function of DEAD-box helicases is limited by a lack of information about their cellular roles. DEAD-box helicases are essential for many aspects of RNA biogenesis, but in many cases the details of their roles in these pathways have yet to be fully elucidated, and specific targets, activities, and cofactors are not well defined. Our limited knowledge of DEAD-box helicase function in both contexts hampers our ability to understand determinants governing RNA substrate selection and helicase activity and certainly our ability to predict whether or how they might interact with viral infection. We speculate that, in many cases, the viral substrates of these proteins might resemble their endogenous targets, whether through commonalities in target RNA secondary structure or in interactions with the variable N- and C-terminal domains that disrupt or coopt their cellular roles. As further examples and details of DEAD-box helicases in viral infection are characterized, complementary studies of their normal cellular functions will be necessary to determine how their interactions with viral infection resemble or differ from their normal biology. Ultimately, a better understanding of the virus–host interactions that affect the outcome of infection could lead to better antiviral therapeutics. Virus research has a long history of providing insight into cellular function, and these mechanisms will also teach us about cellular RNA regulation in broader contexts. This is an exciting time to explore the interface between viruses and cellular helicases.
Author Contributions
Writing Reviewing & Editing, S.C. and F.T. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the National Institutes of Health to S.C. (R01AI074951, R01AI122749, R01AI140539) and F.T. (T32GM07229). S.C. is a recipient of the Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Award.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Linder, P.; Lasko, P.F.; Ashburner, M.; Leroy, P.; Nielsen, P.J.; Nishi, K.; Schnier, J.; Slonimski, P.P. Birth of the d-e-a-d box. Nature 1989, 337, 121–122. [Google Scholar] [CrossRef] [PubMed]
- Tanner, N.K.; Linder, P. DExD/H box RNA helicases. Mol. Cell 2001, 8, 251–262. [Google Scholar] [CrossRef]
- Tanner, N.K.; Cordin, O.; Banroques, J.; Doère, M.; Linder, P. The Q motif: A newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell 2003, 11, 127–138. [Google Scholar] [CrossRef]
- Cordin, O.; Banroques, J.; Tanner, N.K.; Linder, P. The dead-box protein family of rna helicases. Gene 2006, 367, 17–37. [Google Scholar] [CrossRef] [PubMed]
- Sengoku, T.; Nureki, O.; Nakamura, A.; Kobayashi, S.; Yokoyama, S. Structural basis for rna unwinding by the dead-box protein drosophila vasa. Cell 2006, 125, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Schutz, P.; Karlberg, T.; van den Berg, S.; Collins, R.; Lehtio, L.; Hogbom, M.; Holmberg-Schiavone, L.; Tempel, W.; Park, H.W.; Hammarstrom, M.; et al. Comparative structural analysis of human dead-box RNA helicases. PLoS ONE 2010, 5, e12791. [Google Scholar] [CrossRef]
- Hilbert, M.; Karow, A.R.; Klostermeier, D. The mechanism of atp-dependent RNA unwinding by dead box proteins. Biol. Chem. 2009, 390, 1237–1250. [Google Scholar] [CrossRef]
- Talwar, T.; Vidhyasagar, V.; Qing, J.; Guo, M.; Kariem, A.; Lu, Y.; Singh, R.S.; Lukong, K.E.; Wu, Y. The dead-box protein ddx43 (hage) is a dual rna-DNA helicase and has a k-homology domain required for full nucleic acid unwinding activity. J. Biol. Chem. 2017, 292, 10429–10443. [Google Scholar] [CrossRef]
- Fairman, M.E.; Maroney, P.A.; Wang, W.; Bowers, H.A.; Gollnick, P.; Nilsen, T.W.; Jankowsky, E. Protein displacement by dexh/d “RNA helicases” without duplex unwinding. Science 2004, 304, 730–734. [Google Scholar] [CrossRef]
- Linder, P.; Jankowsky, E. From unwinding to clamping—The dead box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011, 12, 505–516. [Google Scholar] [CrossRef]
- Li, L.; Monckton, E.A.; Godbout, R. A role for dead box 1 at DNA double-strand breaks. Mol. Cell. Biol. 2008, 28, 6413–6425. [Google Scholar] [CrossRef]
- Shih, J.W.; Lee, Y.H. Human DExD/H RNA helicases: Emerging roles in stress survival regulation. Clin. Chim. Acta 2014, 436, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Takeuchi, O.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Matsui, K.; Uematsu, S.; Jung, A.; Kawai, T.; Ishii, K.J.; et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006, 441, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Loo, Y.M.; Gale, M., Jr. Immune signaling by RIG-I-like receptors. Immunity 2011, 34, 680–692. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Takeuchi, O.; Mikamo-Satoh, E.; Hirai, R.; Kawai, T.; Matsushita, K.; Hiiragi, A.; Dermody, T.S.; Fujita, T.; Akira, S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008, 205, 1601–1610. [Google Scholar] [CrossRef]
- Berke, I.C.; Modis, Y. MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA. EMBO J. 2012, 31, 1714–1726. [Google Scholar] [CrossRef]
- Peisley, A.; Wu, B.; Yao, H.; Walz, T.; Hur, S. RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol. Cell 2013, 51, 573–583. [Google Scholar] [CrossRef]
- Zhou, Y.; Wu, W.; Xie, L.; Wang, D.; Ke, Q.; Hou, Z.; Wu, X.; Fang, Y.; Chen, H.; Xiao, S.; et al. Cellular RNA helicase DDX1 is involved in transmissible gastroenteritis virus NSP 14-induced interferon-β production. Front. Immunol. 2017, 8, 940. [Google Scholar] [CrossRef]
- Ishaq, M.; Ma, L.; Wu, X.; Mu, Y.; Pan, J.; Hu, J.; Hu, T.; Fu, Q.; Guo, D. The dead-box RNA helicase DDX1 interacts with rela and enhances nuclear factor kappab-mediated transcription. J. Cell. Biochem. 2009, 106, 296–305. [Google Scholar] [CrossRef]
- Dong, Y.; Ye, W.; Yang, J.; Han, P.; Wang, Y.; Ye, C.; Weng, D.; Zhang, F.; Xu, Z.; Lei, Y. Ddx21 translocates from nucleus to cytoplasm and stimulates the innate immune response due to dengue virus infection. Biochem. Biophys. Res. Commun. 2016, 473, 648–653. [Google Scholar] [CrossRef]
- Zhang, Z.; Kim, T.; Bao, M.; Facchinetti, V.; Jung, S.Y.; Ghaffari, A.A.; Qin, J.; Cheng, G.; Liu, Y.J. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsrna in dendritic cells. Immunity 2011, 34, 866–878. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.Y.; Segovia, J.A.; Chang, T.H.; Morris, I.R.; Berton, M.T.; Tessier, P.A.; Tardif, M.R.; Cesaro, A.; Bose, S. DAMP molecule S100A9 acts as a molecular pattern to enhance inflammation during influenza A virus infection: Role of DDX21-TRIF-TLR4-MyD88 pathway. PLoS Pathog. 2014, 10, e1003848. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013, 339, 786–791. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Sun, L.; Chen, X.; Du, F.; Shi, H.; Chen, C.; Chen, Z.J. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 2013, 339, 826–830. [Google Scholar] [CrossRef]
- Zhang, Z.; Yuan, B.; Bao, M.; Lu, N.; Kim, T.; Liu, Y.J. The helicase DDX41 senses intracellular DNA mediated by the adaptor sting in dendritic cells. Nat. Immunol. 2011, 12, 959–965. [Google Scholar] [CrossRef]
- Soulat, D.; Burckstummer, T.; Westermayer, S.; Goncalves, A.; Bauch, A.; Stefanovic, A.; Hantschel, O.; Bennett, K.L.; Decker, T.; Superti-Furga, G. The dead-box helicase DDX3x is a critical component of the tank-binding kinase 1-dependent innate immune response. EMBO J. 2008, 27, 2135–2146. [Google Scholar] [CrossRef]
- Fullam, A.; Gu, L.; Hohn, Y.; Schroder, M. DDX3 directly facilitates ikkalpha activation and regulates downstream signalling pathways. Biochem. J. 2018, 475, 3595–3607. [Google Scholar] [CrossRef]
- Gu, L.; Fullam, A.; Brennan, R.; Schroder, M. Human dead box helicase 3 couples ikappab kinase epsilon to interferon regulatory factor 3 activation. Mol. Cell. Biol. 2013, 33, 2004–2015. [Google Scholar] [CrossRef]
- Grunvogel, O.; Esser-Nobis, K.; Reustle, A.; Schult, P.; Muller, B.; Metz, P.; Trippler, M.; Windisch, M.P.; Frese, M.; Binder, M.; et al. DDX60l is an interferon-stimulated gene product restricting hepatitis C virus replication in cell culture. J. Virol. 2015, 89, 10548–10568. [Google Scholar] [CrossRef]
- Merino, E.J.; Wilkinson, K.A.; Coughlan, J.L.; Weeks, K.M. RNA structure analysis at single nucleotide resolution by selective 2’-hydroxyl acylation and primer extension (shape). J. Am. Chem. Soc. 2005, 127, 4223–4231. [Google Scholar] [CrossRef]
- Miyashita, M.; Oshiumi, H.; Matsumoto, M.; Seya, T. DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Mol. Cell. Biol. 2011, 31, 3802–3819. [Google Scholar] [CrossRef] [PubMed]
- Oshiumi, H.; Miyashita, M.; Okamoto, M.; Morioka, Y.; Okabe, M.; Matsumoto, M.; Seya, T. DDX60 is involved in RIG-I-dependent and independent antiviral responses, and its function is attenuated by virus-induced egfr activation. Cell Rep. 2015, 11, 1193–1207. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Moore, R.; Xu, X.; Barber, G.N. DDX24 negatively regulates cytosolic RNA-mediated innate immune signaling. PLoS Pathog. 2013, 9, e1003721. [Google Scholar] [CrossRef] [PubMed]
- Schroder, M.; Baran, M.; Bowie, A.G. Viral targeting of dead box protein 3 reveals its role in TBK1/ikkepsilon-mediated IRF activation. EMBO J. 2008, 27, 2147–2157. [Google Scholar] [CrossRef]
- Wang, H.; Kim, S.; Ryu, W.S. DDX3 dead-box RNA helicase inhibits hepatitis B virus reverse transcription by incorporation into nucleocapsids. J. Virol. 2009, 83, 5815–5824. [Google Scholar] [CrossRef]
- Wang, H.; Ryu, W.S. Hepatitis B virus polymerase blocks pattern recognition receptor signaling via interaction with DDX3: Implications for immune evasion. PLoS Pathog. 2010, 6, e1000986. [Google Scholar] [CrossRef]
- Li, Q.; Pene, V.; Krishnamurthy, S.; Cha, H.; Liang, T.J. Hepatitis C virus infection activates an innate pathway involving ikk-alpha in lipogenesis and viral assembly. Nat. Med. 2013, 19, 722–729. [Google Scholar] [CrossRef]
- Loureiro, M.E.; Zorzetto-Fernandes, A.L.; Radoshitzky, S.; Chi, X.; Dallari, S.; Marooki, N.; Leger, P.; Foscaldi, S.; Harjono, V.; Sharma, S.; et al. DDX3 suppresses type I interferons and favors viral replication during arenavirus infection. PLoS Pathog. 2018, 14, e1007125. [Google Scholar] [CrossRef]
- Ahmad, S.; Hur, S. Helicases in antiviral immunity: Dual properties as sensors and effectors. Trends Biochem. Sci. 2015, 40, 576–585. [Google Scholar] [CrossRef]
- Hopkins, K.C.; McLane, L.M.; Maqbool, T.; Panda, D.; Gordesky-Gold, B.; Cherry, S. A genome-wide RNAi screen reveals that mrna decapping restricts bunyaviral replication by limiting the pools of DCP2-accessible targets for CAP-snatching. Genes Dev. 2013, 27, 1511–1525. [Google Scholar] [CrossRef]
- Moy, R.H.; Cole, B.S.; Yasunaga, A.; Gold, B.; Shankarling, G.; Varble, A.; Molleston, J.M.; tenOever, B.R.; Lynch, K.W.; Cherry, S. Stem-loop recognition by DDX17 facilitates miRNA processing and antiviral defense. Cell 2014, 158, 764–777. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Chen, G.; Jia, H.; He, X.; Jing, Z. DDX5 RNA helicases: Emerging roles in viral infection. Int. J. Mol. Sci. 2018, 19, 1122. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Guo, X.; Lv, F.; Xu, Y.; Gao, G. P72 dead box RNA helicase is required for optimal function of the zinc-finger antiviral protein. Proc. Natl. Acad Sci. USA 2008, 105, 4352–4357. [Google Scholar] [CrossRef]
- Guo, X.; Ma, J.; Sun, J.; Gao, G. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc. Natl. Acad Sci. USA 2007, 104, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.Y.; Nagy, P.D. Blocking tombusvirus replication through the antiviral functions of DDX17-like RH30 dead-box helicase. PLoS Pathog. 2019, 15, e1007771. [Google Scholar] [CrossRef]
- Li, C.; Ge, L.L.; Li, P.P.; Wang, Y.; Sun, M.X.; Huang, L.; Ishag, H.; Di, D.D.; Shen, Z.Q.; Fan, W.X.; et al. The DEAD-box RNA helicase DDX5 acts as a positive regulator of Japanese encephalitis virus replication by binding to viral 3’ UTR. Antiviral Res. 2013, 100, 487–499. [Google Scholar] [CrossRef]
- Li, C.; Ge, L.-L.; Li, P.-P.; Wang, Y.; Dai, J.-J.; Sun, M.-X.; Huang, L.; Shen, Z.-Q.; Hu, X.-C.; Ishag, H.; et al. Cellular DDX3 regulates Japanese encephalitis virus replication by interacting with viral un-translated regions. Virology 2014, 449, 70–81. [Google Scholar] [CrossRef]
- Ruan, J.; Cao, Y.; Ling, T.; Li, P.; Wu, S.; Peng, D.; Wang, Y.; Jia, X.; Chen, S.; Xu, A.; et al. DDX23, an evolutionary conserved dsRNA sensor, participates in innate antiviral responses by pairing with TRIF or MAVS. Front. Immunol. 2019, 10, 2202. [Google Scholar] [CrossRef]
- Boulon, S.; Westman, B.J.; Hutten, S.; Boisvert, F.M.; Lamond, A.I. The nucleolus under stress. Mol. Cell 2010, 40, 216–227. [Google Scholar] [CrossRef]
- Feric, M.; Vaidya, N.; Harmon, T.S.; Mitrea, D.M.; Zhu, L.; Richardson, T.M.; Kriwacki, R.W.; Pappu, R.V.; Brangwynne, C.P. Coexisting liquid phases underlie nucleolar subcompartments. Cell 2016, 165, 1686–1697. [Google Scholar] [CrossRef]
- Martin, R.; Straub, A.U.; Doebele, C.; Bohnsack, M.T. DExD/H-box RNA helicases in ribosome biogenesis. RNA Biol. 2013, 10, 4–18. [Google Scholar] [CrossRef] [PubMed]
- Rawlinson, S.M.; Moseley, G.W. The nucleolar interface of RNA viruses. Cell Microbiol. 2015, 17, 1108–1120. [Google Scholar] [CrossRef] [PubMed]
- Zirwes, R.F.; Eilbracht, J.; Kneissel, S.; Schmidt-Zachmann, M.S. A novel helicase-type protein in the nucleolus: Protein NOH61. Mol. Biol. Cell 2000, 11, 1153–1167. [Google Scholar] [CrossRef] [PubMed]
- Reid, C.R.; Hobman, T.C. The nucleolar helicase DDX56 redistributes to West Nile Virus assembly sites. Virology 2017, 500, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Anderson, R.; Hobman, T.C. The capsid-binding nucleolar helicase DDX56 is important for infectivity of West Nile Virus. J. Virol. 2011, 85, 5571–5580. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Hobman, T.C. The helicase activity of DDX56 is required for its role in assembly of infectious West Nile Virus particles. Virology 2012, 433, 226–235. [Google Scholar] [CrossRef]
- Reed, J.C.; Molter, B.; Geary, C.D.; McNevin, J.; McElrath, J.; Giri, S.; Klein, K.C.; Lingappa, J.R. HIV-1 Gag co-opts a cellular complex containing DDX6, a helicase that facilitates capsid assembly. J. Cell. Biol. 2012, 198, 439–456. [Google Scholar] [CrossRef]
- Xing, Y.-H.; Yao, R.-W.; Zhang, Y.; Guo, C.-J.; Jiang, S.; Xu, G.; Dong, R.; Yang, L.; Chen, L.-L. Slert regulates DDX21 rings associated with pol I transcription. Cell 2017, 169, 664–678.e16. [Google Scholar] [CrossRef]
- Watanabe, Y.; Ohtaki, N.; Hayashi, Y.; Ikuta, K.; Tomonaga, K. Autogenous translational regulation of the borna disease virus negative control factor X from polycistronic mRNA using host RNA helicases. PLoS Pathog. 2009, 5, e1000654. [Google Scholar] [CrossRef]
- Chen, G.; Liu, C.-H.; Zhou, L.; Krug, R.M. Cellular DDX21 RNA helicase inhibits influenza A virus replication but is counteracted by the viral NS1 protein. Cell Host Microbe 2014, 15, 484–493. [Google Scholar] [CrossRef]
- Standart, N.; Weil, D. P-bodies: Cytosolic droplets for coordinated mRNA storage. Trends Genet. 2018, 34, 612–626. [Google Scholar] [CrossRef]
- Beckham, C.J.; Parker, R. P bodies, stress granules, and viral life cycles. Cell Host Microbe 2008, 3, 206–212. [Google Scholar] [CrossRef]
- Ozgur, S.; Stoecklin, G. Role of Rck-Pat1b binding in assembly of processing-bodies. RNA Biol. 2013, 10, 528–539. [Google Scholar] [CrossRef][Green Version]
- Minshall, N.; Kress, M.; Weil, D.; Standart, N. Role of p54 RNA helicase activity and its C-terminal domain in translational repression, p-body localization and assembly. Mol. Biol. Cell. 2009, 20, 2464–2472. [Google Scholar] [CrossRef] [PubMed]
- Nunez, R.D.; Budt, M.; Saenger, S.; Paki, K.; Arnold, U.; Sadewasser, A.; Wolff, T. The RNA helicase DDX6 associates with RIG-I to augment induction of antiviral signaling. Int. J. Mol. Sci. 2018, 19, 1877. [Google Scholar] [CrossRef] [PubMed]
- Lumb, J.H.; Li, Q.; Popov, L.M.; Ding, S.; Keith, M.T.; Merrill, B.D.; Greenberg, H.B.; Li, J.B.; Carette, J.E. DDX6 represses aberrant activation of interferon-stimulated genes. Cell Rep. 2017, 20, 819–831. [Google Scholar] [CrossRef] [PubMed]
- Goertz, G.P.; van Bree, J.W.M.; Hiralal, A.; Fernhout, B.M.; Steffens, C.; Boeren, S.; Visser, T.M.; Vogels, C.B.F.; Abbo, S.R.; Fros, J.J.; et al. Subgenomic flavivirus RNA binds the mosquito dead/H-box helicase ME31b and determines zika virus transmission by Aedes Aegypti. Proc. Natl. Acad Sci. USA 2019, 116, 19136–19144. [Google Scholar] [CrossRef]
- Michalski, D.; Ontiveros, J.G.; Russo, J.; Charley, P.A.; Anderson, J.R.; Heck, A.M.; Geiss, B.J.; Wilusz, J. Zika virus noncoding sfRNAs sequester multiple host-derived RNA-binding proteins and modulate mRNA decay and splicing during infection. J. Biol. Chem. 2019, 294, 16282–16296. [Google Scholar] [CrossRef]
- Ward, A.M.; Bidet, K.; Yinglin, A.; Ler, S.G.; Hogue, K.; Blackstock, W.; Gunaratne, J.; Garcia-Blanco, M.A. Quantitative mass spectrometry of Denv-2 RNA-interacting proteins reveals that the dead-box RNA helicase DDX6 binds the DB1 and DB2 3’ utr structures. RNA Biol. 2011, 8, 1173–1186. [Google Scholar] [CrossRef]
- Chahar, H.S.; Chen, S.; Manjunath, N. P-body components lsm1, GW182, DDX3, DDX6 and XRN1 are recruited to WNV replication sites and positively regulate viral replication. Virology 2013, 436, 1–7. [Google Scholar] [CrossRef]
- Buchan, J.R.; Parker, R. Eukaryotic stress granules: The ins and outs of translation. Mol. Cell 2009, 36, 932–941. [Google Scholar] [CrossRef]
- Thulasi Raman, S.N.; Liu, G.; Pyo, H.M.; Cui, Y.C.; Xu, F.; Ayalew, L.E.; Tikoo, S.K.; Zhou, Y. DDX3 interacts with influenza A virus NS1 and NP proteins and exerts antiviral function through regulation of stress granule formation. J. Virol. 2016, 90, 3661. [Google Scholar] [CrossRef]
- Chuang, C.; Prasanth, K.R.; Nagy, P.D. Coordinated function of cellular dead-box helicases in suppression of viral RNA recombination and maintenance of viral genome integrity. PLoS Pathog. 2015, 11, e1004680. [Google Scholar] [CrossRef]
- Yedavalli, V.S.; Neuveut, C.; Chi, Y.H.; Kleiman, L.; Jeang, K.T. Requirement of DDX3 dead box RNA helicase for HIV-1 rev-rre export function. Cell 2004, 119, 381–392. [Google Scholar] [CrossRef]
- Edgcomb, S.P.; Carmel, A.B.; Naji, S.; Ambrus-Aikelin, G.; Reyes, J.R.; Saphire, A.C.S.; Gerace, L.; Williamson, J.R. DDX1 is an RNA-dependent ATPase involved in HIV-1 rev function and virus replication. J. Mol. Biol. 2012, 415, 61–74. [Google Scholar] [CrossRef]
- Xue, Q.; Liu, H.; Zeng, Q.; Zheng, H.; Xue, Q.; Cai, X. The dead-box RNA helicase DDX1 interacts with the viral protein 3D and inhibits foot-and-mouth disease virus replication. Virol. Sin. 2019, 34, 610–617. [Google Scholar] [CrossRef]
- Fang, J.; Kubota, S.; Yang, B.; Zhou, N.; Zhang, H.; Godbout, R.; Pomerantz, R.J. A dead box protein facilitates HIV-1 replication as a cellular co-factor of rev. Virology 2004, 330, 471–480. [Google Scholar] [CrossRef]
- Hammond, J.A.; Lamichhane, R.; Millar, D.P.; Williamson, J.R. A dead-box helicase mediates an RNA structural transition in the HIV-1 rev response element. J. Mol. Biol. 2017, 429, 697–714. [Google Scholar] [CrossRef][Green Version]
- Robertson-Anderson, R.M.; Wang, J.; Edgcomb, S.P.; Carmel, A.B.; Williamson, J.R.; Millar, D.P. Single-molecule studies reveal that dead box protein DDX1 promotes oligomerization of HIV-1 rev on the rev response element. J. Mol. Biol. 2011, 410, 959–971. [Google Scholar] [CrossRef]
- Sherpa, C.; Rausch, J.W.; Le Grice, S.F.; Hammarskjold, M.L.; Rekosh, D. The HIV-1 rev response element (rre) adopts alternative conformations that promote different rates of virus replication. Nucleic Acids Res. 2015, 43, 4676–4686. [Google Scholar] [CrossRef]
- Xu, L.; Khadijah, S.; Fang, S.; Wang, L.; Tay, F.P.; Liu, D.X. The cellular RNA helicase DDX1 interacts with coronavirus nonstructural protein 14 and enhances viral replication. J. Virol. 2010, 84, 8571–8583. [Google Scholar] [CrossRef] [PubMed]
- Kovalev, N.; Barajas, D.; Nagy, P.D. Similar roles for yeast dbp2 and arabidopsis rh20 dead-box RNA helicases to ded1 helicase in tombusvirus plus-strand synthesis. Virology 2012, 432, 470–484. [Google Scholar] [CrossRef]
- Li, M.; Johnson, J.R.; Truong, B.; Kim, G.; Weinbren, N.; Dittmar, M.; Shah, P.S.; Von Dollen, J.; Newton, B.W.; Jang, G.M.; et al. Identification of antiviral roles for the exon–junction complex and nonsense-mediated decay in flaviviral infection. Nature Microbiol. 2019, 4, 985–995. [Google Scholar] [CrossRef]
- Naji, S.; Ambrus, G.; Cimermančič, P.; Reyes, J.R.; Johnson, J.R.; Filbrandt, R.; Huber, M.D.; Vesely, P.; Krogan, N.J.; Yates, J.R.; et al. Host cell interactome of HIV-1 rev includes RNA helicases involved in multiple facets of virus production. Mol. Cell. Proteom. 2012, 11, M111.015313. [Google Scholar] [CrossRef]
- Knoener, R.A.; Becker, J.T.; Scalf, M.; Sherer, N.M.; Smith, L.M. Elucidating the in vivo interactome of HIV-1 RNA by hybridization capture and Mass spectrometry. Sci. Rep. 2017, 7, 16965. [Google Scholar] [CrossRef]
- Yasuda-Inoue, M.; Kuroki, M.; Ariumi, Y. Distinct DDX dead-box RNA helicases cooperate to modulate the HIV-1 rev function. Biochem. Biophys. Res. Commun. 2013, 434, 803–808. [Google Scholar] [CrossRef]
- Hoffmann, D.; Schwarck, D.; Banning, C.; Brenner, M.; Mariyanna, L.; Krepstakies, M.; Schindler, M.; Millar, D.P.; Hauber, J. Formation of trans-activation competent HIV-1 rev:Rre complexes requires the recruitment of multiple protein activation domains. PLoS ONE 2012, 7, e38305. [Google Scholar] [CrossRef]
- Rahman, M.M.; Bagdassarian, E.; Ali, M.A.M.; McFadden, G. Identification of host dead-box RNA helicases that regulate cellular tropism of oncolytic myxoma virus in human cancer cells. Sci. Rep. 2017, 7, 15710. [Google Scholar] [CrossRef]
- Krishnan, V.; Zeichner, S.L. Alterations in the expression of dead-box and other RNA binding proteins during HIV-1 replication. Retrovirology 2004, 1, 42. [Google Scholar] [CrossRef]
- Krishnan, V.; Zeichner, S.L. Host cell gene expression during human immunodeficiency virus type 1 latency and reactivation and effects of targeting genes that are differentially expressed in viral latency. J. Virol. 2004, 78, 9458. [Google Scholar] [CrossRef]
- Dethoff, E.A.; Boerneke, M.A.; Gokhale, N.S.; Muhire, B.M.; Martin, D.P.; Sacco, M.T.; McFadden, M.J.; Weinstein, J.B.; Messer, W.B.; Horner, S.M.; et al. Pervasive tertiary structure in the dengue virus RNA genome. Proc. Natl. Acad. Sci. USA 2018, 115, 11513–11518. [Google Scholar] [CrossRef]
- Huber, R.G.; Lim, X.N.; Ng, W.C.; Sim, A.Y.L.; Poh, H.X.; Shen, Y.; Lim, S.Y.; Sundstrom, K.B.; Sun, X.; Aw, J.G.; et al. Structure mapping of dengue and Zika viruses reveals functional long-range interactions. Nat. Commun. 2019, 10, 1408. [Google Scholar] [CrossRef]
- Ko, C.; Lee, S.; Windisch, M.P.; Ryu, W.S. DDX3 dead-box RNA helicase is a host factor that restricts hepatitis B virus replication at the transcriptional level. J. Virol. 2014, 88, 13689–13698. [Google Scholar] [CrossRef]
- Sithole, N.; Williams, C.A.; Vaughan, A.M.; Kenyon, J.C.; Lever, A.M.L. Ddx17 specifically, and independently of DDX5, controls use of the HIV a4/5 splice acceptor cluster and is essential for efficient replication of HIV. J. Mol. Biol. 2018, 430, 3111–3128. [Google Scholar] [CrossRef]
- Wisskirchen, C.; Ludersdorfer, T.H.; Muller, D.A.; Moritz, E.; Pavlovic, J. The cellular rna helicase uap56 is required for prevention of double-stranded RNA formation during influenza A virus infection. J. Virol. 2011, 85, 8646–8655. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).