Characterization of Salmonella Isolates from Various Geographical Regions of the Caucasus and Their Susceptibility to Bacteriophages
Abstract
:1. Introduction
2. Materials and Methods
2.1. Initial Isolation and Identification of Salmonella Isolates
2.2. MALDI-TOF MS Identification
2.3. Serological Characterization of Salmonella Isolates
2.4. Molecular Typing of Salmonella Isolates
2.5. Antimicrobial Susceptibility Profiles of Salmonella Isolates
2.6. Bacteriophages and the Propagation Bacterial Strains Used in the Study
2.7. Phage Isolation
2.8. Preparation of High-Titer Phage Stocks
2.9. Bacteriophage Susceptibility Test
2.10. Sequencing and Analysis of Phage Genomes
3. Results and Discussion
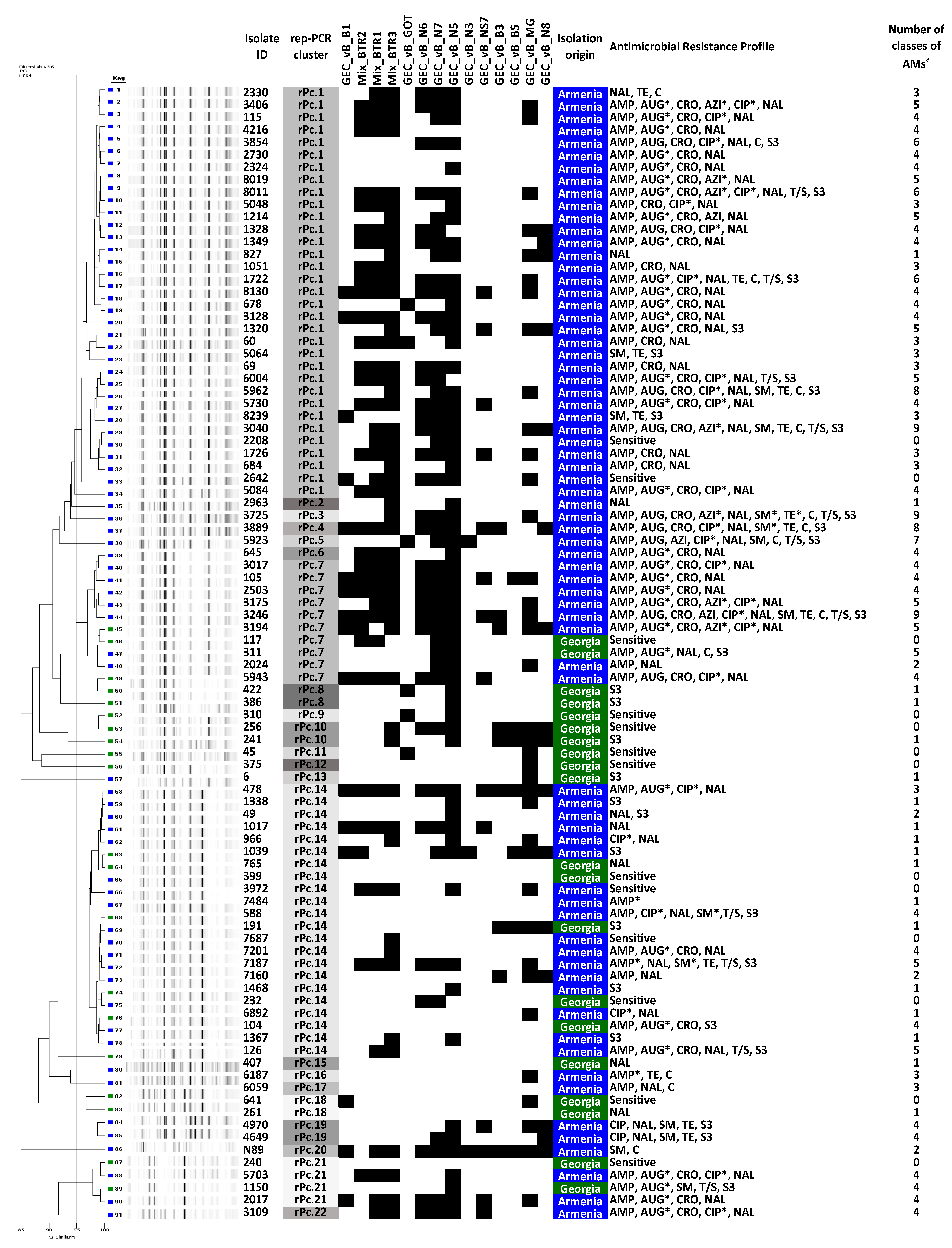
3.1. Isolation, Identification and Characterization of Salmonella Isolates
3.2. Antimicrobial Susceptibility
3.3. Susceptibility of Salmonella Isolates to Different Phages
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Salmonella (Non-Typhoidal). Newsroom. 20 February 2018. Available online: https://www.who.int/en/news-room/fact-sheets/detail/salmonella-(non-typhoidal) (accessed on 10 October 2020).
- Wang, X.; Biswas, S.; Paudyal, N.; Pan, H.; Li, X.; Fang, W.; Yue, M. Antibiotic Resistance in Salmonella Typhimurium Isolates Recovered from the Food Chain Through National Antimicrobial Resistance Monitoring System Between 1996 and 2016. Front. Microbiol. 2019, 10, 985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States. 2019. Available online: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf (accessed on 11 October 2020).
- Shrivastava, S.R.; Shrivastava, P.S.; Ramasamy, J. World Health Organization releases global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. J. Med. Soc. 2018, 32, 76. [Google Scholar] [CrossRef]
- Al, F.M.E.; Gu, W.; Mahon, B.E.; Judd, M.; Folster, J.; Griffin, P.M.; Hoekstra, R.M. Estimated incidence of antimicrobial drug–resistant nontyphoidal Salmonella infections, United States, 2004–2012. Emerg. Infect. Dis. 2017, 23, 29. [Google Scholar]
- Elashvili, E.; Mikadze, G.; Vepkhvadze, N.; Zurashvili, B.; Giorgobiani, M.; Lashkarashvili, M. Food related Salmonellosis poisoning cases in Georgia and modern preventive methods. J. Exp. Clin. Med. 2018, 4, 13–16. [Google Scholar]
- Cisek, A.A.; Dąbrowska, I.; Gregorczyk, K.P.; Wyżewski, Z. Phage therapy in bacterial infections treatment: One hundred years after the discovery of bacteriophages. Curr. Microbiol. 2017, 74, 277–283. [Google Scholar] [CrossRef] [Green Version]
- Sillankorva, S.M.; Oliveira, H.; Azeredo, J. Bacteriophages and their role in food safety. Int. J. Microbiol. 2012. [Google Scholar] [CrossRef] [Green Version]
- Nagel, T.E.; Chan, B.K.; De Vos, D.; El-Shibiny, A.; Kang’Ethe, E.K.; Makumi, A.; Pirnay, J.-P. The developing world urgently needs phages to combat pathogenic bacteria. Front. Microbiol. 2016, 7, 882. [Google Scholar] [CrossRef]
- Jamshidi, A.; Kalidari, G.A.; Hedayati, M. Isolation and identification of Salmonella Enteritidis and Salmonella Typhimurium from the eggs of retail stores in Mashhad, Iran using conventional culture method and multiplex PCR assay. J. Food Safety 2010, 30, 558–568. [Google Scholar]
- Public Health England. Identification of Enterobacteriaceae. UK Standards for Microbiology Investigations. ID16, Issue No 4; 2015. Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/423601/ID_16i4.pdf (accessed on 11 October 2020).
- Wattal, C.; Oberoi, J.K.; Goel, N.; Raveendran, R.; Khanna, S. Matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS) for rapid identification of micro-organisms in the routine clinical microbiology laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 807–812. [Google Scholar] [CrossRef]
- Grimont, P.A.; Weill, F.X. Antigenic Formulas of the Salmonella Serovars, 9th ed.; World Health Organization Collaborating Centre for Reference and Research on Salmonella: Paris, France, 2007; Available online: https://www.pasteur.fr/sites/default/files/veng_0.pdf (accessed on 11 October 2020).
- Doléans-Jordheim, A.; Cournoyer, B.; Bergeron, E.; Croizé, J.; Salord, H.; André, J.; Mazoyer, M.-A.; Renaud, F.N.R.; Freney, J. Reliability of Pseudomonas aeruginosa semi-automated rep-PCR genotyping in various epidemiological situations. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 1105–1111. [Google Scholar] [CrossRef]
- Serrano, I.; De Vos, D.; Santos, J.P.; Bilocq, F.; Leitão, A.; Tavares, L.; Pirnay, J.-P.; Oliveira, M. Antimicrobial resistance and genomic rep-PCR fingerprints of Pseudomonas aeruginosa strains from animals on the background of the global population structure. BMC Vet. Res. 2016, 13, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauer, A.W. Antibiotic susceptibility testing by a standardized single disc method. Am. J. Clin. Pathol. 1966, 45, 149–158. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 29th ed.; CLSI Supplement. M100S; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019. [Google Scholar]
- Adams, M.H. Bacteriophages; Interscience Publishers: New York, NY, USA, 1959. [Google Scholar]
- Van Twest, R.; Kropinski, A.M. Bacteriophage enrichment from water and soil. In Bacteriophages; Clokie, M.R., Kropinski, A.M., Eds.; Humana Press: Totowa, NJ, USA, 2009; Volume 501, pp. 15–21. [Google Scholar] [CrossRef]
- Merabishvili, M.; Pirnay, J.-P.; Verbeken, G.; Chanishvili, N.; Tediashvili, M.; Lashkhi, N.; Glonti, T.; Krylov, V.; Mast, J.; Van Parys, L.; et al. Quality-controlled small-scale production of a well-defined bacteriophage cocktail for use in human clinical trials. PLoS ONE 2009, 4, e4944. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D. Molecular Cloning: A Laboratory Manual; Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017, 45, D535–D542. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.J.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Grubaugh, N.D.; Gangavarapu, K.; Quick, J.; Matteson, N.L.; De Jesus, J.G.; Main, B.J.; Tan, A.L.; Paul, L.M.; Brackney, D.E.; Grewal, S.; et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019, 20, 8. [Google Scholar] [CrossRef] [Green Version]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.; Carmeli, Y.; Falagas, M.; Giske, C.; Harbarth, S.; Hindler, J.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Makalatia, K.; Kakabadze, E.; Bakuradze, N.; Grdzelishvili, N.; Natroshvili, G.; Kusradze, I.; Goderdzishvili, M.; Sedrakyan, A.; Arakelova, K.; Mkrtchyan, M.; et al. Activity of bacteriophages to multiply resistant strains of salmonella and their various serotypes. Vet. Biotech. 2018, 32, 500–508. [Google Scholar] [CrossRef] [Green Version]
- Gambino, M.; Sørensen, A.N.; Ahern, S.; Smyrlis, G.; Gencay, Y.E.; Hendrix, H.; Neve, H.; Noben, J.P.; Lavigne, R.; Brøndsted, L. Phage S144, A New Polyvalent Phage Infecting Salmonella spp. and Cronobacter sakazakii. Int. J. Mol. Sci. 2020, 21, 5196. [Google Scholar] [CrossRef] [PubMed]
- Tolen, T.N.; Xie, Y.; Hernandez, A.C.; Everett, G.F.K. Complete genome sequence of Salmonella enterica serovar Typhimurium myophage Mushroom. Genome Announc. 2015, 3, e00154-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whichard, J.M.; Weigt, L.A.; Borris, D.J.; Li, L.; Zhang, Q.; Kapur, V.; Pierson, F.W.; Lingohr, E.J.; She, Y.-M.; Kropinski, A.M.; et al. Complete genomic sequence of bacteriophage felix o1. Viruses 2010, 2, 710–730. [Google Scholar] [CrossRef]
- Santos, S.B.; Kropinski, A.M.; Ceyssens, P.-J.; Ackermann, H.-W.; Villegas, A.; Lavigne, R.; Krylov, V.N.; Carvalho, C.; Ferreira, E.C.; Azeredo, J. Genomic and proteomic characterization of the broad-host-range Salmonella phage PVP-SE1: Creation of a new phage genus. J. Virol. 2011, 85, 11265–11273. [Google Scholar] [CrossRef] [Green Version]
- Hooton, S.P.; Timms, A.R.; Rowsell, J.; Wilson, R.; Connerton, I.F. Salmonella Typhimurium-specific bacteriophage ΦSH19 and the origins of species specificity in the Vi01-like phage family. Virol. J. 2011, 8, 498. [Google Scholar] [CrossRef] [Green Version]
- Luna, A.J.; Wood, T.L.; Chamakura, K.R.; Everett, G.F.K. Complete genome of Salmonella enterica serovar Enteritidis myophage Marshall. Genome Announc. 2013, 1, e00867-13. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Ryu, S. Characterization of a T5-like coliphage, SPC35, and differential development of resistance to SPC35 in Salmonella enterica serovar Typhimurium and Escherichia coli. Appl. Environ. Microbiol. 2011, 77, 2042–2050. [Google Scholar] [CrossRef] [Green Version]
- Goodridge, L.D. Designing phage therapeutics. Curr. Pharm. Biotechnol. 2010, 11, 15–27. [Google Scholar] [CrossRef]
- Schmerer, M.; Molineux, I.J.; Bull, J.J. Synergy as a rationale for phage therapy using phage cocktails. PeerJ 2014, 2, 590. [Google Scholar] [CrossRef] [Green Version]
- Merabishvili, M.; Pirnay, J.P.; De Vos, D. Guidelines to Compose an Ideal Bacteriophage Cocktail. Methods. Mol. Biol. 2018, 1693, 99–110. [Google Scholar] [CrossRef] [PubMed]




| Phage Name | Genus (Morphology) | Size of Phage Head/Tail (nm) | Genome Size (kb) | NCBI Closest Match | NCBI Query Coverage (%) | NCBI Percent Identity (%) | Isolation Year | Isolation Source/Place |
|---|---|---|---|---|---|---|---|---|
| GEC_vB_N3 | Tequintavirus (Siphoviridae) | 68/140 | 110 | Salmonella phage 1-29 | 79 | 97 | 2013 | River Mtkvari, Tbilisi, Georgia |
| GEC_vB_N5 | Tequintavirus (Siphoviridae) | 90/231 | 149 | E. coli phage T5 | 86 | 97 | 2013 | River Mtkvari, Tbilisi, Georgia |
| GEC_vB_N7 | Tequintavirus (Siphoviridae) | 87/136 | 112 | Salmonella phage 1-29 | 81 | 95 | 2013 | River Mtkvari, Tbilisi, Georgia |
| GEC_vB_N8 | Tequintavirus (Siphoviridae) | 77/168 | 51 | E. coli phage SPC35 | 84 | 92 | 2013 | River Mtkvari, Tbilisi, Georgia |
| GEC_vB_N6 | Viunavirus (Myoviridae) | 104/140 | 158 | Salmonella phage STML-13-1 | 90 | 98 | 2013 | River Mtkvari, Tbilisi, Georgia |
| GEC_vB_NS7 | Felixounavirus (Myoviridae) | 63/109 | 55 | Salmonella phage Mushroom | 96 | 99 | 2015 | Cow raw milk, Tbilisi, Georgia |
| GEC_vB_MG | Seunavirus (Myoviridae) | 95/104 | 142 | Salmonella phage PVP-SE1 | 89 | 96 | 2013 | Sewage water, Tbilisi, Georgia |
| GEC_vB_BS | Felixounavirus (Myoviridae) | 77/118 | 86 | Salmonella phage Mushroom | 98 | 98 | 2013 | Black Sea, Batumi, Georgia |
| GEC_vB_B1 | Felixounavirus (Myoviridae) | 81/122 | 87 | Salmonella phage Mushroom | 96 | 98 | 2013 | River Mtkvari, Tbilisi, Georgia |
| GEC_vB_GOT | Viunavirus (Myoviridae) | 90/119 | 157 | Salmonella phage STML-13-1 | 90 | 99 | 2013 | Sewage water, Tbilisi, Georgia |
| GEC_vB_B3 | Felixounavirus (Myoviridae) | 72/113 | 87 | Salmonella phage Mushroom | 96 | 98 | 2013 | River Mtkvari, Tbilisi, Georgia |
| Name of Cocktail | Name of Phages |
|---|---|
| Mix_BTR1 | GEC_vB_MG, GEC_vB__N7, GEC_vB_N5 |
| Mix_BTR2 | GEC_vB_N7, GEC_vB_N5, GEC_vB_N6 |
| Mix_BTR3 | GEC_vB_MG, GEC_vB_N7, GEC_vB_N6 |
| Strain ID | Species/Serotype | Isolation Source | Isolation Place | Isolation Year | Propagated Phages |
|---|---|---|---|---|---|
| SeE.3 | S. enterica Enteritidis | Pig feces | Pig farm, South Korea | 2011 | GEC_vB_N3, GEC_vB_N8, GEC_vB_N5, GEC_vB_MG |
| SeT.4 | S. enterica Typhimurium | Pig feces | Pig farm, South Korea | 2011 | GEC_vB_N7, GEC_vB_GOT, GEC_vB_BS |
| SeT.6 | S. enterica Typhimurium | Pig feces | Pig farm, South Korea | 2011 | GEC_vB_N6, GEC_vB_NS7, GEC_vB_B3, GEC_vB_B1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Makalatia, K.; Kakabadze, E.; Wagemans, J.; Grdzelishvili, N.; Bakuradze, N.; Natroshvili, G.; Macharashvili, N.; Sedrakyan, A.; Arakelova, K.; Ktsoyan, Z.; et al. Characterization of Salmonella Isolates from Various Geographical Regions of the Caucasus and Their Susceptibility to Bacteriophages. Viruses 2020, 12, 1418. https://doi.org/10.3390/v12121418
Makalatia K, Kakabadze E, Wagemans J, Grdzelishvili N, Bakuradze N, Natroshvili G, Macharashvili N, Sedrakyan A, Arakelova K, Ktsoyan Z, et al. Characterization of Salmonella Isolates from Various Geographical Regions of the Caucasus and Their Susceptibility to Bacteriophages. Viruses. 2020; 12(12):1418. https://doi.org/10.3390/v12121418
Chicago/Turabian StyleMakalatia, Khatuna, Elene Kakabadze, Jeroen Wagemans, Nino Grdzelishvili, Nata Bakuradze, Gulnara Natroshvili, Nino Macharashvili, Anahit Sedrakyan, Karine Arakelova, Zhanna Ktsoyan, and et al. 2020. "Characterization of Salmonella Isolates from Various Geographical Regions of the Caucasus and Their Susceptibility to Bacteriophages" Viruses 12, no. 12: 1418. https://doi.org/10.3390/v12121418
APA StyleMakalatia, K., Kakabadze, E., Wagemans, J., Grdzelishvili, N., Bakuradze, N., Natroshvili, G., Macharashvili, N., Sedrakyan, A., Arakelova, K., Ktsoyan, Z., Zakharyan, M., Gevorgyan, Z., Mnatsakanyan, A., Tishkova, F., Lood, C., Vandenheuvel, D., Lavigne, R., Pirnay, J.-P., De Vos, D., ... Merabishvili, M. (2020). Characterization of Salmonella Isolates from Various Geographical Regions of the Caucasus and Their Susceptibility to Bacteriophages. Viruses, 12(12), 1418. https://doi.org/10.3390/v12121418










