Characterization of Novel Lytic Bacteriophages of Achromobacter marplantensis Isolated from a Pneumonia Patient
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Bacterial Cultures and Identification
2.3. Isolation and Purification of Bacteriophages
2.4. Extraction of Bacteriophage DNA
2.5. Host Range Analysis
2.6. One Step Growth Analysis
2.7. Transmission Electron Microscopy
2.8. Genomic Characterization and Phylogeny
2.9. Codon Usage Analysis
3. Results
3.1. Bacterial Identification
3.2. Isolation and Phenotypic Characterization of Novel A. marplatensis Bacteriophages
3.3. Genome Analysis of A. marplatensis Bacteriophages AMA1 and AMA2
3.4. Presence of tRNA Genes in AMA1 and AMA2 Genomes
3.5. Bacteriophage Phylogeny
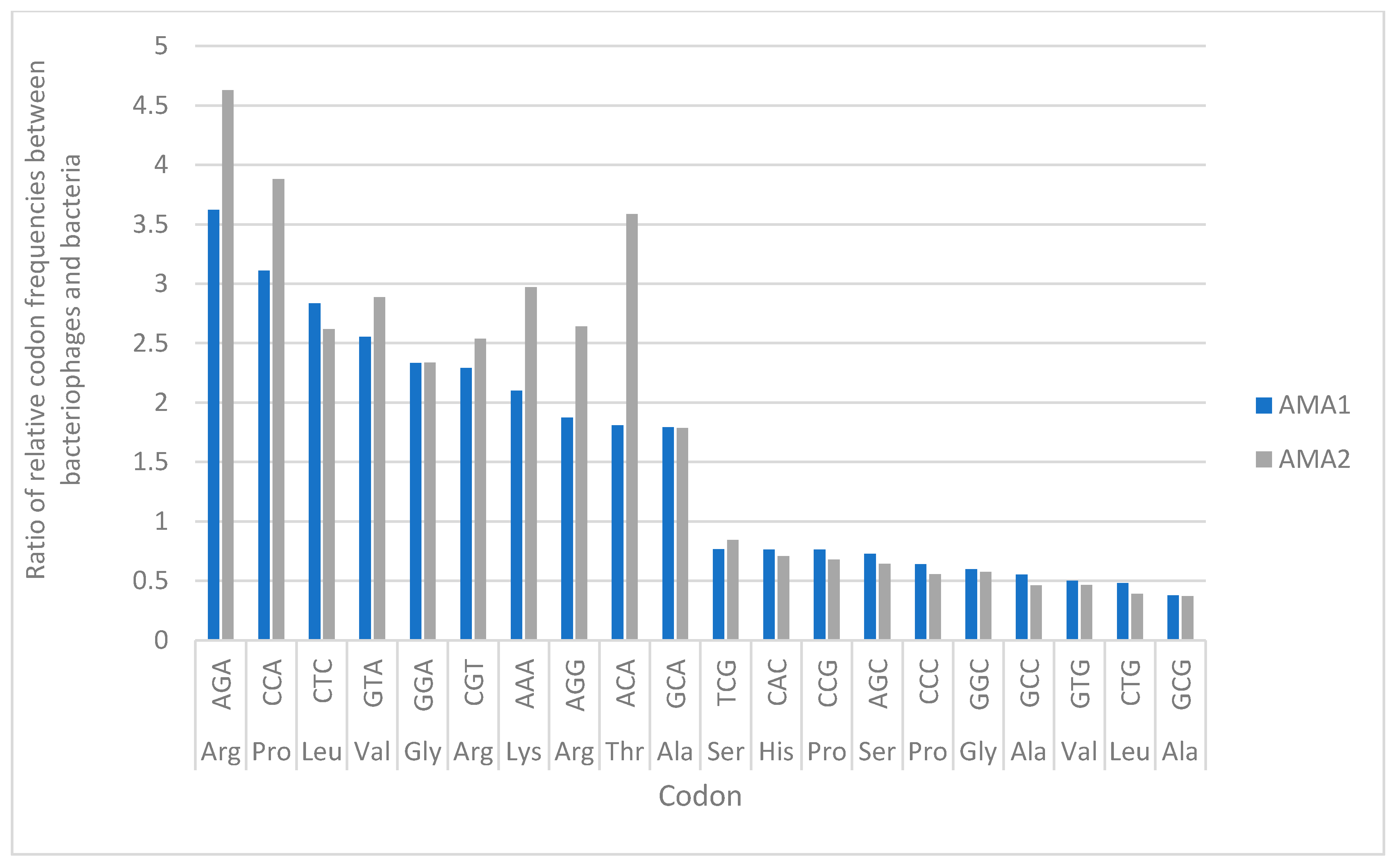
3.6. Codon Usage Analysis in AMA1, AMA2 and A. marplantensis
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Swenson, C.E.; Sadikot, R.T. Achromobacter Respiratory Infections. Ann. Am. Thorac. Soc. 2015, 12, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Marion-Sanchez, K.; Pailla, K.; Olive, C.; Le Coutour, X.; Derancourt, C. Achromobacter spp. healthcare associated infections in the French West Indies: A longitudinal study from 2006 to 2016. BMC Infect. Dis. 2019, 19, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Huen, A.O.; Kestenbaum, L.A.; Sarezky, M.D.; Coughlin, C.C.; Yan, A.C. Achromobacter xylosoxidans Bacteremia and Cellulitis: A Report of a Case. Pediatr. Dermatol. 2015, 32, 186–187. [Google Scholar] [CrossRef]
- Villegas, V.M.; Emanuelli, A.; Flynn, H.W., Jr.; Berrocal, A.M.; Miller, D.; Kao, A.A.; Dubovy, S.R.; Alfonso, E. Endophthalmitis caused by Achromobacter xylosoxidans after cataract surgery. Retina 2014, 34, 583–586. [Google Scholar] [CrossRef] [PubMed]
- Panchal, B.; Tyagi, M.; Pathengay, A.; Sharma, S.; Dave, V.P.; Gandhi, U.; Balakrishnan, D.; Pappuru, R.R.; Joseph, J.; Kekunnaya, R.; et al. Endophthalmitis following Suture Removal—Clinical Outcomes and Microbiological Profile. Semin. Ophthalmol. 2019, 34, 115–123. [Google Scholar] [CrossRef]
- Bellissimo, F.; Pinzone, M.R.; Tosto, S.; Nunnari, G.; Cacopardo, B. Achromobacter xylosoxidans meningitis in an immunosuppressed patient. Qjm Int. J. Med. 2013, 107, 65–66. [Google Scholar] [CrossRef]
- Edwards, B.D.; Greysson-Wong, J.; Somayaji, R.; Waddell, B.; Whelan, F.J.; Storey, D.G.; Rabin, H.R.; Surette, M.G.; Parkins, M.D. Prevalence and Outcomes of Achromobacter Species Infections in Adults with Cystic Fibrosis: A North American Cohort Study. J. Clin. Microbiol. 2017, 55, 2074–2085. [Google Scholar] [CrossRef]
- Somayaji, R.; Stanojevic, S.; Tullis, E.; Stephenson, A.; Ratjen, F.; Waters, V.J. Clinical Outcomes Associated with Achromobacter Species Infection in Patients with Cystic Fibrosis. Ann. Am. Thorac. Soc. 2017, 14, 1412–1418. [Google Scholar] [CrossRef]
- Recio, R.; Brañas, P.; Martinez, M.T.; Chaves, F.; Orellana, M.Á. Effect of respiratory Achromobacter spp. infection on pulmonary function in patients with cystic fibrosis. J. Med Microbiol. 2018, 67, 952–956. [Google Scholar] [CrossRef]
- Lo, C.I.; Fall, B.; Sambe-Ba, B.; Diawara, S.; Gueye, M.W.; Mediannikov, O.; Sokhna, C.; Faye, N.; Diémé, Y.; Wade, B.; et al. MALDI-TOF Mass Spectrometry: A Powerful Tool for Clinical Microbiology at Hôpital Principal de Dakar, Senegal (West Africa). PLoS ONE 2015, 10, e0145889. [Google Scholar] [CrossRef]
- Spilker, T.; Vandamme, P.; Lipuma, J.J. A Multilocus Sequence Typing Scheme Implies Population Structure and Reveals Several Putative Novel Achromobacter Species. J. Clin. Microbiol. 2012, 50, 3010–3015. [Google Scholar] [CrossRef] [PubMed]
- Amoureux, L.; Bador, J.; Zouak, F.B.; Chapuis, A.; De Curraize, C.; Neuwirth, C. Distribution of the species of Achromobacter in a French Cystic Fibrosis Centre and multilocus sequence typing analysis reveal the predominance of A. xylosoxidans and clonal relationships between some clinical and environmental isolates. J. Cyst. Fibros. 2016, 15, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Gomila, M.; Prince-Manzano, C.; Svensson-Stadler, L.; Busquets, A.; Erhard, M.; Martínez, D.L.; Lalucat, J.; Moore, E.R.B. Genotypic and Phenotypic Applications for the Differentiation and Species-Level Identification of Achromobacter for Clinical Diagnoses. PLoS ONE 2014, 9, e114356. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.K.; Von Keudell, A.; Moroder, P.; Appleton, P.; Wigmore, R.; Rodriguez, E.K. Recurrent Septic Arthritis Due to Achromobacter xylosoxidans in a Patient with Granulomatosis with Polyangiitis. Open Forum Infect. Dis. 2015, 2, 145. [Google Scholar] [CrossRef] [PubMed]
- Vandamme, P.; Peeters, C.; Cnockaert, M.; Gomila, M.; Moore, E.R.B.; Spilker, T.; Lipuma, J.J. Reclassification of Achromobacter spiritinus Vandamme et al. 2013 as a later heterotypic synonym of Achromobacter marplatensis Gomila et al. 2011. Int. J. Syst. Evol. Microbiol. 2016, 66, 1641–1644. [Google Scholar] [CrossRef] [PubMed]
- Gomila, M.; Tvrzová, L.; Teshim, A.; Sedláček, I.; Gonzalez-Escalona, N.; Zdráhal, Z.; Šedo, O.; Gonzalez, J.F.; Bennasar, A.; Moore, E.R.B.; et al. Achromobacter marplatensis sp. nov., isolated from a pentachlorophenol-contaminated soil. Int. J. Syst. Evol. Microbiol. 2011, 61, 2231–2237. [Google Scholar] [CrossRef]
- Papalia, M.; Steffanowski, C.; Traglia, G.; Almuzara, M.; Martina, P.; Galanternik, L.; Vay, C.; Gutkind, G.; Ramírez, M.S.; Radice, M.A. Diversity of Achromobacter species recovered from patients with cystic fibrosis, in Argentina. Rev. Argent. Microbiol. 2020, 52, 13–18. [Google Scholar] [CrossRef]
- Filipic, B.; Malesevic, M.; Vasiljevic, Z.; Lukic, J.; Novovic, K.; Kojic, M.; Jovcic, B. Uncovering Differences in Virulence Markers Associated with Achromobacter Species of CF and Non-CF Origin. Front. Cell. Infect. Microbiol. 2017, 7, 224. [Google Scholar] [CrossRef]
- Gade, S.S.; Nørskov-Lauritsen, N.; Ridderberg, W. Prevalence and species distribution of Achromobacter sp. cultured from cystic fibrosis patients attending the Aarhus centre in Denmark. J. Med Microbiol. 2017, 66, 686–689. [Google Scholar] [CrossRef]
- Coward, A.; Kenna, D.T.; Perry, C.; Martin, K.; Doumith, M.; Turton, J.F.; Information, P.E.K.F.C. Use of nrdA gene sequence clustering to estimate the prevalence of different Achromobacter species among Cystic Fibrosis patients in the UK. J. Cyst. Fibros. 2016, 15, 479–485. [Google Scholar] [CrossRef]
- Tiirola, M.A.; Männistö, M.K.; Puhakka, J.A.; Kulomaa, M.S. Isolation and Characterization of Novosphingobium sp. Strain MT1, a Dominant Polychlorophenol-Degrading Strain in a Groundwater Bioremediation System. Appl. Environ. Microbiol. 2002, 68, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef] [PubMed]
- Schooley, R.T.; Biswas, B.; Gill, J.J.; Hernandez-Morales, A.; Lancaster, J.; Lessor, L.; Barr, J.J.; Reed, S.L.; Rohwer, F.; Benler, S.; et al. Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails To Treat a Patient with a Disseminated Resistant Acinetobacter baumannii Infection. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Duckworth, D.H.; Gulig, P.A. Bacteriophages. BioDrugs 2002, 16, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, J.; Yu, P.; Zuo, P.; Da Silva, M.L.B.; Alvarez, P.J.J. Going Viral: Emerging Opportunities for Phage-Based Bacterial Control in Water Treatment and Reuse. Accounts Chem. Res. 2019, 52, 849–857. [Google Scholar] [CrossRef]
- Dyson, Z.A.; Tucci, J.; Seviour, R.J.; Petrovski, S. Lysis to Kill: Evaluation of the Lytic Abilities, and Genomics of Nine Bacteriophages Infective for Gordonia spp. and Their Potential Use in Activated Sludge Foam Biocontrol. PLoS ONE 2015, 10, e0134512. [Google Scholar] [CrossRef][Green Version]
- Wittmann, J.; Dreiseikelmann, B.; Rohde, C.; Rohde, M.; Sikorski, J. Isolation and Characterization of Numerous Novel Phages Targeting Diverse Strains of the Ubiquitous and Opportunistic Pathogen Achromobacter xylosoxidans. PLoS ONE 2014, 9, e86935. [Google Scholar] [CrossRef]
- Dreiseikelmann, B.; Bunk, B.; Sproer, C.; Rohde, M.; Nimtz, M.; Wittmann, J. Characterization and genome comparisons of three Achromobacter phages of the family Siphoviridae. Arch. Virol. 2017, 162, 2191–2201. [Google Scholar] [CrossRef]
- Ma, Y.; Li, E.; Qi, Z.; Li, H.; Wei, X.; Lin, W.; Zhao, R.; Jiang, A.; Yang, H.; Yin, Z.; et al. Isolation and molecular characterisation of Achromobacter phage phiAxp-3, an N4-like bacteriophage. Sci. Rep. 2016, 6, 24776. [Google Scholar] [CrossRef]
- Li, E.; Zhao, J.; Ma, Y.; Wei, X.; Li, H.; Lin, W.; Wang, X.; Li, C.; Shen, Z.; Zhao, R.; et al. Characterization of a novel Achromobacter xylosoxidans specific siphoviruse: phiAxp-1. Sci. Rep. 2016, 6, 21943. [Google Scholar] [CrossRef]
- Hoyle, N.; Zhvaniya, P.; Balarjishvili, N.; Bolkvadze, D.; Nadareishvili, L.; Nizharadze, D.; Wittmann, J.; Rohde, C.; Kutateladze, M. Phage therapy against Achromobacter xylosoxidans lung infection in a patient with cystic fibrosis: A case report. Res. Microbiol. 2018, 169, 540–542. [Google Scholar] [CrossRef] [PubMed]
- Turner, S.; Pryer, K.M.; Miao, V.P.W.; Palmer, J.D. Investigating Deep Phylogenetic Relationships among Cyanobacteria and Plastids by Small Subunit rRNA Sequence Analysis. J. Eukaryot. Microbiol. 1999, 46, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Hodkinson, B.P.; Lutzoni, F. A microbiotic survey of lichen-associated bacteria reveals a new lineage from the Rhizobiales. Symbiosis 2009, 49, 163–180. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Brown, T.L.; Petrovski, S.; Dyson, Z.A.; Seviour, R.; Tucci, J. The Formulation of Bacteriophage in a Semi Solid Preparation for Control of Propionibacterium acnes Growth. PLoS ONE 2016, 11, e0151184. [Google Scholar] [CrossRef]
- Karumidze, N.; Kusradze, I.; Rigvava, S.; Goderdzishvili, M.; Rajakumar, K.; Alavidze, Z. Isolation and Characterisation of Lytic Bacteriophages of Klebsiella pneumoniae and Klebsiella oxytoca. Curr. Microbiol. 2012, 66, 251–258. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2019, 48, D265–D268. [Google Scholar] [CrossRef]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. ViPTree: The viral proteomic tree server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Laslett, D. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, 54–57. [Google Scholar] [CrossRef]
- Stothard, P. The Sequence Manipulation Suite: JavaScript Programs for Analyzing and Formatting Protein and DNA Sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef]
- Yoshikawa, G.; Askora, A.; Blanc-Mathieu, R.; Kawasaki, T.; Li, Y.; Nakano, M.; Ogata, H.; Yamada, T. Xanthomonas citri jumbo phage XacN1 exhibits a wide host range and high complement of tRNA genes. Sci. Rep. 2018, 8, 4486. [Google Scholar] [CrossRef]
- Ahern, S.; Das, M.; Bhowmick, T.S.; Young, R.F.; Gonzalez, C.F. Characterization of Novel Virulent Broad-Host-Range Phages of Xylella fastidiosa and Xanthomonas. J. Bacteriol. 2013, 196, 459–471. [Google Scholar] [CrossRef]
- Li, E.; Yin, Z.; Ma, Y.; Li, H.; Lin, W.; Wei, X.; Zhao, R.; Jiang, A.; Yuan, J.; Zhao, X. Identification and molecular characterization of bacteriophage phiAxp-2 of Achromobacter xylosoxidans. Sci. Rep. 2016, 6, 34300. [Google Scholar] [CrossRef]
- Lynch, K.H.; Stothard, P.; Dennis, J.J. Comparative analysis of two phenotypically-similar but genomically-distinct Burkholderia cenocepacia-specific bacteriophages. BMC Genom. 2012, 13, 223. [Google Scholar] [CrossRef]
- Santamaría, R.I.; Bustos, P.; Sepúlveda-Robles, O.; Lozano, L.; Rodríguez, C.; Fernández, J.L.; Juárez, S.; Kameyama, L.; Guarneros, G.; Dávila, G.; et al. Narrow-Host-Range Bacteriophages That Infect Rhizobium etli Associate with Distinct Genomic Types. Appl. Environ. Microbiol. 2013, 80, 446–454. [Google Scholar] [CrossRef]
- Lal, T.M.; Ransangan, J. Complete Genome Sequence of VpKK5, a Novel Vibrio parahaemolyticus Lytic Siphophage. Genome Announc. 2015, 3, 01381-14. [Google Scholar] [CrossRef] [PubMed]
- Kulikov, E.E.; Golomidova, A.K.; Letarova, M.A.; Kostryukova, E.S.; Zelenin, A.S.; Prokhorov, N.S.; Letarov, A.V. Genomic Sequencing and Biological Characteristics of a Novel Escherichia Coli Bacteriophage 9g, a Putative Representative of a New Siphoviridae Genus. Viruses 2014, 6, 5077–5092. [Google Scholar] [CrossRef] [PubMed]
- Carstens, A.B.; Kot, W.P.; Hansen, L.H. Complete Genome Sequences of Four Novel Escherichia coli Bacteriophages Belonging to New Phage Groups. Genome Announc. 2015, 3, e00741-15. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, W.N.; Concepción-Acevedo, J.; Park, T.; Andleeb, S.; Bull, J.J.; Levin, B.R. Synergy and Order Effects of Antibiotics and Phages in Killing Pseudomonas aeruginosa Biofilms. PLoS ONE 2017, 12, e0168615. [Google Scholar] [CrossRef]
- Sepúlveda-Robles, O.; Kameyama, L.; Guarneros, G. High Diversity and Novel Species of Pseudomonas aeruginosa Bacteriophages. Appl. Environ. Microbiol. 2012, 78, 4510–4515. [Google Scholar] [CrossRef]
- Essoh, C.; Vernadet, J.-P.; Vergnaud, G.; Coulibaly, A.; Kakou-N’Douba, A.; N’Guetta, A.S.-P.; Ouassa, T.; Pourcel, C. Characterization of sixteen Achromobacter xylosoxidans phages from Abidjan, Côte d’Ivoire, isolated on a single clinical strain. Arch. Virol. 2020, 165, 725–730. [Google Scholar] [CrossRef]
- Ankrah, N.Y.D.; Budinoff, C.R.; Wilson, W.H.; Wilhelm, S.W.; Buchan, A. Genome Sequences of Two Temperate Phages, CB2047-A and CB2047-C, Infecting Sulfitobacter sp. Strain 2047. Genome Announc. 2014, 2, 00108–00114. [Google Scholar] [CrossRef]
- Miyata, R.; Yamaguchi, K.; Uchiyama, J.; Shigehisa, R.; Takemura-Uchiyama, I.; Kato, S.-I.; Ujihara, T.; Sakaguchi, Y.; Daibata, M.; Matsuzaki, S. Characterization of a novel Pseudomonas aeruginosa bacteriophage, KPP25, of the family Podoviridae. Virus Res. 2014, 189, 43–46. [Google Scholar] [CrossRef]
- Schmerer, M.; Molineux, I.J.; Bull, J.J. Synergy as a rationale for phage therapy using phage cocktails. PeerJ 2014, 2, 590. [Google Scholar] [CrossRef]
- Adriaenssens, E.M.; Ceyssens, P.-J.; Dunon, V.; Ackermann, H.-W.; Van Vaerenbergh, J.; Maes, M.; De Proft, M.; Lavigne, R. Bacteriophages LIMElight and LIMEzero of Pantoea agglomerans, Belonging to the “phiKMV-Like Viruses”. Appl. Environ. Microbiol. 2011, 77, 3443–3450. [Google Scholar] [CrossRef]
- Ceyssens, P.-J.; Lavigne, R.; Mattheus, W.; Chibeu, A.; Hertveldt, K.; Mast, J.; Robben, J.; Volckaert, G. Genomic Analysis of Pseudomonas aeruginosa Phages LKD16 and LKA1: Establishment of the φKMV Subgroup within the T7 Supergroup. J. Bacteriol. 2006, 188, 6924–6931. [Google Scholar] [CrossRef] [PubMed]
- Paterson, S.; Vogwill, T.; Buckling, A.; Benmayor, R.; Spiers, A.J.; Thomson, N.R.; Quail, M.; Smith, F.; Walker, D.; Libberton, B.; et al. Antagonistic coevolution accelerates molecular evolution. Nat. Cell Biol. 2010, 464, 275–278. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T. Bacteriophages of Ralstonia solanacearum: Their Diversity and Utilization as Biocontrol Agents in Agriculture. In Bacteriophages; IntechOpen: Rijeka, Croatia, 2012. [Google Scholar]
- Lee, J.-H.; Shin, H.; Kim, H.; Ryu, S. Complete Genome Sequence of Salmonella Bacteriophage SPN3US. J. Virol. 2011, 85, 13470–13471. [Google Scholar] [CrossRef] [PubMed]
- Volozhantsev, N.V.; Oakley, B.B.; Morales, C.A.; Verevkin, V.V.; Bannov, V.A.; Krasilnikova, V.M.; Popova, A.V.; Zhilenkov, E.L.; Garrish, J.K.; Schegg, K.M.; et al. Molecular Characterization of Podoviral Bacteriophages Virulent for Clostridium perfringens and Their Comparison with Members of the Picovirinae. PLoS ONE 2012, 7, e38283. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Satoh, S.; Ishikawa, H.; Fujiwara, A.; Kawasaki, T.; Fujie, M.; Ogata, H. A jumbo phage infecting the phytopathogen Ralstonia solanacearum defines a new lineage of the Myoviridae family. Virology 2010, 398, 135–147. [Google Scholar] [CrossRef]
- Łobocka, M.; Rose, D.J.; Plunkett, G.; Rusin, M.; Samojedny, A.; Lehnherr, H.; Yarmolinsky, M.B.; Blattner, F.R. Genome of Bacteriophage P1. J. Bacteriol. 2004, 186, 7032–7068. [Google Scholar] [CrossRef]
- Adriaenssens, E.M.; Mattheus, W.; Cornelissen, A.; Shaburova, O.; Krylov, V.N.; Kropinski, A.M.; Lavigne, R. Complete Genome Sequence of the Giant Pseudomonas Phage Lu11. J. Virol. 2012, 86, 6369–6370. [Google Scholar] [CrossRef]
- Sykilinda, N.; Bondar, A.A.; Gorshkova, A.S.; Kurochkina, L.P.; Kulikov, E.E.; Shneider, M.M.; Kadykov, V.A.; Solovjeva, N.V.; Kabilov, M.R.; Mesyanzhinov, V.V.; et al. Complete Genome Sequence of the Novel Giant Pseudomonas Phage PaBG. Genome Announc. 2014, 2, 00929-13. [Google Scholar] [CrossRef]
- Gan, H.M.; Sieo, C.C.; Tang, S.G.H.; Omar, A.R.; Ho, Y.W. The complete genome sequence of EC1-UPM, a novel N4-like bacteriophage that infects Escherichia coli O78:K80. Virol. J. 2013, 10, 308. [Google Scholar] [CrossRef]
- Nho, S.-W.; Ha, M.-A.; Kim, K.-S.; Kim, T.-H.; Jang, H.-B.; Cha, I.-S.; Park, S.-B.; Kim, Y.-K.; Jung, T.S. Complete Genome Sequence of the Bacteriophages ECBP1 and ECBP2 Isolated from Two Different Escherichia coli Strains. J. Virol. 2012, 86, 12439–12440. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wittmann, J.; Dreiseikelmann, B.; Rohde, M.; Meier-Kolthoff, J.P.; Bunk, B.; Rohde, C. First genome sequences of Achromobacter phages reveal new members of the N4 family. Virol. J. 2014, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Rocha, E.P.C.; Danchin, A. Base composition bias might result from competition for metabolic resources. Trends Genet. 2002, 18, 291–294. [Google Scholar] [CrossRef]
- Bailly-Bechet, M.; Vergassola, M.; Rocha, E.P.C. Causes for the intriguing presence of tRNAs in phages. Genome Res. 2007, 17, 1486–1495. [Google Scholar] [CrossRef]
- Chang, R.Y.K.; Wallin, M.; Lin, Y.; Leung, S.S.Y.; Wang, H.; Morales, S.; Chan, H.-K. Phage therapy for respiratory infections. Adv. Drug Deliv. Rev. 2018, 133, 76–86. [Google Scholar] [CrossRef]
- Wienhold, S.-M.; Lienau, J.; Witzenrath, M. Towards Inhaled Phage Therapy in Western Europe. Viruses 2019, 11, 295. [Google Scholar] [CrossRef]
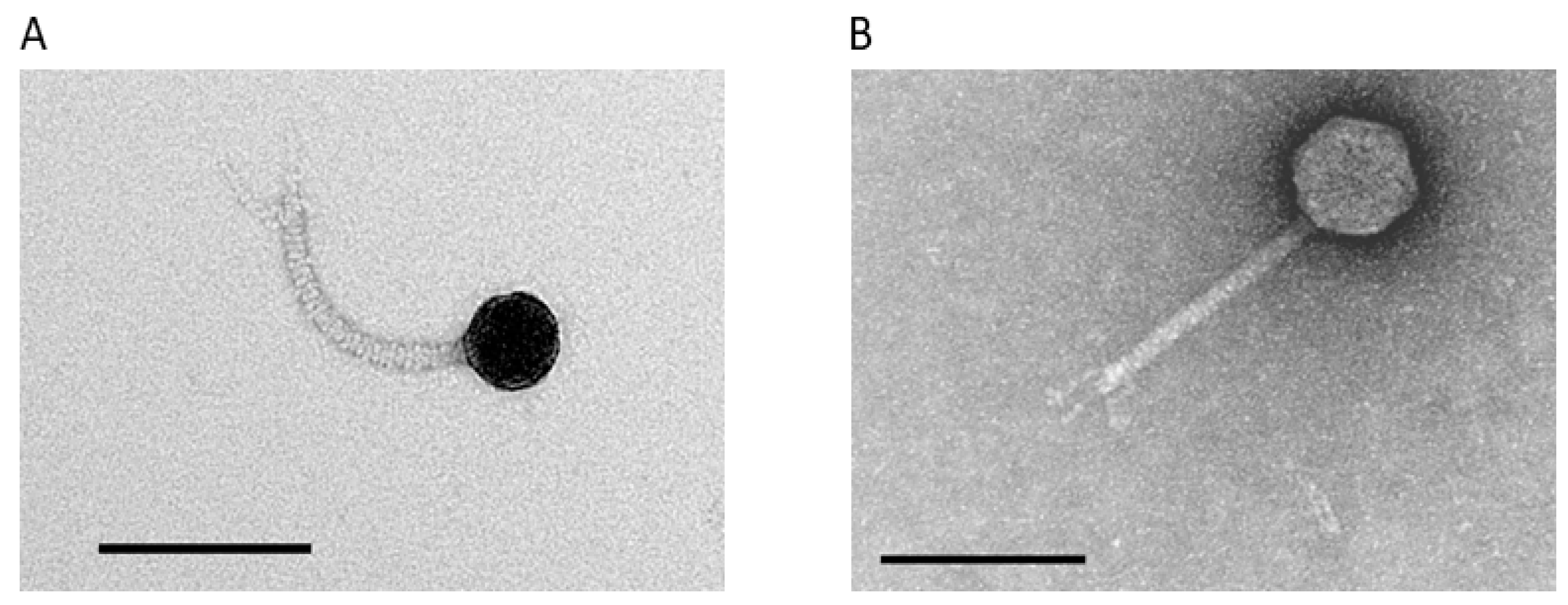
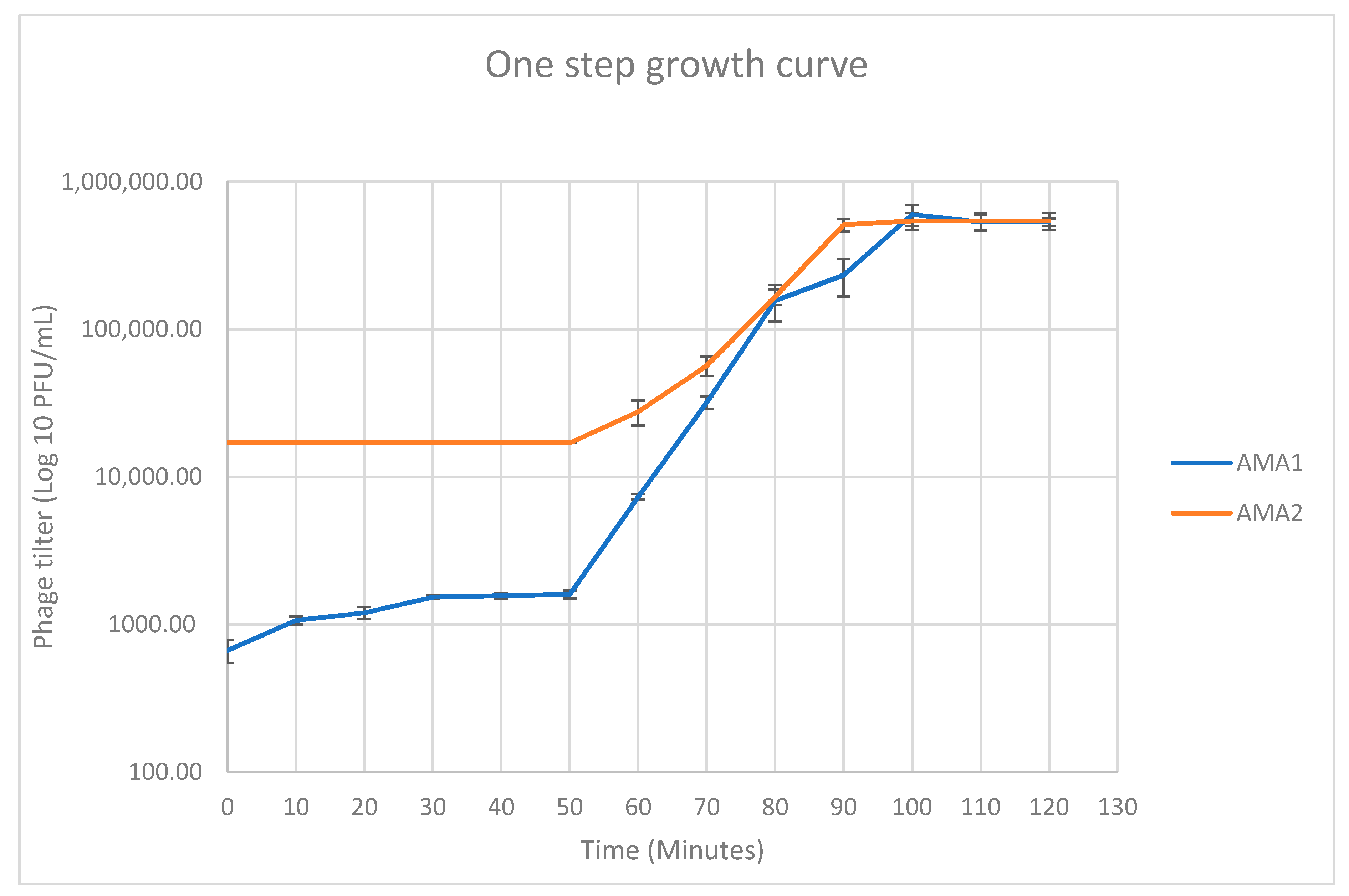
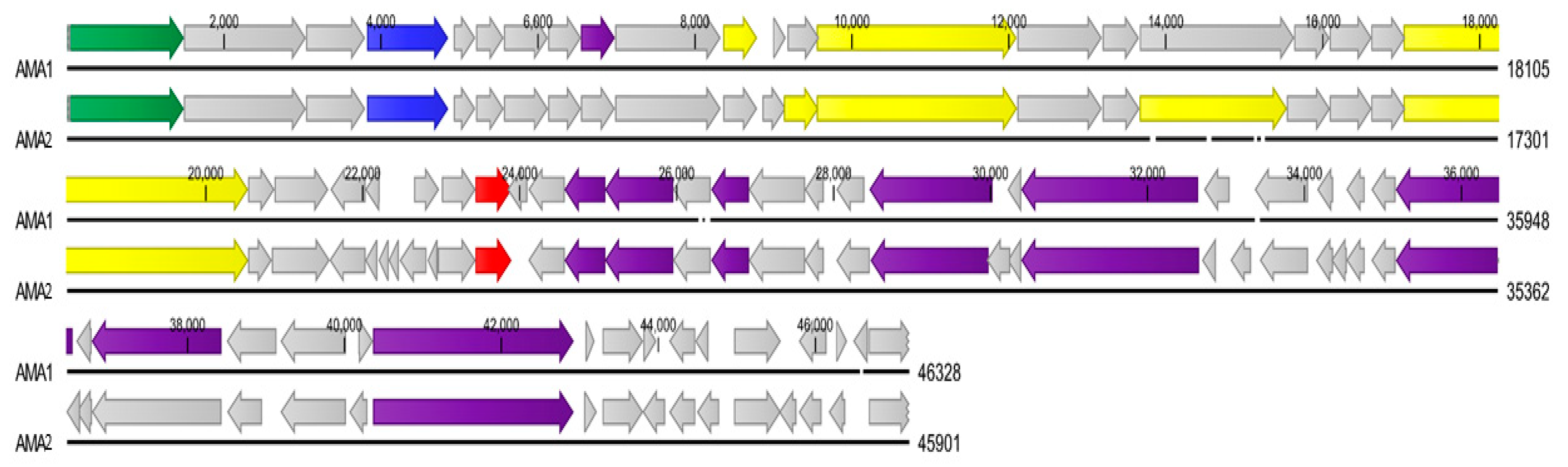
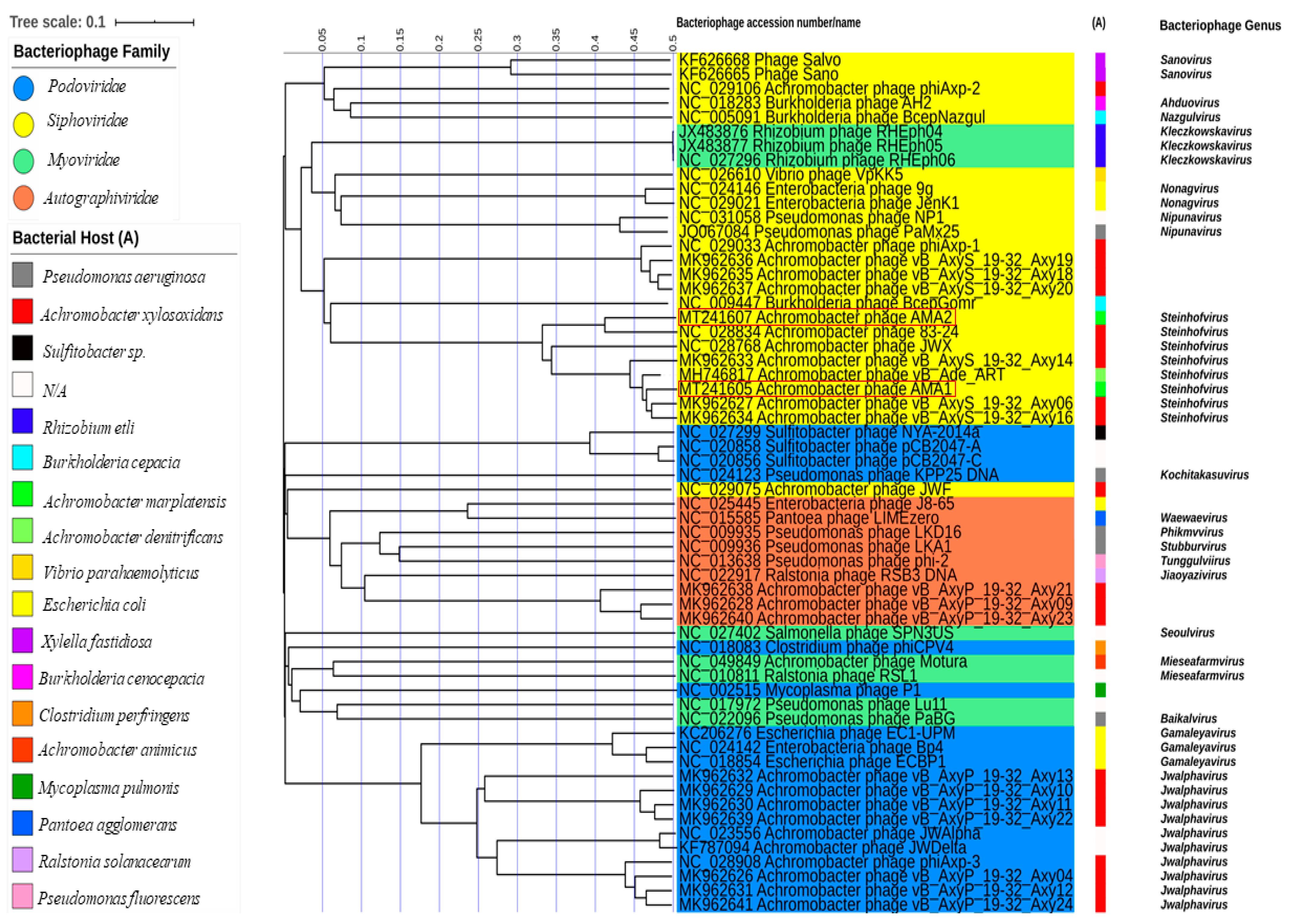
| Bacteriophage | Genome Size | GC Content | Accession Number | Closest Organism Match and Nucleotide Similarity (%) |
|---|---|---|---|---|
| AMA1 | 46,328 bp | 56.30% | MT241605 | Achromobacter phage vB_AxyS_19-32_Axy06, (91.3%) |
| AMA2 | 46,155 bp | 54.50% | MT241606 | Achromobacter phage 83-24, (79.8%) |
| Bacteriophage and ORF Number | Putative Protein Product | Pfam |
|---|---|---|
| AMA1_49 | Autophagy protein Apg6/FlgN protein | Pfam17675 |
| AMA1_5; AMA2_58 | Capsid protein* | |
| AMA1_39; AMA2_26 | DNA helicase | Pfam00271 |
| AMA1_41; AMA2_29 | DNA polymerase I | Pfam00476 |
| AMA1_53; AMA2_44 | DNA primase/polymerase | Pfam09250 |
| AMA1_2; AMA2_55 | Terminase large subunit | Pfam04466 |
| AMA1_12;AMA2_7 | Tail protein | Pfam13550 |
| AMA1_15; AMA2_68 | Tail length tape-measure protein | Pfam09718 |
| AMA1_29; AMA2_17 | Peptidase M15 | Pfam08291 |
| AMA1_32; AMA2_19 | Deoxycytidylate deaminase/MafB19-like deaminase | Pfam00383/Pfam14437 |
| AMA1_33;AMA2_20 | Thymidylate | Pfam00303 |
| AMA1_35; AMA2_22 | Phosphoribosyl-ATP pyrophosphohydrolase | Pfam01503 |
| AMA1_47; AMA2_37 | PD-(D/E)XK nuclease | Pfam12705 |
| AMA1_53; AMA2_44 | Primase C terminal 2 | Pfam08707 |
| AMA2_67 | Tail component | Pfam04883 |
| AMA2_3 | Bacteriophage tail collar protein | Pfam07484 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chan, H.T.; Ku, H.; Low, Y.P.; Batinovic, S.; Kabwe, M.; Petrovski, S.; Tucci, J. Characterization of Novel Lytic Bacteriophages of Achromobacter marplantensis Isolated from a Pneumonia Patient. Viruses 2020, 12, 1138. https://doi.org/10.3390/v12101138
Chan HT, Ku H, Low YP, Batinovic S, Kabwe M, Petrovski S, Tucci J. Characterization of Novel Lytic Bacteriophages of Achromobacter marplantensis Isolated from a Pneumonia Patient. Viruses. 2020; 12(10):1138. https://doi.org/10.3390/v12101138
Chicago/Turabian StyleChan, Hiu Tat, Heng Ku, Ying Ping Low, Steven Batinovic, Mwila Kabwe, Steve Petrovski, and Joseph Tucci. 2020. "Characterization of Novel Lytic Bacteriophages of Achromobacter marplantensis Isolated from a Pneumonia Patient" Viruses 12, no. 10: 1138. https://doi.org/10.3390/v12101138
APA StyleChan, H. T., Ku, H., Low, Y. P., Batinovic, S., Kabwe, M., Petrovski, S., & Tucci, J. (2020). Characterization of Novel Lytic Bacteriophages of Achromobacter marplantensis Isolated from a Pneumonia Patient. Viruses, 12(10), 1138. https://doi.org/10.3390/v12101138






