NMR Studies of Retroviral Genome Packaging
Abstract
1. Introduction
2. Protein Components Important for Genome Packaging
2.1. Nucleocapsid Domain of Gag
2.2. MA Domain of Gag
2.3. Multi-Domain Gag Fragments
3. RNA Components Important for Genome Packaging
3.1. HIV-1
3.1.1. DIS
3.1.2. Ψ-hairpin
3.1.3. AUG and U5:AUG
3.1.4. TAR
3.1.5. PBS
3.1.6. Core Encapsidation Signal of HIV-1
3.1.7. NMR Studies of the Intact HIV-1 Leader
3.2. Other Retroviruses
3.2.1. Moloney Murine Leukemia Virus (MoMuLV)
3.2.2. Rous Sarcoma Virus (RSV)
4. NC–RNA Interactions
4.1. HIV-1
4.2. Other Retroviruses
4.2.1. MoMuLV
4.2.2. Rous Sarcoma Virus
4.3. Chaperone Activity of NC
5. Inhibition of Genome Packaging
5.1. Inhibitors that Target NC
5.2. Inhibitors that Target the RNA Packaging Signal
6. NMR-Based Hybrid Approaches
7. Summary and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Berkowitz, R.; Fisher, J.; Goff, S.P. RNA packaging. Curr. Top. Microbiol. Immun. 1996, 214, 177–218. [Google Scholar]
- Rein, A. Retroviral RNA packaging: A review. Arch. Virol. 1994, 9, 513–522. [Google Scholar]
- Greatorex, J.; Lever, A. Retroviral RNA dimer linkage. J. Gen. Virol. 1998, 79, 2877–2882. [Google Scholar] [CrossRef] [PubMed]
- Paillart, J.-C.; Marquet, R.; Skripkin, E.; Ehresmann, C.; Ehresmann, B. Dimerization of retroviral genomic RNAs: Structural and functional implications. Biochimie 1996, 78, 639–653. [Google Scholar] [CrossRef]
- Jewell, N.A.; Mansky, L.M. In the beginning: Genome recognition, RNA encapsidation and the initiation of complex retrovirus assembly. J. Gen. Virol. 2000, 81, 1889–1899. [Google Scholar] [CrossRef]
- Paillart, J.-C.; Shehu-Xhilaga, M.; Marquet, R.; Mak, J. Dimerization of retroviral RNA genomes: An inseparable pair. Nat. Rev. Microbiol. 2004, 2, 461–472. [Google Scholar] [CrossRef]
- Russell, R.S.; Liang, C.; Wainberg, M.A. Is HIV-1 RNA dimerization a prerequisite for packaging? Yes, no, probably? Retrovirology 2004, 1, 23. [Google Scholar] [CrossRef]
- Greatorex, J. The retroviral RNA dimer linkage: Different structures may reflect different roles. Retrovirology 2004, 1, 22. [Google Scholar] [CrossRef]
- D’Souza, V.; Summers, M.F. How retroviruses select their genomes. Nat. Rev. Microbiol. 2005, 3, 643–655. [Google Scholar] [CrossRef]
- Johnson, S.F.; Telesnitsky, A. Retroviral RNA dimerization and packaging: The what, how, when, where, and why. PLoS Pathog. 2010, 6. [Google Scholar] [CrossRef]
- Kuzembayeva, M.; Dilley, K.; Sardo, L.; Hu, W.-S. Life of psi: How full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology 2014, 454, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Heng, X.; Summers, M.F. Structural determinants and mechanism of HIV-1 genome packaging. J. Mol. Biol. 2011, 410, 609–633. [Google Scholar] [CrossRef] [PubMed]
- Hellmund, C.; Lever, A.M. Coordination of genomic RNA packaging with viral assembly in HIV-1. Viruses 2016, 8, 192. [Google Scholar] [CrossRef] [PubMed]
- Mailler, E.; Bernacchi, S.; Marquet, R.; Paillart, J.C.; Vivet-Boudou, V.; Smyth, R.P. The life-cycle of the HIV-1 Gag-RNA complex. Viruses 2016, 8, 248. [Google Scholar] [CrossRef]
- Maldonado, R.K.; Parent, L.J. Orchestrating the selection and packaging of genomic RNA by retroviruses: An ensemble of viral and host factors. Viruses 2016, 8, 257. [Google Scholar] [CrossRef]
- Bieniasz, P.; Telesnitsky, A. Multiple, Switchable protein: RNA interactions regulate human immunodeficiency virus type 1 assembly. Annu. Rev. Virol. 2018, 5, 165–183. [Google Scholar] [CrossRef]
- Rein, A. RNA Packaging in HIV. Trends Microbiol. 2019, 27, 715–723. [Google Scholar] [CrossRef]
- Newman, J.L.; Butcher, E.W.; Patel, D.T.; Mikhaylenko, Y.; Summers, M.F. Flexibility in the P2 domain of the HIV-1 Gag polyprotein. Protein Sci. 2004, 13, 2101–2107. [Google Scholar] [CrossRef]
- Wright, E.R.; Schooler, J.B.; Ding, H.J.; Kieffer, C.; Fillmore, C.; Sundquist, W.I.; Jensen, G.J. Electron crytomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 2007, 26, 2218–2226. [Google Scholar] [CrossRef]
- Schur, F.K.; Hagen, W.J.; Rumlova, M.; Ruml, T.; Muller, B.; Krausslich, H.G.; Briggs, J.A. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 A resolution. Nature 2015, 517, 505–508. [Google Scholar] [CrossRef]
- Schur, F.K.; Obr, M.; Hagen, W.J.; Wan, W.; Jakobi, A.J.; Kirkpatrick, J.M.; Sachse, C.; Krausslich, H.G.; Briggs, J.A. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science 2016, 353, 506–508. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.M.; Zadrozny, K.K.; Chrustowicz, J.; Purdy, M.D.; Yeager, M.; Ganser-Pornillos, B.K.; Pornillos, O. Crystal structure of an HIV assembly and maturation switch. ELife 2016, 5, 5. [Google Scholar] [CrossRef]
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Dempsey, D.M.; Dutilh, B.E.; Harrach, B.; Harrison, R.L.; Hendrickson, R.C.; Junglen, S.; et al. Changes to virus taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses. Arch. Virol. 2019, 164, 2417–2429. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, T.A.; Panganiban, A.T. Simian immunodeficiency Virus-Rna is efficiently encapsidated by human-immunodeficiency-virus type-1 particles. J. Virol. 1993, 67, 2681–2688. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.D.; Hu, W.-S. RNAs from genetically distinct retroviruses can copackage and exchange genetic information in vivo. J. Virol. 1997, 71, 6237–6242. [Google Scholar] [CrossRef] [PubMed]
- Kaye, J.F.; Lever, A.M. Nonreciprocal packaging of human immunodeficiency virus type 1 and type 2 RNA: A possible role for the p2 domain of gag in RNA encapsidation. J. Virol. 1998, 72, 5877–5885. [Google Scholar] [CrossRef]
- Berkowitz, R.D.; Ohagen, A.; Hoglund, S.; Goff, S.P. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J. Virol. 1995, 69, 6445–6456. [Google Scholar] [CrossRef]
- Zhang, Y.; Barklis, E. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J. Virol. 1995, 69, 5716–5722. [Google Scholar] [CrossRef]
- Jouvenet, N.; Simon, S.M.; Bieniasz, P.D. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc. Natl. Acad. Sci. USA 2009, 106, 19114–19119. [Google Scholar] [CrossRef]
- Chen, J.; Rahman, S.A.; Nikolaitchik, O.A.; Grunwald, D.; Sardo, L.; Burdick, R.C.; Plisov, S.; Liang, E.; Tai, S.; Pathak, V.K.; et al. HIV-1 RNA genome dimerizes on the plasma membrane in the presence of Gag protein. Proc. Natl. Acad. Sci. USA 2016, 113, E201–E208. [Google Scholar] [CrossRef]
- Jouvenet, N.; Bieniasz, P.D.; Simon, S.M. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature 2008, 454, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Ganser-Pornillos, B.K.; Yeager, M.; Sundquist, W.I. The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 2008, 18, 203–217. [Google Scholar] [CrossRef] [PubMed]
- Sundquist, W.I.; Krausslich, H.G. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2012, 2, a006924. [Google Scholar] [CrossRef] [PubMed]
- Levin, J.G.; Grimley, P.M.; Ramseur, J.M.; Berezesky, I.K. Deficiency of 60 to 70S RNA in murine leukemia virus particles assembled in cells treated with actinomycin D. J. Virol. 1974, 14, 152–161. [Google Scholar] [CrossRef]
- Poon, D.T.K.; Wu, J.; Aldovini, A. Charged amino acid residues of human immunodeficiency virus type-1 nucleocapsid P7 protein involved in RNA pacakaging and infectivity. J. Virol. 1996, 70, 6607–6617. [Google Scholar] [CrossRef]
- Cimarelli, A.; Sandin, S.; Hoglund, S.; Luban, J. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 2000, 74, 3046–3057. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-W.; Aldovini, A. RNA incorporation is critical for retroviral particle integrity after cell membrane assembly of Gag complexes. J. Virol. 2002, 76, 11853–11865. [Google Scholar] [CrossRef] [PubMed]
- Muriaux, D.; Mirro, J.; Harvin, D.; Rein, A. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 2001, 98, 5246–5251. [Google Scholar] [CrossRef]
- Muriaux, D.; Mirro, J.; Nagashima, K.; Harvin, D.; Rein, A. Murine leukemia virus nucleocapsid mutant particles lacking viral RNA encapsidate ribosomes. J. Virol. 2002, 76, 11405–11413. [Google Scholar] [CrossRef]
- Heng, X.; Kharytonchyk, S.; Garcia, E.L.; Lu, K.; Divakaruni, S.S.; LaCotti, C.; Edme, K.; Telesnitsky, A.; Summers, M.F. Identification of a minimal region of the HIV-1 5’-leader required for RNA dimerization, NC binding, and packaging. J. Mol. Biol. 2012, 417, 224–239. [Google Scholar] [CrossRef]
- Keane, S.C.; Heng, X.; Lu, K.; Kharytonchyk, S.; Ramakrishnan, V.; Carter, G.; Barton, S.; Hosic, A.; Florwick, A.; Santos, J.; et al. Structure of the HIV-1 RNA packaging signal. Science 2015, 348, 917–921. [Google Scholar] [CrossRef]
- Kharytonchyk, S.; Brown, J.D.; Stilger, K.; Yasin, S.; Iyer, A.S.; Collins, J.; Summers, M.F.; Telesnitsky, A. Influence of gag and RRE sequences on HIV-1 RNA packaging signal structure and function. J. Mol. Biol. 2018, 430, 2066–2079. [Google Scholar] [CrossRef] [PubMed]
- Vogt, V.M. Retroviral virions and genomes. In Retroviruses; Coffin, J.M., Hughes, S.H., Varmus, H.E., Eds.; Cold Spring Harbor Laboratory Press: Plainview, NY, USA, 1997; Volume 1, pp. 27–69. [Google Scholar]
- Hu, W.-S.; Temin, H.M. Genetic consequences of packaging two RNA genomes in one retroviral particle: Pseudodiploidy and high rate of genetic recombination. Proc. Natl. Acad. Sci. USA 1990, 87, 1556–1560. [Google Scholar] [CrossRef]
- Hu, W.S.; Temin, H.M. Retroviral recombination and reverse transcription. Science 1990, 250, 1227–1233. [Google Scholar] [CrossRef] [PubMed]
- Onafuwa-Nuga, A.; Telesnitsky, A. The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol. Mol. Biol. Rev. 2009, 73, 451–480. [Google Scholar] [CrossRef] [PubMed]
- Bender, W.; Chien, Y.-H.; Chattopadhyay, S.; Vogt, P.K.; Gardner, M.B.; Davidson, N. High-molecular-weight RNAs of AKR, NZB and wild mouse viruses and avian reticuloendotheliosis virus all have similar dimer structures. J. Virol. 1978, 25, 888–896. [Google Scholar] [CrossRef]
- Kung, H.-J.; Hu, S.; Bender, W.; Baily, J.M.; Davidson, N.; Nicolson, M.O.; McAllister, R.M. RD-114, baboon and wolly monkey viral RNAs compared in size and structure. Cell 1976, 7, 609–620. [Google Scholar] [CrossRef]
- Maisel, J.; Bender, W.; Hu, S.; Duesberg, P.H.; Davidson, N. Structure of 50 to 70S RNA from Moloney sarcoma viruses. J. Virol. 1978, 25, 384–394. [Google Scholar] [CrossRef] [PubMed]
- Murti, K.G.; Bondurant, M.; Tereba, A. Secondary structural features in the 70S RNAs of Moloney murine leukemia and Rous sarcoma viruses as observed by electron microscopy. J. Virol. 1981, 37, 411–419. [Google Scholar] [CrossRef]
- Fu, W.; Rein, A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 1993, 67, 5443–5449. [Google Scholar] [CrossRef]
- Oritz-Conde, B.A.; Hughes, S.H. Studies of the genomic RNA of leukosis viruses: Implications for RNA dimerization. J. Virol. 1999, 73, 7165–7174. [Google Scholar] [CrossRef]
- Flynn, J.A.; Telesnitsky, A. Two distinct Moloney murine leukemia virus RNAs produced from a single locus dimerize at random. Virology 2006, 344, 391–400. [Google Scholar] [CrossRef]
- Onafuwa, A.; An, W.; Robson, N.D.; Telesnitsky, A. Human immunodeficiency virus Type 1 genetic recombination is more frequent than that of Moloney Murine Leukemia Virus despite similar template switching rates. J. Virol. 2003, 77, 4577–4587. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.A.; King, S.R.; Telesnitsky, A. Nonrandom dimerization of murine leukemia virus genomic RNAs. J. Virol. 2004, 78, 12129–12139. [Google Scholar] [CrossRef] [PubMed]
- Kharytonchyk, S.A.; Kireyeva, A.I.; Osipovich, A.B.; Fomin, I.K. Evidence for preferential copackaging of Moloney murine leukemia virus genomic RNAs transcribed in the same chromosomal site. Retrovirology 2005, 2, 3. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, S.V.; Pedersen, F.S. Co-localization of gammaretroviral RNAs at their transcription site favours co-packaging. J. Gen. Virol. 2006, 87, 2279–2289. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, T.; Wargo, H.; Hu, W.-S. High rates of human immunodeficiency virus type 1 recombination: Near-random segregation of markers one kilobase apart in one round of viral replication. J. Virol. 2003, 77, 11193–11200. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.D.; Nikolaitchik, O.A.; Chen, J.; Hammarskjold, M.L.; Rekosh, D.; Hu, W.S. Probing the HIV-1 genomic RNA trafficking pathway and dimerization by genetic recombination and single virion analyses. PLoS Pathog. 2009, 5, e1000627. [Google Scholar] [CrossRef] [PubMed]
- Coffin, J.M.; Hughes, S.H.; Varmus, H.E. Retroviruses; Cold Spring Harbor Laboratory Press: Plainview, NY, USA, 1997. [Google Scholar]
- Lever, A.M. HIV-1 RNA packaging. Adv. Pharmacol. 2007, 55, 1–32. [Google Scholar] [PubMed]
- Adam, M.A.; Miller, A.D. Identification of a signal in a murine retrovirus that is sufficient for packaging of nonretroviral RNA into virions. J. Virol. 1988, 62, 3802–3806. [Google Scholar] [CrossRef] [PubMed]
- Mougel, M.; Barklis, E. A role for two hairpin structures as a core RNA encapsidation signal in murine leukemia virus virions. J. Virol. 1997, 71, 8061–8065. [Google Scholar] [CrossRef] [PubMed]
- Aronoff, R.; Linial, M. Specificity of retroviral RNA packaging. J. Virol. 1991, 65, 71–80. [Google Scholar] [CrossRef]
- Aronoff, R.; Hajjar, A.M.; Linial, M.L. Avian retroviral RNA encapsidation: Reexamination of functional 5’ RNA sequences and the role of nucleocapsid Cys-His motifs. J. Virol. 1993, 67, 178–188. [Google Scholar] [CrossRef]
- Banks, J.D.; Yeo, A.; Green, K.; Cepeda, F.; Linial, M.L. A minimal avian retroviral packaging sequence has a complex structure. J. Virol. 1998, 72, 6190–6194. [Google Scholar] [CrossRef]
- Doria-Rose, N.A.; Vogt, V.M. In vivo selection of Rous sarcoma virus mutants with randomized sequences in the packaging signal. J. Virol. 1998, 72, 8073–8082. [Google Scholar] [CrossRef]
- Banks, J.D.; Kealoha, B.O.; Linial, M.L. An MY containing heterologous RNA, but not env mRNA, is efficiently packaged into avian retroviral particles. J. Virol. 1999, 73, 8926–8933. [Google Scholar] [CrossRef] [PubMed]
- Banks, J.D.; Linial, M.L. Secondary structure analysis of a minimal avian leukosis-sarcoma virus packaging signal. J. Virol. 2000, 74, 456–464. [Google Scholar] [CrossRef] [PubMed]
- Prats, A.-C.; Roy, C.; Wang, P.; Erard, M.; Housset, V.; Gabus, C.; Paoletti, C.; Darlix, J.-L. Cis elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. J. Virol. 1990, 64, 774–783. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, V.; Summers, M.F. Structural basis for packaging the dimeric genome of Moloney Murine Leukaemia Virus. Nature 2004, 431, 586–590. [Google Scholar] [CrossRef]
- Miyazaki, Y.; Garcia, E.; King, S.R.; Iyalla, K.; Loeliger, K.; Starck, P.; Syed, S.; Telesnitsky, A.; Summers, M.F. An RNA structural switch regulates diploid genome packaging by Moloney Murine leukemia virus. J. Mol. Biol. 2010, 396, 141–152. [Google Scholar] [CrossRef]
- Gherghe, C.; Lombo, T.; Leonard, C.W.; Datta, S.A.; Bess, J.W.; Gorelick, R.J.; Rein, A.; Weeks, K.M. Definition of a high-affinity Gag recognition structure mediating packaging of a retroviral RNA genome. Proc. Natl. Acad. Sci. USA 2010, 107, 19248–19253. [Google Scholar] [CrossRef]
- Darlix, J.-L.; Gabus, C.; Nugeyre, M.-T.; Clavel, F.; Barre-Sinoussi, F. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J. Mol. Biol. 1990, 216, 689–699. [Google Scholar] [CrossRef]
- Ooms, M.; Huthoff, H.; Russell, R.; Liang, C.; Berkhout, B. A riboswitch regulates RNA dimerization and packaging in human immunodeficiency virus type 1 virions. J. Virol. 2004, 78, 10814–10819. [Google Scholar] [CrossRef]
- Lu, K.; Heng, X.; Garyu, L.; Monti, S.; Garcia, E.; Kharytonchyk, S.; Dorjsuren, B.; Kulandaivel, G.; Jones, S.; Hiremath, A.; et al. NMR detection of structures in the HIV-1 5´-leader RNA that regulate genome packaging. Science 2011, 344, 242–245. [Google Scholar] [CrossRef]
- Masuda, T.; Sato, Y.; Huang, Y.L.; Koi, S.; Takahata, T.; Hasegawa, A.; Kawai, G.; Kannagi, M. Fate of HIV-1 cDNA intermediates during reverse transcription is dictated by transcription initiation site of virus genomic RNA. Sci. Rep. 2015, 5, 17680. [Google Scholar] [CrossRef]
- Kharytonchyk, S.; Monti, S.; Smaldino, P.J.; Van, V.; Bolden, N.C.; Brown, J.D.; Russo, E.; Swanson, C.; Shuey, A.; Telesnitsky, A.; et al. Transcriptional start site heterogeneity modulates the structure and function of the HIV-1 genome. Proc. Natl. Acad. Sci. USA 2016, 113, 13378–13383. [Google Scholar] [CrossRef] [PubMed]
- Esquiaqui, J.M.; Kharytonchyk, S.; Drucker, D.; Telesnitsky, A. HIV-1 spliced RNAs display transcription start site bias. RNA 2020, 26, 708–714. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.D.; Kharytonchyk, S.; Chaudry, I.; Iyer, A.S.; Carter, H.; Becker, G.; Desai, Y.; Glang, L.; Choi, S.H.; Singh, K.; et al. Structural basis for transcriptional start site control of HIV-1 RNA fate. Science 2020, 368, 413–417. [Google Scholar] [PubMed]
- Turner, B.G.; Summers, M.F. Structural Biology of HIV. J. Mol. Biol. 1999, 285, 1–32. [Google Scholar] [CrossRef]
- Wüthrich, K. NMR of Proteins and Nucleic Acids; John Wiley & Sons: New York, NY, USA, 1986. [Google Scholar]
- Allain, F.H.-T.; Varani, G. How accurately and precisely can RNA structure be determined by NMR? J. Mol. Biol. 1997, 267, 338–351. [Google Scholar] [CrossRef] [PubMed]
- Lukavsky, P.J.; Puglisi, J.D. Structure determination of large biological RNAs. Methods Enzymol. 2005, 394, 399–415. [Google Scholar] [PubMed]
- Lu, K.; Miyazaki, Y.; Summers, M.F. Isotope labeling strategies for NMR studies of RNA. J. Biomol. NMR 2010, 46, 113–125. [Google Scholar] [CrossRef]
- Kotar, A.; Foley, H.N.; Baughman, K.M.; Keane, S.C. Advanced approaches for elucidating structures of large RNAs using NMR spectroscopy and complementary methods. Methods 2020. [Google Scholar] [CrossRef]
- Summers, M.F.; Henderson, L.E.; Chance, M.R.; Bess, J.W.J.; South, T.L.; Blake, P.R.; Sagi, I.; Perez-Alvarado, G.; Sowder, R.C.I.; Hare, D.R.; et al. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1992, 1, 563–574. [Google Scholar] [CrossRef]
- Henderson, L.E.; Copeland, T.D.; Sowder, R.C.; Smythers, G.W.; Oroslzan, S. Primary structure of the low molecular weight nucleic acid-binding proteins of murine leukemia viruses. J. Biol. Chem. 1981, 256, 8400–8406. [Google Scholar]
- Miller, J.; McLachlan, A.D.; Klug, A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985, 4, 1609–1614. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.M. Potential metal-binding domains in nucleic acid binding proteins. Science 1986, 232, 485–487. [Google Scholar] [CrossRef] [PubMed]
- Schiff, L.A.; Nibert, M.L.; Fields, B.N. Characterization of a zinc blotting technique: Evidence that a retroviral gag protein binds zinc. Proc. Natl. Acad. Sci. USA 1988, 85, 4195–4199. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.M.; Jentoft, J.E. Conserved cysteine and histidine residues in avian myeloblastosis virus nucleocapsid protein pp12 are not zinc binding ligands. Biophys. J. 1988, 53, 295a. [Google Scholar]
- Gorelick, R.J.; Henderson, L.E.; Hanser, J.P.; Rein, A. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: Evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc. Natl. Acad. Sci. USA 1988, 85, 8420–8424. [Google Scholar] [CrossRef] [PubMed]
- Méric, C.; Goff, S.P. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. J. Virol. 1989, 63, 1558–1568. [Google Scholar] [CrossRef]
- Green, L.M.; Berg, J.M. A retroviral Cys-Xaa2-Cys-Xaa4-His-Xaa4-Cys peptide binds metal ions: Spectroscopic studies and a proposed three-dimensional structure. Proc. Natl. Acad. Sci. USA 1989, 86, 4047–4051. [Google Scholar] [CrossRef]
- South, T.L.; Kim, B.; Summers, M.F. 113Cd NMR studies of a 1:1 Cd adduct with an 18-residue zinc finger peptide from HIV-1 nucleic acid binding protein, p7. J. Am. Chem. Soc. 1989, 111, 395–396. [Google Scholar] [CrossRef]
- Summers, M.F.; South, T.L.; Kim, B.; Hare, D.R. High-resolution structure of an HIV zinc fingerlike domain via a new NMR-based distance geometry approach. Biochemistry 1990, 29, 329–340. [Google Scholar] [CrossRef]
- South, T.L.; Blake, P.R.; Sowder, R.C., 3rd; Arthur, L.O.; Henderson, L.E.; Summers, M.F. The nucleocapsid protein isolated from HIV-1 particles binds zinc and forms retroviral-type zinc fingers. Biochemistry 1990, 29, 7786–7789. [Google Scholar] [CrossRef]
- Chance, M.R.; Sagi, I.; Wirt, M.D.; Frisbie, S.M.; Scheuring, E.; Chen, E.; Bess, J.W.; Henderson, L.E.; Arthur, L.O.; South, T.L. Extended x-ray absorption fine structure studies of a retrovirus: Equine infectious anemia virus cysteine arrays are coordinated to zinc. Proc. Natl. Acad. Sci. USA 1992, 89, 10041–10045. [Google Scholar] [CrossRef] [PubMed]
- South, T.L.; Blake, P.R.; Hare, D.R.; Summers, M.F. C-terminal retroviral-type zinc finger domain from the HIV-1 nucleocapsid protein is structurally similar to the N-terminal zinc finger domain. Biochemistry 1991, 30, 6342–6349. [Google Scholar] [CrossRef] [PubMed]
- Omichinski, J.G.; Clore, G.M.; Sakaguchi, K.; Appella, E.; Gronenborn, A.M. Structural characterization of a 39-residue synthetic peptide containing the two zinc binding domains from the HIV-1 p7 nucleocapsid protein by CD and NMR spectroscopy. Fed. Eur. Biochem. Soc. Lett. 1991, 292, 25–30. [Google Scholar]
- Déméné, H.; Dong, C.Z.; Ottmann, M.; Rouyez, M.C.; Jullian, N.; Morellet, N.; Mely, Y.; Darlix, J.L.; Fournié-Zaluski, M.C.; Saragosti, S.; et al. 1H NMR structure and biological studies of the His23 to Cys mutant nucleocapsid protein of HIV-1 indicate that the conformation of the first zinc finger is critial for virus infectivity. Biochemistry 1994, 33, 11707–11716. [Google Scholar] [CrossRef]
- Klein, D.; Johnson, P.E.; Zollars, E.S.; de Guzman, R.N.; Summers, M.F. The NMR structure of the nucleocapsid protein from the mouse mammary tumor virus reveals unusual folding of the C-terminal zinc knuckle. Biochemistry 2000, 39, 1604–1612. [Google Scholar] [CrossRef]
- Bertola, F.; Manigand, C.; Picard, P.; Belghazi, M.; Precigoux, G. Human T-lymphotrophic virus type I nucleocapsid protein NCp15: Structural study and stability of the N-terminal zinc-finger. Biochem. J. 2000, 352, 293–300. [Google Scholar] [CrossRef]
- Mély, Y.; Piémont, E.; Sorinas-Jimeno, M.; de Rocquigny, H.; Julian, N.; Morellet, N.; Roques, B.P.; Gérard, D. Structural and dynamic characterization of the aromatic amino acids of the human immunodeficiency virus type 1 nucleocapsid protein zinc fingers and their involvement in heterologous tRNAPhe binding: A steady-state and time resolved flourescence study. Biophys. J. 1993, 65, 1513–1522. [Google Scholar] [CrossRef][Green Version]
- Déméné, H.; Jullian, N.; Morellet, N.; de Rocquigny, H.; Cornille, F.; Maigret, B.; Roques, B.P. Three-dimensional 1H NMR structure of the nucleocapsid protein NCp10 of Moloney murine leukemia virus. J. Biomol. NMR 1994, 4, 153–170. [Google Scholar] [CrossRef]
- Morellet, N.; Jullian, N.; de Rocquigny, H.; Maigret, B.; Darlix, J.-L.; Roques, B.P. Determination of the structure of the nucleocapsid protein NCp7 from the human immunodeficiency virus type 1 by 1H NMR. EMBO J. 1992, 11, 3059–3065. [Google Scholar] [CrossRef]
- Morellet, N.; Meudal, H.; Bouaziz, S.; Roques, B.P. Structure of the zinc finger domain encompassing residues 13–51 of the nucleocapsid protein from simian immunodeficiency virus. Biochem. J. 2006, 393, 725–732. [Google Scholar] [CrossRef]
- Kodera, Y.; Sato, K.; Tsukahara, T.; Ksmatsu, H.; Maeda, T.; Kohno, T. High-resolution solution NMR structure of the minimal active domain of the human immunodeficiency virus type-2 nucleocapsid protein. Biochemistry 1998, 37, 17704–17713. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Boyd, J.; Pielak, G.J.; Williams, R.J.P. Proton nuclear magnetic resonance as a probe of differences in structure between the C102T and F82S, C102T Variants of Iso-1-cytochrome c from the Yeast Saccharomyces cerevisiae. Biochemistry 1991, 30, 7033–7040. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Kaluarachchi, K.; Giedroc, D.P. Solution structure and backbone dynamics of Mason-Pfizer monkey virus (MPMV) nucleocapsid protein. Protein Sci. 1998, 7, 2265–2280. [Google Scholar] [CrossRef] [PubMed]
- South, T.L.; Summers, M.F. Zinc- and sequence-dependent binding to nucleic acids by the N-terminal zinc finger of the HIV-1 nucleocapsid protein: NMR structure of the complex with the Psi-site analog, dACGCC. Protein Sci. 1993, 2, 3–19. [Google Scholar] [CrossRef]
- De Guzman, R.N.; Turner, R.B.; Summers, M.F. Protein-RNA recognition. Biopolymers 1999, 48, 181–195. [Google Scholar] [CrossRef]
- De Guzman, R.N.; Wu, Z.R.; Stalling, C.C.; Pappalardo, L.; Borer, P.N.; Summers, M.F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 Y-RNA recognition element. Science 1998, 279, 384–388. [Google Scholar] [CrossRef]
- Amarasinghe, G.K.; De Guzman, R.N.; Turner, R.B.; Chancellor, K.; Wu, Z.-R.; Summers, M.F. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the Y-RNA packaging signal. J. Mol. Biol. 2000, 301, 491–511. [Google Scholar] [CrossRef]
- Morellet, N.; Demene, H.; Teilleux, V.; Huynh-Dinh, T.; de Rocquigny, H.; Fournie-Zaluski, M.C.; Roques, B.P. Structure of the complex between the HIV-1 nucleocapsid protein NCp7 and the single-stranded pentanucleotide d(ACGCC). J. Mol. Biol. 1998, 283, 419–434. [Google Scholar] [CrossRef]
- Zhou, J.; Bean, R.L.; Vogt, V.M.; Summers, M.F. Solution structure of the Rous sarcoma virus nucleocapsid protein:uY RNA packaging signal complex. J. Mol. Biol. 2007, 365, 453–467. [Google Scholar] [CrossRef]
- Dey, A.; York, D.; Smalls-Mantey, A.; Summers, M.F. Composition and sequence dependent binding of RNA to the nucleocapsid protein of Moloney Murine Leukemia Virus. Biochemistry 2005, 44, 3735–3744. [Google Scholar] [CrossRef]
- Spriggs, S.; Garyu, L.; Connor, R.; Summers, M.F. Potential intra- and intermolecular interactions involving the Unique-5′ Region of the HIV-1 5′-UTR. Biochemistry 2008, 46, 13064–13073. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, B.M.; de Guzman, R.N.; Turner, B.G.; Tjandra, N.; Summers, M.F. Dynamical behavior of the HIV-1 nucleocapsid protein. J. Mol. Biol. 1998, 279, 633–649. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, L.; Ghirlando, R.; Clore, G.M. Conformation and dynamics of the Gag polyprotein of the human immunodeficiency virus 1 studied by NMR spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, 3374–3379. [Google Scholar] [CrossRef] [PubMed]
- Matthews, S.; Barlow, P.; Boyd, J.; Barton, G.; Russell, R.; Mills, H.; Cunningham, M.; Meyers, N.; Burns, N.; Clark, N.; et al. Structural similarity between the p17 matrix protein of HIV-1 and interferon-g. Nature 1994, 370, 666–668. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Ndassa, Y.; Summers, M.F. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat. Struct. Biol. 2002, 9, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Loeliger, E.; Luncsford, P.; Kinde, I.; Beckett, D.; Summers, M.F. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc. Natl. Acad. Sci. USA 2004, 101, 517–522. [Google Scholar] [CrossRef]
- Saad, J.S.; Loeliger, E.; Luncsford, P.; Liriano, M.; Tai, J.; Kim, A.; Miller, J.; Joshi, A.; Freed, E.O.; Summers, M.F. Point mutations in the HIV-1 matrix protein turn off the myristyl switch. J. Mol. Biol. 2007, 366, 574–585. [Google Scholar] [CrossRef]
- Saad, J.S.; Ablan, S.D.; Ghanam, R.H.; Kim, A.; Andrews, K.; Nagashima, K.; Soheilian, F.; Freed, E.O.; Summers, M.F. Structure of the myristylated human immunodeficiency virus type 2 matrix protein and the role of phosphatidylinositol-(4,5)-bisphosphate in membrane targeting. J. Mol. Biol. 2008, 382, 434–447. [Google Scholar] [CrossRef]
- Mercredi, P.Y.; Bucca, N.; Loeliger, B.; Gaines, C.R.; Mehta, M.; Bhargava, P.; Tedbury, P.R.; Charlier, L.; Floquet, N.; Muriaux, D.; et al. Structural and molecular determinants of membrane binding by the HIV-1 matrix protein. J. Mol. Biol. 2016, 428, 1637–1655. [Google Scholar] [CrossRef]
- Saad, J.S.; Miller, J.; Tai, J.; Kim, A.; Ghanam, R.H.; Summers, M.F. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. USA 2006, 103, 11364–11369. [Google Scholar] [CrossRef]
- Saad, J.S.; Kim, A.; Ghanam, R.H.; Dalton, A.K.; Vogt, M.V.; Wu, Z.; Lu, W.; Summers, M.F. Mutations that mimic phosphorylation of the HIV-1 matrix protein: Implications for trafficking. Protein Sci. 2007, 16, 1793–1797. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.E.; Smal, A.B.; Blach, J.; Mas, V.; Prevelige, P.E.; Saad, J.S. Structural and biophysical characterizations of HIV-1 matrix trimer binding to lipid nanodiscs shed light on virus assembly. J. Biol. Chem. 2019, 294, 18600–18612. [Google Scholar] [CrossRef] [PubMed]
- Gaines, C.R.; Tkacik, E.; Rivera-Oven, A.; Somani, P.; Achimovich, A.; Alabi, T.; Zhu, A.; Getachew, N.; Yang, A.L.; McDonough, M.; et al. HIV-1 Matrix protein interactions with tRNA: Implications for membrane targeting. J. Mol. Biol. 2018, 430, 2113–2127. [Google Scholar] [CrossRef] [PubMed]
- Jouvenet, N.; Simon, S.M.; Bieniasz, P.D. Visualizing HIV-1 assembly. J. Mol. Biol. 2011, 410, 501–511. [Google Scholar] [CrossRef]
- Freed, E.O. HIV-1 assembly, release and maturation. Nat. Rev. Microbiol. 2015, 13, 484–496. [Google Scholar] [CrossRef]
- Carlson, L.A.; Briggs, J.A.; Glass, B.; Riches, J.D.; Simon, M.N.; Johnson, M.C.; Muller, B.; Grunewald, K.; Krausslich, H.G. Three-dimensional analysis of budding sites and released virus suggests a revised model for HIV-1 morphogenesis. Cell Host Microbe 2008, 4, 592–599. [Google Scholar] [CrossRef]
- Massiah, M.A.; Starich, M.R.; Paschall, C.; Summers, M.F.; Christensen, A.M.; Sundquist, W.I. Three dimensional structure of the human immunodeficiency virus type 1 matrix protein. J. Mol. Biol. 1994, 244, 198–223. [Google Scholar] [CrossRef]
- Matthews, S.; Barlow, P.; Clark, N.; Kingsman, S.; Kingsman, A.; Campbell, I. Refined solution structure of p17, the HIV matrix protein. Biochem. Soc. Trans. 1995, 23, 725–728. [Google Scholar] [CrossRef]
- Hill, C.P.; Worthylake, D.; Bancroft, D.P.; Christensen, A.M.; Sundquist, W.I. Crystal Structures of the Trimeric HIV-1 Matrix Protein: Implications for Membrane Association. Proc. Natl. Acad. Sci. USA 1996, 93, 3099–3104. [Google Scholar] [CrossRef]
- Massiah, M.A.; Worthylake, D.; Christensen, A.M.; Sundquist, W.I.; Hill, C.P.; Summers, M.F. Comparison of the NMR and X-ray structures of the HIV-1 matrix protein: Evidence for conformational changes during viral assembly. Protein Sci. 1996, 5, 2391–2398. [Google Scholar] [CrossRef]
- Freed, E.O.; Martin, A.M. Domains of the human immonodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J. Virol. 1996, 70, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Tedbury, P.R.; Novikova, M.; Alfadhli, A.; Hikichi, Y.; Kagiampakis, I.; KewalRamani, V.N.; Barklis, E.; Freed, E.O. HIV-1 Matrix trimerization-impaired mutants are rescued by matrix substitutions that enhance envelope glycoprotein incorporation. J. Virol. 2019, 94, 1. [Google Scholar] [CrossRef]
- Tedbury, P.R.; Novikova, M.; Ablan, S.D.; Freed, E.O. Biochemical evidence of a role for matrix trimerization in HIV-1 envelope glycoprotein incorporation. Proc. Natl. Acad. Sci. USA 2016, 113, E182–E190. [Google Scholar] [CrossRef] [PubMed]
- Freed, E.O.; Englund, G.; Martin, A.M. Role of the basic domain of human immondeficiency virus type 1 matrix in macrophage infection. J. Virol. 1995, 69, 3949–3954. [Google Scholar] [CrossRef] [PubMed]
- Deichaite, I.; Casson, L.P.; Ling, H.P.; Resh, M.D. In vitro synthesis of pp60v-src: Myristylation in a cell-free system. Mol. Cell Biol. 1988, 8, 4295–4301. [Google Scholar] [CrossRef]
- Brown, L.A.; Cox, C.; Baptiste, J.; Summers, H.; Button, R.; Bahlow, K.; Spurrier, V.; Kyser, J.; Luttge, B.G.; Kuo, L.; et al. NMR structure of the myristylated feline immunodeficiency virus matrix protein. Viruses 2015, 7, 2210–2229. [Google Scholar] [CrossRef]
- Brown, J.B.; Summers, H.R.; Brown, L.A.; Marchant, J.; Canova, P.N.; O’Hern, C.T.; Abbott, S.T.; Nyaunu, C.; Maxwell, S.; Johnson, T.; et al. Structural and Mechanistic studies of the rare myristoylation signal of the feline immunodeficiency virus. J. Mol. Biol. 2020, 432, 4076–4091. [Google Scholar] [CrossRef]
- Fledderman, E.L.; Fujii, K.; Ghanam, R.H.; Waki, K.; Prevelige, P.E.; Freed, E.O.; Saad, J.S. Myristate exposure in the human immunodeficiency virus type 1 matrix protein is modulated by pH. Biochemistry 2010, 49, 9551–9562. [Google Scholar] [CrossRef]
- Ono, A.; Ablan, S.D.; Lockett, S.J.; Nagashima, K.; Freed, E.O. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl. Acad. Sci. USA 2004, 101, 14889–14894. [Google Scholar] [CrossRef]
- Zimmerman, C.; Klein, K.C.; Kiser, P.K.; Singh, A.R.; Bonnie, F.L.; Riba, S.C.; Lingappa, J.R. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature 2002, 415, 88–92. [Google Scholar] [CrossRef]
- Behnia, R.; Munro, S. Organelle identity and the signposts for membrane traffic. Nature 2005, 438, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Brugger, B.; Glass, B.; Haberkant, P.; Leibrecht, I.; Wieland, F.T.; Krausslich, H.G. The HIV lipidome: A raft with an unusual composition. Proc. Natl. Acad. Sci. USA 2006, 103, 2641–2646. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.H.; Hildreth, J.E. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 2000, 74, 3264–3272. [Google Scholar] [CrossRef] [PubMed]
- Vlach, J.; Saad, J.S. Trio engagement via plasma membrane phospholipids and the myristoyl moiety governs HIV-1 matrix binding to bilayers. Proc. Natl. Acad. Sci. USA 2013, 110, 3525–3530. [Google Scholar] [CrossRef] [PubMed]
- Cannon, P.M.; Matthews, S.; Clark, N.; Byles, E.D.; Iourin, O.; Hockley, D.J.; Kingsman, S.; Kingsman, A. Structure-Function studies of the Human immunodefeciency virus type 1 matrix protein, p17. J. Virol. 1997, 71, 3474–3483. [Google Scholar] [CrossRef]
- Kutluay, S.B.; Zang, T.; Blanco-Melo, D.; Powell, C.; Jannain, D.; Errando, M.; Bieniasz, P.D. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell 2014, 159, 1096–1109. [Google Scholar] [CrossRef]
- Bénas, P.; Bec, G.; Keith, G.; Marquet, R.; Ehresmann, C.; Ehresmann, B.; Dumas, P. The crystal structure of HIV reverse-transcription primer tRNA(Lys,3) shows a canonical anticodon loop. RNA 2000, 6, 1347–1355. [Google Scholar] [CrossRef]
- Kleiman, L.; Jones, C.; Musier-Forsyth, K. Formation of the tRNALys packaging complex in HIV-1. FEBS Lett. 2010, 584, 359–365. [Google Scholar] [CrossRef]
- Kleiman, L. tRNA(Lys3): The primer tRNA for reverse transcription in HIV-1. IUBMB Life 2002, 53, 107–114. [Google Scholar] [CrossRef]
- Chukkapalli, V.; Oh, S.J.; Ono, A. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc. Natl. Acad. Sci. USA 2010, 107, 1600–1605. [Google Scholar] [CrossRef]
- Gamble, T.R.; Yoo, S.; Vajdos, F.F.; von Schwedler, U.K.; Korthylake, D.K.; Wang, H.; McCutcheon, J.P.; Sundquist, W.I.; Hill, C.P. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 1997, 278, 849–853. [Google Scholar] [CrossRef] [PubMed]
- Pervushin, K.; Riek, R.; Wider, G.; Wüthrich, K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. USA 1997, 94, 12366–12371. [Google Scholar] [CrossRef] [PubMed]
- Harrison, G.P.; Lever, A.M.L. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J. Virol. 1992, 66, 4144–4153. [Google Scholar] [CrossRef] [PubMed]
- Baudin, F.; Marquet, R.; Isel, C.; Darlix, J.-L.; Ehresmann, B.; Ehresmann, C. Functional sites in the 5’ region of human immunodeficiency virus type 1 RNA form defined structural domains. J. Mol. Biol. 1993, 229, 382–397. [Google Scholar] [CrossRef] [PubMed]
- Clever, J.; Sassetti, C.; Parslow, T.G. RNA secondary structure and binding sites for gag gene products in the 5’ packaging signal of human immunodeficiency virus type 1. J. Virol. 1995, 69, 2101–2109. [Google Scholar] [CrossRef] [PubMed]
- McBride, M.S.; Panganiban, A.T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J. Virol. 1996, 70, 2963–2973. [Google Scholar] [CrossRef] [PubMed]
- Clever, J.L.; Miranda, J.D.; Parslow, T.G. RNA structure and packaging signals in the 5’ leader region of the human immunodeficiency virus type 1 genome. J. Virol. 2002, 76, 12381–12387. [Google Scholar] [CrossRef]
- Abbink, T.E.M.; Berkhout, B. A novel long distance base-pairing interaction in Human Immunodeficiency Virus Type 1 RNA occludes the Gag start codon. J. Biol. Chem. 2003, 278, 11601–11611. [Google Scholar] [CrossRef]
- Damgaard, C.K.; Andersen, E.S.; Knudsen, B.; Gorodkin, J.; Kjems, J. RNA interactions in the 5’ region of the HIV-1 genome. J. Mol. Biol. 2004, 336, 369–379. [Google Scholar] [CrossRef]
- Paillart, J.C.; Dettenhofer, M.; Yu, X.-F.; Ehresmann, C.; Ehresmann, B.; Marquet, R. First snapshots of the HIV-1 RNA structure in infected cells and in virions. J. Biol. Chem. 2004, 279, 48397–48403. [Google Scholar] [CrossRef]
- Wilkinson, K.A.; Gorelick, R.J.; Vasa, S.M.; Guex, N.; Rein, A.; Mathews, D.H.; Giddings, M.C.; Weeks, K.M. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008, 6, 883–899. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.M.; Dang, K.K.; Gorelick, R.J.; Leonard, C.W.; Bess, J.W., Jr.; Swanstrom, R.; Burch, C.L.; Weeks, K.M. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 2009, 460, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Carroll, R.; Peterlin, B.M.; Derse, D. Inhibition of human immunodeficiency virus type 1 Tat activity by coexpression of heterologous trans activators. J. Virol. 1992, 66, 2000–2007. [Google Scholar] [CrossRef] [PubMed]
- Boeras, I.; Seufzer, B.; Brady, S.; Rendahl, A.; Heng, X.; Boris-Lawrie, K. The basal translation rate of authentic HIV-1 RNA is regulated by 5’UTR nt-pairings at junction of R and U5. Sci. Rep. 2017, 7, 6902. [Google Scholar] [CrossRef] [PubMed]
- Sleiman, D.; Barraud, P.; Brachet, F.; Tisne, C. The Interaction between tRNA(Lys) 3 and the primer activation signal deciphered by NMR spectroscopy. PLoS ONE 2013, 8, e64700. [Google Scholar] [CrossRef]
- Sleiman, D.; Goldschmidt, V.; Barraud, P.; Marquet, R.; Paillart, J.C.; Tisne, C. Initiation of HIV-1 reverse transcription and functional role of nucleocapsid-mediated tRNA/viral genome interactions. Virus Res. 2012, 169, 324–339. [Google Scholar] [CrossRef]
- Rhim, H.; Park, J.; Morrow, C.D. Deletion in the tRNALys primer-binding site of human immunodeficiency virus type 1 identity essential regions for reverse transcription. J. Virol. 1991, 65, 4555–4564. [Google Scholar] [CrossRef]
- Abbink, T.E.M.; Berkhout, B. RNA structure modulates splicing efficiency at the human immunodeficiency virus type 1 major splice donor. J. Virol. 2008, 82, 3090–3098. [Google Scholar] [CrossRef]
- Plank, T.D.; Whitehurst, J.T.; Cencic, R.; Pelletier, J.; Kieft, J.S. Internal translation initiation from HIV-1 transcripts is conferred by a common RNA structure. Translation 2014, 2, e27694. [Google Scholar] [CrossRef]
- Hayashi, T.; Shioda, T.; Iwakura, Y.; Shibuta, H. RNA packaging signal of human immunodeficiency virus type 1. Virology 1992, 188, 590–599. [Google Scholar] [CrossRef]
- Hayashi, T.; Ueno, Y.; Okamoto, T. Elucidation of a conserved RNA stem-loop structure in the packaging signal of human immunodeficiency virus type 1. FEBS Lett. 1993, 327, 213–218. [Google Scholar] [CrossRef]
- McBride, M.S.; Panganiban, A.T. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J. Virol. 1997, 71, 2050–2058. [Google Scholar] [CrossRef] [PubMed]
- Harrison, G.P.; Miele, G.; Hunter, E.; Lever, A.M.L. Functional analysis of the core human immunodeficiency virus type 1 packaging signal in a permissive cell line. J. Virol. 1998, 72, 5886–5896. [Google Scholar] [CrossRef]
- Clever, J.L.; Parslow, T.G. Mutant Human Immunodeficiency Virus type 1 genomes with defects in RNA dimerization or encapsidation. J. Virol. 1997, 71, 3407–3414. [Google Scholar] [CrossRef] [PubMed]
- Berkhout, B. Structural features in TAR RNA of human and simian immunodeficiency viruses: A phylogenetic analysis. Nucleic Acids Res. 1992, 20, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Berkhout, B.; Klaver, B.; Das, A.T. A conserved hairpin structure predicted for the poly(A) signal of human and simian immunodeficiency viruses. Virology 1995, 207, 276–281. [Google Scholar] [CrossRef][Green Version]
- Skripkin, E.; Paillart, J.C.; Marquet, R.; Ehresmann, B.; Ehresmann, C. Identification of the primary site of the human immunodefieiency virus type 1 RNA dimerization in vitro. Proc. Natl. Acad. Sci. USA 1994, 91, 4945–4949. [Google Scholar] [CrossRef]
- Nikolaitchik, O.; Rhodes, T.D.; Ott, D.; Hu, W.-S. Effects of mutations in the Human Immunodeficiency Virus Type 1 gag gene on RNA packaging and recombination. J. Virol. 2006, 80, 4691–4697. [Google Scholar] [CrossRef]
- Song, R.; Kafaie, J.; Laughrea, M. Role of the 5’ TAR stem–loop and the U5-AUG duplex in dimerization of HIV-1 genomic RNA. Biochemistry 2008, 47, 3283–3293. [Google Scholar] [CrossRef]
- Sakuragi, J.-I.; Shioda, T.; Panganiban, A.T. Duplication of the primary encapsidation and dimer linkage region of Human immunodeficiency virus type 1 RNA results in the appearance of monomeric RNA in virions. J. Virol. 2001, 75, 2557–2565. [Google Scholar] [CrossRef]
- Laughrea, M.; Jette, L.; Mak, J.; Kleiman, L.; Liang, C.; Wainberg, M.A. Mutations in the kissing-loop hairpin of human immunodeficiency virus type 1 reduce viral infectivity as well as genomic RNA packaging and dimerization. J. Virol. 1997, 71, 3397–3406. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.D.; Fu, W.; Nikolaitchik, O.; Chen, J.; Ptak, R.G.; Hu, W.-S. Dimer initiation signal of human immunodeficiency virus type 1: Its role in partner selection during RNA copackaging and its effects on recombination. J. Virol. 2007, 81, 4002–4011. [Google Scholar] [CrossRef] [PubMed]
- Shen, N.; Jette, L.; Liang, C.; Wainberg, M.A.; Laughrea, M. Impact of human immunodeficiency virus type 1 RNA dimerization on viral infectivity and of stem-loop B on RNA dimerization and reverse transcription and dissociation of dimerization from packaging. J. Virol. 2000, 74, 5729–5735. [Google Scholar] [CrossRef] [PubMed]
- Paillart, J.C.; Berthoux, L.; Ottmann, M.; Darlix, J.L.; Marquet, R.; Ehresmann, B.; Ehresmann, C. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J. Virol. 1996, 70, 8348–8354. [Google Scholar] [CrossRef]
- Laughrea, M.; Jette, L. A 19-nucleotide sequence upstream of the 5’ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry 1994, 33, 13464–13474. [Google Scholar] [CrossRef]
- Paillart, J.C.; Marquet, R.; Skripkin, E.; Ehresmann, B.; Ehresmann, C. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J. Biol. Chem. 1994, 269, 27486–27493. [Google Scholar]
- Muriaux, D.; de Rocquigny, H.; Roques, B.P.; Paoletti, J. NCp7 activates HIV-1Lai RNA dimerization by converting a transient loop-loop complex into a stable dimer. J. Biol. Chem. 1996, 271, 33686–33692. [Google Scholar] [CrossRef]
- Fosse, P.; Motte, N.; Roumer, A.; Gabus, C.; Muriaux, D.; Darlix, J.-L.; Paoletti, J. A short autocomplementary sequence plays an essential role in avian sarcoma-leukosis virus RNA dimerization. Biochemistry 1996, 35, 16601–16609. [Google Scholar] [CrossRef]
- Song, R.; Kafaie, J.; Yang, L.; Laughrea, M. HIV-1 viral RNA is selected in the form of monomers that dimerize in a three-step protease-dependent process; the DIS of stem-loop 1 initiates viral RNA dimerization. J. Mol. Biol. 2007, 371, 1084–1096. [Google Scholar] [CrossRef]
- Paillart, J.-C.; Skripkin, E.; Ehresmann, B.; Ehresmann, C.; Marquet, R. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl. Acad. Sci. USA 1996, 93, 5572–5577. [Google Scholar] [CrossRef]
- Clever, J.L.; Wong, M.L.; Parslow, T.G. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J. Virol. 1996, 70, 5902–5908. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Nikolaitchik, O.; Singh, J.; Wright, A.; Bencsics, C.E.; Coffin, J.M.; Ni, N.; Lockett, S.; Pathak, V.K.; Hu, W.S. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc. Natl. Acad. Sci. USA 2009, 106, 13535–13540. [Google Scholar] [CrossRef] [PubMed]
- Muriaux, D.; Girard, P.-M.; Bonnet-Mathoniere, B.; Paoletti, J. Dimerization of HIV-1Lai RNA at low ionic strength: An autocomplementary sequence in the 5’ leader region is evidence dby an antisense oligonucleotide. J. Biol. Chem. 1995, 270, 8209–8216. [Google Scholar] [CrossRef] [PubMed]
- Windbichler, N.; Werner, M.; Schroeder, R. Kissing complex-mediated dimerisation of HIV-1 RNA: Coupling extended duplex formation to ribozyme cleavage. Nucleic Acids Res. 2003, 31, 6419–6427. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Laughrea, M.; Jette, L. Kissing-loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248–271 are dispensable for dimer formation. Biochemistry 1996, 35, 1589–1598. [Google Scholar] [CrossRef]
- Laughrea, M.; Jette, L. HIV-1 genome dimerization: Kissing-loop hairpin dictates whether nucleotides downstream of the 5’ splice junction contribute to loose and tight dimerization of human immunodeficiency virus RNA. Biochemistry 1997, 36, 9501–9508. [Google Scholar] [CrossRef]
- Sakuragi, S.; Yokoyama, M.; Shioda, T.; Sato, H.; Sakuragi, J.-I. SL1 revisited: Functional analysis of the structure and conformation of HIV-1 genome RNA. Retrovirology 2016, 13, 1–13. [Google Scholar] [CrossRef][Green Version]
- Greatorex, J.; Gallego, J.; Varani, G.; Lever, A. Structure and stability of wild-type and mutant RNA internal loops from the SL1 domain of the HIV-1 packaging signal. J. Mol. Biol. 2002, 322, 543–557. [Google Scholar] [CrossRef]
- Mujeeb, A.; Clever, J.L.; Billeci, T.M.; James, T.L.; Parslow, T.G. Structure of the dimer initiation complex of HIV-1 genomic RNA. Nat. Struct. Biol. 1998, 5, 432–436. [Google Scholar] [CrossRef]
- Ulyanov, N.B.; Mujeeb, A.; Du, Z.; Tonelli, M.; Parslow, T.G.; James, T.L. NMR structure of the full-length linear dimer of stem-loop-1 RNA in the HIV-1 dimer initiation site. J. Biol. Chem. 2006, 281, 16168–16177. [Google Scholar] [CrossRef]
- Zhang, K.; Keane, S.C.; Su, Z.; Irobalieva, R.N.; Chen, M.; Van, V.; Sciandra, C.A.; Marchant, J.; Heng, X.; Schmid, M.F.; et al. Structure of the 30 kDa HIV-1 RNA dimerization signal by a hybrid Cryo-EM, NMR, and molecular dynamics approach. Structure 2018, 26, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Kieken, F.; Paquet, F.; Brule, F.; Paoletti, J.; Lancelot, G. A new NMR solution structure of the SL1 HIV-1Lai loop-loop dimer. Nucleic Acids Res. 2006, 34, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Dardel, F.; Marquet, R.; Ehresmann, C.; Ehresmann, B.; Blanquet, S. Solution studies of the dimerization initiation site of HIV-1 genomic RNA. Nucleic Acids Res. 1998, 26, 3567–3571. [Google Scholar] [CrossRef] [PubMed]
- Baba, S.; Takahashi, K.; Noguchi, S.; Takaku, H.; Koyanagi, Y.; Yamamoto, N.; Kawai, G. Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers. J. Biochem. 2005, 138, 583–592. [Google Scholar] [CrossRef]
- Mihailescu, M.-R.; Marino, J.P. A proton-coupled dynamic conformational switch in the HIV-1 dimerization initiation site kissing complex. Proc. Natl. Acad. Sci. USA 2004, 101, 1189–1194. [Google Scholar] [CrossRef]
- Lawrence, D.C.; Stover, C.C.; Noznitsky, J.; Wu, Z.-R.; Summers, M.F. Structure of the intact stem and bulge of HIV-1 ψ-RNA stem loop SL1. J. Mol. Biol. 2003, 326, 529–542. [Google Scholar] [CrossRef]
- Ennifar, E.; Walter, P.; Ehresmann, B.; Ehresmann, C.; Dumas, P. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat. Struct. Biol. 2001, 8, 1064–1068. [Google Scholar] [CrossRef]
- Muriaux, D.; Fosse, P.; Paoletti, J. A kissing complex together with a stable dimer is involved in the HIV-1Lai RNA dimerization process in vitro. Biochemistry 1996, 35, 5075–5082. [Google Scholar] [CrossRef]
- Takahashi, K.-I.; Baba, S.; Hayashi, Y.; Koyanagi, Y.; Yamamoto, N.; Takaku, H.; Kawai, G. NMR analysis of intra- and inter-molecular stems in dimerization initiation site. J. Biochem. 2000, 127, 681–686. [Google Scholar] [CrossRef]
- Mujeeb, A.; Parslow, T.G.; Zarrinpar, A.; Das, C.; James, T.L. NMR structure of the mature dimer initiation complex of HIV-1 genomic RNA. FEBS Lett. 1999, 458, 387–392. [Google Scholar] [CrossRef]
- Feng, Y.-X.; Copeland, T.D.; Henderson, L.E.; Gorelick, R.J.; Bosche, W.J.; Levin, J.G.; Rein, A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc. Natl. Acad. Sci. USA 1996, 93, 7577–7581. [Google Scholar] [CrossRef] [PubMed]
- Ennifar, E.; Yusupov, M.; Walter, P.; Marquet, R.; Ehresmann, B.; Ehresmann, C.; Dumas, P. The crystal structure of the dimerization initiation site of genomic HIV-1 RNA reveals an extended duplex with two adenine bulges. Structure 1999, 7, 1439–1449. [Google Scholar] [CrossRef]
- Girard, F.; Barbault, F.; Gouyett, C.; Huynh-dinh, T.; Paoletti, J.; Lamcelot, G. Dimer initiation sequence of HIV-1 Lai genomic RNA: NMR solution structure of the extended nucleus. J. Biomol. Struct. Dyn. 1999, 16, 1145–1157. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Kerwood, D.J.; Paoletti, A.C.; Shubsda, M.F.; Borer, P.N. Stem of SL1 RNA in HIV-1: Structure and nucleocapsid protein binding for a 1 × 3 internal loop. Biochemistry 2003, 42, 5259–5269. [Google Scholar] [CrossRef] [PubMed]
- Battiste, J.L.; Mao, H.; Rao, N.S.; Tan, R.; Muhandiram, D.R.; Kay, L.E.; Frankel, A.D.; Williamson, J.R. Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex. Science 1996, 273, 1547–1551. [Google Scholar] [CrossRef] [PubMed]
- Peterson, R.D.; Feigon, J. Structural change in Rev responsive element RNA of HIV-1 on binding Rev peptide. J. Mol. Biol. 1996, 264, 863–877. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, Q.; Al-Hashimi, H.M. Resolving fast and slow motions in the internal loop containing stem-loop 1 of HIV-1 that are modulated by Mg2+ binding: Role in the kissing-duplex structural transition. Nucleic Acids Res. 2007, 35, 1698–1713. [Google Scholar] [CrossRef]
- Keane, S.C.; Van, V.; Frank, H.M.; Sciandra, C.A.; McCowin, S.; Santos, J.; Heng, X.; Summers, M.F. NMR detection of intermolecular interaction sites in the dimeric 5’-leader of the HIV-1 genome. Proc. Natl. Acad. Sci. USA 2016, 113, 13033–13038. [Google Scholar] [CrossRef]
- Ganser, L.R.; Al-Hashimi, H.M. HIV-1 leader RNA dimeric interface revealed by NMR. Proc. Natl. Acad. Sci. USA 2016, 113, 13263–13265. [Google Scholar] [CrossRef]
- Mujeeb, A.; Ulyanov, N.B.; Georgantis, S.; Smirnov, I.; Chung, J.; Parslow, T.G.; James, T.L. Nucleocapsid protein-mediated maturation of dimer initiation complex of full-length SL1 stemloop of HIV-1: Sequence effects and mechanism of RNA refolding. Nucleic Acids Res. 2007, 35, 2026–2034. [Google Scholar] [CrossRef]
- Takahashi, K.; Baba, S.; Koyanagi, Y.; Yamamoto, N.; Takaku, H.; Kawai, G. Two basic regions of NCp7 are sufficient for conformational conversion of HIV-1 dimerization initiation site from kissing-loop dimer to extended-duplex dimer. J. Biol. Chem. 2001, 276, 31274–31278. [Google Scholar] [CrossRef]
- Aduri, R.; Briggs, K.T.; Gorelick, R.J.; Marino, J.P. Molecular determinants of HIV-1 NCp7 chaperone activity in maturation of the HIV-1 dimerization initiation site. Nucleic Acids Res. 2013, 41, 2565–2580. [Google Scholar] [CrossRef] [PubMed]
- Aldovini, A.; Young, R.A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J. Virol. 1990, 64, 1920–1926. [Google Scholar] [CrossRef] [PubMed]
- Poznansky, M.; Lever, A.M.L.; Bergeron, L.; Haseltine, W.; Sodroski, J. Gene transfer into human lymphocytes by a defective human immunodeficiency virus type 1 vector. J. Virol. 1991, 65, 532–536. [Google Scholar] [CrossRef] [PubMed]
- Lever, A.M.L.; Göttlinger, H.G.; Haseltine, W.A.; Sodroski, J.G. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J. Virol. 1989, 63, 4085–4087. [Google Scholar] [CrossRef] [PubMed]
- Clavel, F.; Orenstein, J.M. A mutant of human immunodeficiency virus with reduced RNA packaging and abnormal particle morphology. J. Virol. 1990, 64, 5230–5234. [Google Scholar] [CrossRef] [PubMed]
- Sakuragi, J.; Sakuragi, S.; Shioda, T. Minimal region sufficient for genome dimerization in the human immunodeficiency virus type 1 virion and its potential roles in the early stages of viral replication. J. Virol. 2007, 81, 7985–7992. [Google Scholar] [CrossRef]
- Sakaguchi, K.; Zambrano, N.; Baldwin, E.T.; Shapiro, B.A.; Erickson, J.W.; Omichinski, J.G.; Clore, G.M.; Gronenborn, A.M.; Appella, E. Identification of a binding site for the human immunodeficiency virus type 1 nucleocapsid protein. Proc. Natl. Acad. Sci. USA 1993, 90, 5219–5223. [Google Scholar] [CrossRef]
- Rong, L.; Russell, R.S.; Hu, J.; Laughrea, M.; Wainberg, M.A.; Liang, C. Deletion of stem-loop 3 is compensated by second-site mutations within the Gag protein of human immunodeficiency virus type 1. Virology 2003, 314, 221–228. [Google Scholar] [CrossRef][Green Version]
- Russell, R.S.; Hu, J.; Beriault, V.; Mouland, A.J.; Kleiman, L.; Wainberg, M.A.; Liang, C. Sequences downstream of the 5’ splice donor site are required for both packaging and dimerization of human immunodeficiency virus type-1 RNA. J. Virol. 2003, 77, 84–96. [Google Scholar] [CrossRef]
- Webb, J.A.; Jones, C.P.; Parent, L.J.; Rouzina, I.; Musier-Forsyth, K. Distinct binding interactions of HIV-1 Gag to Psi and non-Psi RNAs: Implications for viral genomic RNA packaging. RNA 2013, 19, 1078–1088. [Google Scholar] [CrossRef] [PubMed]
- Comas-Garcia, M.; Datta, S.A.; Baker, L.; Varma, R.; Gudla, P.R.; Rein, A. Dissection of specific binding of HIV-1 Gag to the “packaging signal” in viral RNA. Elife 2017, 6, 6. [Google Scholar] [CrossRef] [PubMed]
- Ding, P.; Kharytonchyk, S.; Waller, A.; Mbaekwe, U.; Basappa, S.; Kuo, N.; Frank, H.M.; Quasney, C.; Kidane, A.; Swanson, C.; et al. Identification of the initial nucleocapsid recognition element in the HIV-1 RNA packaging signal. Proc. Natl. Acad. Sci. USA 2020, 117, 17737–17746. [Google Scholar] [CrossRef] [PubMed]
- Pappalardo, L.; Kerwood, D.J.; Pelczer, I.; Borer, P.N. Three-dimensional folding of an RNA hairpin required for packaging HIV-1. J. Mol. Biol. 1998, 282, 801–818. [Google Scholar] [CrossRef] [PubMed]
- Zeffman, A.; Hassard, S.; Varani, G.; Lever, A. The major HIV-1 packaging signal is an extended bulged stem loop whose structure is altered on interaction with the Gag polyprotein. J. Mol. Biol. 2000, 297, 877–893. [Google Scholar] [CrossRef] [PubMed]
- McBride, M.S.; Schwartz, M.D.; Panganiban, A.T. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J. Virol. 1997, 71, 4544–4554. [Google Scholar] [CrossRef]
- Luban, J.; Goff, S.P. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J. Virol. 1994, 68, 3784–3793. [Google Scholar] [CrossRef]
- Parolin, C.; Dorfman, T.; Palu, G.; Gottlinger, H.G.; Sodroski, J. Analysis in human immunodeficiency virus type 1 vectors of cis-acting sequences that affect gene transfer into human lymphocytes. J. Virol. 1994, 68, 3888–3895. [Google Scholar] [CrossRef]
- Kerwood, D.J.; Cavaluzzi, M.J.; Borer, P.N. Structure of SL4 RNA from the HIV-1 packaging signal. Biochemistry 2001, 40, 14518–14529. [Google Scholar] [CrossRef]
- Amarasinghe, G.K.; Zhou, J.; Miskimon, M.; Chancellor, K.J.; McDonald, J.A.; Matthews, A.G.; Miller, R.A.; Rouse, M.D.; Summers, M.F. Stem-loop SL4 of the HIV-1 Ψ-RNA packaging signal exhibits weak affinity for the nucleocapsid protein. Structural studies and implications for genome recognition. J. Mol. Biol. 2001, 314, 961–969. [Google Scholar] [CrossRef]
- Wimmer, J.; Fujinaga, K.; Taube, R.; Cujec, T.P.; Zhu, Y.; Peng, J.; Price, D.H.; Peterlin, B.M. Interactions between Tat and TAR and human immunodeficiency virus replication are facilitated by human cyclin T1 but not cyclins T2a or T2b. Virology 1999, 255, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Brady, J.; Kashanchi, F. Tat gets the “green” light on transcription initiation. Retrovirology 2005, 2, 69. [Google Scholar] [CrossRef] [PubMed]
- Laspia, M.F.; Rice, A.P.; Mathews, M.B. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell 1989, 59, 283–292. [Google Scholar] [CrossRef]
- Chulze-Gahmen, U.; Hurley, J.H. Structural mechanism for HIV-1 TAR loop recognition by Tat and the super elongation complex. Proc. Natl. Acad. Sci. USA 2018, 115, 12973–12978. [Google Scholar] [CrossRef] [PubMed]
- Chavali, S.S.; Bonn-Breach, R.; Wedekind, J.E. Face-time with TAR: Portraits of an HIV-1 RNA with diverse modes of effector recognition relevant for drug discovery. J. Biol. Chem. 2019, 294, 9326–9341. [Google Scholar] [CrossRef]
- Jalalirad, M.; Saadatmand, J.; Laughrea, M. Dominant role of the 5’ TAR bulge in dimerization of HIV-1 genomic RNA, but no evidence of TAR-TAR kissing during in vivo virus assembly. Biochemistry 2012, 51, 3744–3758. [Google Scholar] [CrossRef]
- Andersen, E.S.; Contera, S.A.; Knudsen, B.; Damgaard, C.K.; Besenbacher, F.; Kjems, J. Role of the trans-activation response element in dimerization of HIV-1 RNA. J. Biol. Chem. 2004, 279, 22243–22249. [Google Scholar] [CrossRef]
- Berkhout, B.; Vastenhouw, N.L.; Klasens, B.I.; Huthoff, H. Structural features in the HIV-1 repeat region facilitate strand transfer during reverse transcription. RNA 2001, 7, 1097–1114. [Google Scholar] [CrossRef]
- Helga-Maria, C.; Hammarskjold, M.-L.; Rekosh, D. An intact TAR element and cytoplasmic localization are necessary for efficient packaging of human immunodeficiency virus type-1 genomic RNA. J. Virol. 1999, 73, 4127–4135. [Google Scholar] [CrossRef]
- Clever, J.L.; Eckstein, D.A.; Parslow, T.G. Genetic dissociation of the encapsidation and reverse transcription functions in the 5’ R region of human immunodeficiency virus type 1. J. Virol. 1999, 73, 101–109. [Google Scholar] [CrossRef]
- Das, A.T.; Klaver, B.; Berkhout, B. The 5´ and 3´ TAR elements of human immunodeficiency virus exert effects at several points in the virus life cycle. J. Virol. 1998, 72, 9217–9223. [Google Scholar] [CrossRef]
- Bennasser, Y.; Le, S.Y.; Yeung, M.L.; Jeang, K.T. HIV-1 encoded candidate micro-RNAs and their cellular targets. Retrovirology 2004, 1, 43. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Feng, Z.; Yue, H.; Bazdar, D.; Mbonye, U.; Zender, C.; Harding, C.V.; Bruggeman, L.; Karn, J.; Sieg, S.F.; et al. Exosomes derived from HIV-1-infected cells promote growth and progression of cancer via HIV TAR RNA. Nat. Commun. 2018, 9, 4585. [Google Scholar] [CrossRef] [PubMed]
- Das, A.T.; Harwig, A.; Vrolijk, M.M.; Berkhout, B. The TAR hairpin of human immunodeficiency virus Type 1 can be deleted when not required for Tat-mediated activation of transcription. J. Virol. 2007, 81, 7742–7748. [Google Scholar] [CrossRef] [PubMed]
- Brandt, S.; Blissenbach, M.; Grewe, B.; Konietzny, R.; Grunwald, T.; Uberla, K. Rev proteins of human and simian immunodeficiency virus enhance RNA encapsidation. PLoS Pathog. 2007, 3, e54. [Google Scholar] [CrossRef] [PubMed]
- O’Carroll, I.P.; Soheilian, F.; Kamata, A.; Nagashima, K.; Rein, A. Elements in HIV-1 Gag contributing to virus particle assembly. Virus Res. 2013, 171, 341–345. [Google Scholar] [CrossRef]
- Rulli, J.S.J.; Hibbert, C.S.; Mirro, J.; Pederson, T.; Biswal, S.; Rein, A. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J. Virol. 2007, 81, 6623–6631. [Google Scholar] [CrossRef]
- Aboul-Ela, F.; Karn, J.; Varani, G. The structure of the human immunodeficiency virus type-1 TAR RNA reveals principles of RNA recognition by Tat protein. J. Mol. Biol. 1995, 253, 313–332. [Google Scholar] [CrossRef]
- Hauber, J.; Malim, M.H.; Cullen, B.R. Mutational analysis of the conserved basic domain of human immunodeficiency virus tat protein. J. Virol. 1989, 63, 1181–1187. [Google Scholar] [CrossRef]
- Delling, U.; Reid, L.S.; Barnett, R.W.; Ma, M.Y.; Climie, S.; Sumner-Smith, M.; Sonenberg, N. Conserved nucleotides in the TAR RNA stem of human immunodeficiency virus type 1 are critical for Tat binding and trans activation: Model for TAR RNA tertiary structure. J. Virol. 1992, 66, 3018–3025. [Google Scholar] [CrossRef]
- Dingwall, C.; Ernberg, I.; Gait, M.J.; Green, S.M.; Heaphy, S.; Karn, J.; Lowe, A.D.; Singh, M.; Skinner, M.A. HIV-1 tat protein stimulates transcription by binding to a U-rich bulge in the stem of the TAR RNA structure. EMBO J. 1990, 9, 4145–4153. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.; Garber, M.E.; Fang, S.-M.; Fischer, W.H.; Jones, K.A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 1998, 92, 451–462. [Google Scholar] [CrossRef]
- Richter, S.; Ping, Y.H.; Rana, T.M. TAR RNA loop: A scaffold for the assembly of a regulatory switch in HIV replication. Proc. Natl. Acad. Sci. USA 2002, 99, 7928–7933. [Google Scholar] [CrossRef] [PubMed]
- Puglisi, J.D.; Tan, R.; Calnan, B.J.; Frankel, A.D.; Williamson, J.R. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science 1992, 257, 76–80. [Google Scholar] [CrossRef] [PubMed]
- Puglisi, J.D.; Chen, L.; Frankel, A.D.; Williamson, J.R. Role of RNA structure in arginine recognition of TAR RNA. Proc. Natl. Acad. Sci. USA 1993, 90, 3680–3684. [Google Scholar] [CrossRef]
- Aboul-Ela, F.; Karn, J.; Varani, G. Structure of HIV-1 TAR RNA in the absence of ligands reveals a novel conformation of the trinucleotide bulge. Nucleic Acids Res. 1996, 24, 3974–3981. [Google Scholar] [CrossRef] [PubMed]
- Long, K.S.; Crothers, D.M. Characterization of the solution conformations of unbound and Tat peptide-bound forms of HIV-1 TAR RNA. Biochemistry 1999, 38, 10059–10069. [Google Scholar] [CrossRef]
- Davidson, A.; Leeper, T.C.; Athanassiou, Z.; Patora-Komisarska, K.; Karn, J.; Robinson, J.A.; Varani, G. Simultaneous recognition of HIV-1 TAR RNA bulge and loop sequences by cyclic peptide mimics of Tat protein. Proc. Natl. Acad. Sci. USA 2009, 106, 11931–11936. [Google Scholar] [CrossRef]
- Pitt, S.W.; Majumdar, A.; Serganov, A.; Patel, D.J.; Al-Hashimi, H.M. Argininamide binding arrests global motions in HIV-1 TAR RNA: Comparison with Mg2+-induced conformational stabilization. J. Mol. Biol. 2004, 338, 7–16. [Google Scholar] [CrossRef][Green Version]
- Shortridge, M.D.; Wille, P.T.; Jones, A.N.; Davidson, A.; Bogdanovic, J.; Arts, E.; Karn, J.; Robinson, J.A.; Varani, G. An ultra-high affinity ligand of HIV-1 TAR reveals the RNA structure recognized by P-TEFb. Nucleic Acids Res. 2019, 47, 1523–1531. [Google Scholar] [CrossRef]
- Pham, V.V.; Salguero, C.; Khan, S.N.; Meagher, J.L.; Brown, W.C.; Humbert, N.; de Rocquigny, H.; Smith, J.L.; D’Souza, V.M. HIV-1 Tat interactions with cellular 7SK and viral TAR RNAs identifies dual structural mimicry. Nat. Commun. 2018, 9, 4266. [Google Scholar] [CrossRef] [PubMed]
- Dethoff, E.A.; Petzold, K.; Chugh, J.; Casiano-Negroni, A.; Al-Hashimi, H.M. Visualizing transient low-populated structures of RNA. Nature 2012, 491, 724–728. [Google Scholar] [CrossRef] [PubMed]
- Ganser, L.R.; Kelly, M.L.; Patwardhan, N.N.; Hargrove, A.E.; Al-Hashimi, H.M. Demonstration that small molecules can bind and stabilize Low-abundance Short-lived RNA excited conformational states. J. Mol. Biol. 2020, 432, 1297–1304. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Stelzer, A.C.; Fisher, C.K.; Al-Hashimi, H.M. Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature 2007, 450, 1263–1267. [Google Scholar] [CrossRef] [PubMed]
- Tisne, C.; Roques, B.P.; Dardel, F. The annealing mechanism of HIV-1 reverse transcription primer onto the viral genome. J. Biol. Chem. 2004, 279, 3588–3595. [Google Scholar] [CrossRef]
- Beerens, N.; Groot, F.; Berkhout, B. Initiation of HIV-1 reverse transcription is regulated by a primer activation signal. J. Biol. Chem. 2001, 276, 31247–31256. [Google Scholar] [CrossRef]
- Puglisi, E.V.; Puglisi, J.D. Secondary structure of the HIV reverse transcription initiation complex by NMR. J. Mol. Biol. 2011, 410, 863–874. [Google Scholar] [CrossRef]
- Goldschmidt, V.; Rigourd, M.; Ehresmann, C.; Le Grice, S.F.; Ehresmann, B.; Marquet, R. Direct and indirect contributions of RNA secondary structure elements to the initiation of HIV-1 reverse transcription. J. Biol. Chem. 2002, 277, 43233–43242. [Google Scholar] [CrossRef]
- Xing, L.; Liang, C.; Kleiman, L. Coordinate Roles of Gag and RNA Helicase A in Promoting the Annealing of tRNALys3 to HIV-1 RNA. J. Virol. 2011, 85, 1847–1860. [Google Scholar] [CrossRef]
- Jeang, K.-T.; Yedavalli, V. Role of RNA helicases in HIV-1 replication. Nucleic Acids Res. 2006, 34, 4198–4205. [Google Scholar] [CrossRef]
- Roy, B.B.; Hu, J.; Guo, X.; Russell, R.S.; Guo, F.; Kleiman, L.; Liang, C. Association of RNA Helicase A with human immunodeficiency virus type 1 particles. J. Biol. Chem. 2006, 281, 12625–12635. [Google Scholar] [CrossRef] [PubMed]
- Bolinger, C.; Sharma, A.; Singh, D.; Yu, L.; Boris-Lawrie, K. RNA helicase A modulates translation of HIV-1 and infectivity of progeny virions. Nucleic Acids Res. 2010, 38, 1686–1696. [Google Scholar] [CrossRef] [PubMed]
- Boeras, I.; Song, Z.; Moran, A.; Franklin, J.; Brown, W.C.; Johnson, M.; Boris-Lawrie, K.; Heng, X. DHX9/RHA Binding to the PBS-Segment of the genomic RNA during HIV-1 assembly bolsters virion infectivity. J. Mol. Biol. 2016, 428, 2418–2429. [Google Scholar] [CrossRef] [PubMed]
- Barton, S.; Heng, X.; Johnson, B.A.; Summers, M.F. Database proton NMR chemical shifts for RNA signal assignment and validation. J. Biomol. NMR 2013, 55, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.D.; Summers, M.F.; Johnson, B.A. Prediction of hydrogen and carbon chemical shifts from RNA using database mining and support vector regression. J. Biomol. NMR 2015, 63, 39–52. [Google Scholar] [CrossRef]
- Johnson, B.A.; Blevins, R.A. NMRview: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 1994, 4, 603–614. [Google Scholar] [CrossRef]
- Marchant, J.; Summers, M.F.; Johnson, B.A. Assigning NMR spectra of RNA, peptides and small organic molecules using molecular network visualization software. J. Biomol. NMR 2019, 73, 525–529. [Google Scholar] [CrossRef]
- Tolbert, B.S.; Miyazaki, Y.; Barton, S.; Kinde, B.; Starck, P.; Singh, R.; Bax, A.; Case, D.A.; Summers, M.F. Major groove width variations in RNA structures determined by NMR and impact of 13C residual chemical shift anisotropy and 1H-13C residual dipolar coupling on refinement. J. Biomol. NMR 2010, 47, 205–219. [Google Scholar] [CrossRef]
- Brigham, B.S.; Kitzrow, J.P.; Reyes, J.C.; Musier-Forsyth, K.; Munro, J.B. Intrinsic conformational dynamics of the HIV-1 genomic RNA 5’UTR. Proc. Natl. Acad. Sci. USA 2019, 116, 10372–10381. [Google Scholar] [CrossRef]
- Tran, T.; Liu, Y.; Marchant, J.; Monti, S.; Seu, M.; Zaki, J.; Yang, A.L.; Bohn, J.; Ramakrishnan, V.; Singh, R.; et al. Conserved determinants of lentiviral genome dimerization. Retrovirology 2015, 12, 83. [Google Scholar] [CrossRef]
- Chiu, Y.L.; Coronel, E.; Ho, C.K.; Shuman, S.; Rana, T.M. HIV-1 Tat protein interacts with mammalian capping enzyme and stimulates capping of TAR RNA. J. Biol. Chem. 2001, 276, 12959–12966. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Deng, L.; Kashanchi, F.; Brady, J.N.; Shatkin, A.J.; Kumar, A. The Tat/TAR-dependent phosphorylation of RNA polymerase II C-terminal domain stimulates cotranscriptional capping of HIV-1 mRNA. Proc. Natl. Acad. Sci. USA 2003, 100, 12666–12671. [Google Scholar] [CrossRef] [PubMed]
- Menees, T.M.; Muller, B.; Krausslich, H.G. The major 5’ end of HIV type 1 RNA corresponds to G456. AIDS Res. Hum. Retrovir. 2007, 23, 1042–1048. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Yilmaz, A.; Marsh, K.; Cochrane, A.; Boris-Lawrie, K. Thriving under stress: Selective translation of HIV-1 structural protein mRNA during Vpr-mediated impairment of eIF4E translation activity. PLoS Pathog. 2012, 8, e1002612. [Google Scholar] [CrossRef]
- Muesing, M.A.; Smith, D.H.; Cabradilla, C.D.; Benton, C.V.; Lasky, L.A.; Capon, D.J. Nucleic acid structure and expression of the human AIDS/lymphadenopathy retrovirus. Nature 1985, 313, 450–458. [Google Scholar] [CrossRef]
- Starcich, B.; Ratner, L.; Josephs, S.F.; Okamoto, T.; Gallo, R.C.; Wong-Staal, F. Characterization of long terminal repeat sequences of HTLV-III. Science 1985, 227, 538–540. [Google Scholar] [CrossRef]
- Vogt, V. Proteolytic processing and particle maturation. Curr. Top. Microbiol. Immunol. 1996, 214, 95–132. [Google Scholar]
- Mann, R.; Mulligan, R.C.; Baltimore, D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell 1983, 33, 153–159. [Google Scholar] [CrossRef]
- Murphy, J.E.; Goff, S.P. Construction and analysis of deletion mutations in the U5 region of Moloney murine leukemia virus: Effects on RNA packaging and reverse transcription. J. Virol. 1989, 63, 319–327. [Google Scholar] [CrossRef]
- Yu, S.S.; Kim, J.-M.; Kim, S. The 17 nucleotides downstream from the env gene stop codon are important for Murine Leukemia Virus packaging. J. Virol. 2000, 74, 8775–8780. [Google Scholar]
- Bender, M.A.; Palmer, T.D.; Gelinas, R.E.; Miller, A.D. Evidence that the packaging signal of Moloney murine leukemia virus extends into the gag region. J. Virol. 1987, 61, 1639–1646. [Google Scholar] [CrossRef] [PubMed]
- Alford, R.L.; Honda, S.; Lawrence, C.B.; Belmont, J.W. RNA secondary structure analysis of the packing signal for Moloney murine leukemia virus. Virology 1991, 183, 611–619. [Google Scholar] [CrossRef]
- Fisher, J.; Goff, S.P. Mutational analysis of stem-loops in the RNA packaging signal of the moloney murine leukemia virus. Virology 1998, 244, 133–145. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Konings, D.A.M.; Nash, M.A.; Maizel, J.V.; Arlinghaus, R.B. Novel GACG-hairpin pair motif in the 5’ untranslated region of type C retrovirus related to murine leukemia virus. J. Virol. 1992, 66, 632–640. [Google Scholar] [CrossRef]
- Mougel, M.; Tounekti, N.; Darlix, J.-L.; Paoletti, J.; Ehresmann, B.; Ehresmann, C. Conformational analysis of the 5’ leader and the gag initiation site of Mo-MuLV RNA and allosteric transitions induced by dimerization. Nucleic Acids Res. 1993, 21, 4677–4684. [Google Scholar] [CrossRef]
- Mougel, M.; Zhang, Y.; Barklis, E. Cis-active structural motifs involved in specific encapsidation of Moloney Murine leukemia virus RNA. J. Virol. 1996, 70, 5043–5050. [Google Scholar] [CrossRef]
- Tounekti, N.; Mougel, M.; Roy, C.; Marquet, R.; Darlix, J.-L.; Paoletti, J.; Ehresmann, B.; Ehresmann, C. Effect of dimerization on the conformation of the encapsidation psi domain of Moloney murine leukemia virus RNA. J. Mol. Biol. 1992, 223, 205–220. [Google Scholar] [CrossRef]
- Shinnick, T.M.; Lerner, R.A.; Sutcliffe, J.G. Nucleotide sequence of Moloney murine leukaemia virus. Nature 1981, 293, 543–548. [Google Scholar] [CrossRef]
- Beasley, B.E.; Hu, W.S. Cis-acting elements imporant for retroviral RNA packaging specificity. J. Virol. 2002, 76, 4950–4960. [Google Scholar] [CrossRef]
- Evans, M.J.; Bacharach, E.; Goff, S.P. RNA sequences in the Moloney murine leukemia virus genome bound by the Gag precursor protein in the Yeast three-hybrid system. J. Virol. 2004, 78, 7677–7684. [Google Scholar] [CrossRef][Green Version]
- D’Souza, V.; Melamed, J.; Habib, D.; Pullen, K.; Wallace, K.; Summers, M.F. Identification of a high-affinity nucleocapsid protein binding site within the Moloney Murine Leukemia Virus Y-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 2001, 314, 217–232. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, Y.; Irobalieva, R.N.; Tolbert, B.S.; Smalls-Manty, A.; Iyalla, K.; Loeliger, K.; D’Souza, V.; Khant, H.; Schmid, M.F.; Garcia, E.; et al. Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography. J. Mol. Biol. 2010, 404, 751–772. [Google Scholar] [CrossRef]
- Kim, C.-H.; Tinoco, I., Jr. A retroviral RNA kissing complex containing only two G-C base pairs. Proc. Natl. Acad. Sci. USA 2000, 97, 9396–9401. [Google Scholar] [CrossRef] [PubMed]
- Oroudjev, E.M.; Kang, P.C.E.; Kohlstaedt, L.A. An additional dimer linkage structure in Moloney Murine Leukemia Virus RNA. J. Mol. Biol. 1999, 291, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Girard, P.-M.; Bonnet-Mathoniere, B.; Mauriaux, D.; Paoletti, J. A short autocomplementary sequence in the 5’ leader region is responsible for dimerization of MoMuLV genomic RNA. Biochemistry 1995, 34, 9785–9794. [Google Scholar] [CrossRef]
- De Tapia, M.; Metzler, V.; Mougel, M.; Ehresmann, B.; Ehresmann, C. Dimerization of MoMuLV genomic RNA: Redefinition of the role of the palindromic stem-loop H1 (278–303) and new roles for stem-loops H2 (310-352) and H3 (355-374). Biochemistry 1998, 37, 6077–6085. [Google Scholar] [CrossRef]
- Ly, H.; Parslow, T.G. Bipartite signal for genomic RNA dimerization in the Moloney Murine Leukemia Virus. J. Virol. 2002, 76, 3135–3144. [Google Scholar] [CrossRef][Green Version]
- Chen, A.A.; Garcia, A.E. Mechanism of enhanced mechanical stability of a minimal RNA kissing complex elucidated by nonequilibrium molecular dynamics simulations. Proc. Natl. Acad. Sci. USA 2012, 109, E1530–E1539. [Google Scholar] [CrossRef]
- D’Souza, V.; Dey, A.; Habib, D.; Summers, M.F. NMR structure of the 101-nucleotide core encapsidation signal of the Moloney murine leukemia virus. J. Mol. Biol. 2004, 337, 427–442. [Google Scholar] [CrossRef]
- Merino, E.J.; Wilkinson, K.A.; Coughlan, J.L.; Weeks, K.M. RNA structure analysis at single nucleotide resolution by selective 2’-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 2005, 127, 4223–4231. [Google Scholar] [CrossRef]
- Badorrek, C.S.; Weeks, K.M. Flexibility in the dimerization domain of a gamma retrovirus. Nat. Chem. Biol. 2005, 1, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Brickell, P.M. The p60c-src family of protein-tyrosine kinases: Structure, regulation and function. Crit. Rev. Oncog. 1992, 3, 401–446. [Google Scholar] [PubMed]
- Hackett, P.B.; Swanstrom, R.; Varmus, H.E.; Bishop, J.M. The leader sequence of the subgenomic mRNA’s of Rous sarcoma virus is approximately 390 nucleotides. J. Virol. 1982, 41, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Hackett, P.; Dalton, M.W.; Johnson, D.P.; Peterson, R.B. Phylogenetic and physical analysis of the 5’ leader RNA sequences of avian retroviruses. Nucleic Acids Res. 1991, 19, 6929–6934. [Google Scholar] [CrossRef]
- Sonstegard, T.S.; Hackett, P.B. Autogenous regulation of RNA translation and packaging by Rous sarcoma virus Pr76gag. J. Virol. 1996, 70, 6642–6652. [Google Scholar] [CrossRef]
- Donze, O.; Damay, P.; Spahr, P.-F. The first and third uORFs in RSV leader RNA are efficiently translated: Implications for translational regulation and viral RNA packaging. Nucleic Acids Res. 1995, 23, 861–868. [Google Scholar] [CrossRef]
- Moustakas, A.; Sonstegard, T.S.; Hackett, P.B. Alterations of the three short open reading frames in the Rous sarcoma virus leader RNA modulate viral replication and gene expression. J. Virol. 1993, 67, 4337–4349. [Google Scholar] [CrossRef]
- Donze, O.; Spahr, P.-F. Role of the open reading frames of Rous sarcoma virus leader RNA in translation and genome packaging. EMBO J. 1992, 11, 3747–3757. [Google Scholar] [CrossRef]
- Anderson, D.J.; Lee, P.; Levine, K.L.; Sang, J.S.; Shah, S.A.; Yang, O.O.; Shank, P.R.; Linial, M.L. Molecular cloning and characterization of the RNA packaging-defective retrovirus SE21Q1b. J. Virol. 1992, 66, 204–216. [Google Scholar] [CrossRef]
- Gallis, B.; Linial, M.; Eisenman, R. An avian oncovirus mutant deficient in genomic RNA: Characterization of the packaged RNA as cellular messenger RNA. Virology 1979, 94, 146–161. [Google Scholar] [CrossRef]
- Kawai, S.; Koyama, T. Characterization of a Rouse Sarcoma Virus mutant defective in packaging it’s own genomic RNA: Biological properties of mutant TK15 and mutant-induced transformation. J. Virol. 1984, 51, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Koyama, T.; Harada, F.; Kawai, S. Characterization of a Rous sarcoma virus mutant defectie in packaging its own genomic RNA: Biochemical properties of mutant TK15 and mutant-induced transformants. J. Virol. 1984, 51, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Linial, M.; Medeiros, E.; Hayward, W.S. An avian oncovirus mutant (SE 21Q1b) deficient in genomic RNA: Biological and biochemical characterization. Cell 1978, 15, 1371–1381. [Google Scholar] [CrossRef]
- Nishizawa, M.; Koyama, T.; Kawai, S. Unusual features of the leader sequence of Rous sarcoma virus packaging mutant TK15. J. Virol. 1985, 55, 881–885. [Google Scholar] [CrossRef]
- Katz, R.A.; Terry, R.W.; Skalka, A.M. A conserved cis-acting sequence in the 5’ leader of avian sarcoma virus RNA is required for packaging. J. Virol. 1986, 59, 163–167. [Google Scholar] [CrossRef]
- Stoker, A.W.; Bissell, M.J. Development of avian sarcoma and leukosis virus based vector packaging cell lines. J. Virol. 1988, 16, 1161–1170. [Google Scholar] [CrossRef]
- Shank, P.R.; Linial, M. Avian oncovirus mutant (SE21Q1b) deficient in genomic RNA: Characterization of a deletion in the provirus. J. Virol. 1980, 36, 450–456. [Google Scholar] [CrossRef]
- Cobrink, D.; Aiyar, A.; Ge, Z.; Katzman, M.; Huang, H.; Leis, J. Overlapping retrovirus U5 sequence elements are required for efficient integration and initiation of reverse transcriptiuon. J. Virol. 1991, 65, 3864–3872. [Google Scholar] [CrossRef]
- Cobrink, D.; Soskey, L.; Leis, J. A retroviral RNA secondary structure required for efficient initiation of reverse transcription. J. Virol. 1988, 62, 3622–3630. [Google Scholar] [CrossRef]
- Norton, P.A.; Coffin, J.M. Bacterial b-galactosidase as a marker of Rous sarcoma virus gene expression and replication. Mol. Cell Biol. 1985, 5, 281–290. [Google Scholar] [CrossRef]
- Liu, S.; Maldonado, R.K.; Rye-McCurdy, T.; Binkley, C.; Bah, A.; Chen, E.C.; Rice, B.L.; Parent, L.J.; Musier-Forsyth, K. Rous Sarcoma Virus genomic RNA Dimerization capability in vitro is not a prerequisite for viral infectivity. Viruses 2020, 12, 568. [Google Scholar] [CrossRef] [PubMed]
- Knight, J.B.; Hai, S.Z.; Stoltzfus, M.C. A base-paired sequence in the avian sarcoma virus 5’ leader is required for efficient encapsidation of RNA. J. Virol. 1994, 68, 4493–4502. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; McAllen, K.; Tailor, Y.; Summers, M.F. High affinity nucleocapsid protein binding to the μΨ RNA packaging signal of Rous sarcoma virus. J. Mol. Biol. 2005, 349, 976–988. [Google Scholar] [CrossRef] [PubMed]
- Lear, A.L.; Haddrick, M.; Heaply, S. A study of the dimerization of the Rous sarcoma virus RNA in vitro and in vivo. Virology 1995, 212, 47–57. [Google Scholar] [CrossRef]
- Canaani, E.; Helm, K.V.D.; Duesberg, P. Evidence for a 30–40s RNA as a precursor for the 60-70s RNA of the Rous Sarcoma Virus. Proc. Natl. Acad. Sci. USA 1973, 70, 401–405. [Google Scholar] [CrossRef]
- Cheung, K.S.; Smith, R.E.; Stone, M.P.; Joklik, W.K. Comparison of immature (rapid harvest) and mature Rous Sarcoma Virus. Virology 1972, 50, 851–864. [Google Scholar] [CrossRef]
- Oertle, S.; Spahr, P.-F. Role of the Gag polyprotein precursor in packaging and maturation of Rous sarcoma virus genomic RNA. J. Virol. 1990, 64, 5757–5763. [Google Scholar] [CrossRef]
- Polge, E.; Darlix, J.-L.; Paoletti, J.; Fosse, P. Characterization of the loose and tight dimer forms of avian leukosis virus RNA. J. Mol. Biol. 2000, 300, 41–56. [Google Scholar] [CrossRef]
- Bieth, E.; Gabus, C.; Darlix, J.-L. A study of the dimer formation of Rous sarcoma virus RNA and of its effect on viral protein synthesis in vitro. Nucleic Acids Res. 1990, 18, 119–127. [Google Scholar] [CrossRef]
- Delahunty, M.; South, T.L.; Summers, M.F.; Karpel, R.L. Nucleic acid interactive properties of a peptide corresponding to the N-terminal zinc finger domain of HIV-1 nucleocapsid protein. Biochemistry 1992, 31, 6461–6469. [Google Scholar] [CrossRef]
- Summers, M.F. Zinc finger motif for single-stranded nucleic acids? Investigations by nuclear magnetic resonance. J. Cell. Biochem. 1991, 45, 41–48. [Google Scholar] [CrossRef] [PubMed]
- South, T.L.; Kim, B.; Hare, D.R.; Summers, M.F. Zinc fingers and molecular recognition. Structure and nucleic acid binding studies of an HIV zinc finger-like domain. Biochem. Pharm. 1990, 40, 123–129. [Google Scholar] [CrossRef]
- Schuler, W.; Dong, C.; Wecker, K.; Roques, B.P. NMR structure of the complex between the zinc finger protein NCp10 of Moloney murine leukemia virus and the single-stranded pentanucleotide d(ACGCC): Comparison with HIV-NCp7 complexes. Biochemistry 1999, 38, 12984–12994. [Google Scholar] [CrossRef] [PubMed]
- Deffaud, C.; Darlix, J.-L. Rous sarcoma virus translation revisited: Characterization of an internal ribosome entry segment in the 5’ leader of the genomic RNA. J. Virol. 2000, 74, 11581–11588. [Google Scholar] [CrossRef][Green Version]
- Levin, J.G.; Guo, J.; Rouzina, I.; Musier-Forsyth, K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: Critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 2005, 80, 217–286. [Google Scholar]
- Fu, W.; Ortiz-Conde, B.A.; Gorelick, R.J.; Hughes, S.H.; Rein, A. Placement of tRNA primer on the primer-binding site requires pol gene expression in avian but not murine retroviruses. J. Virol. 1997, 71, 6940–6946. [Google Scholar] [CrossRef]
- Feng, Y.-X.; Campbell, S.; Harvin, D.; Ehresmann, B.; Ehresmann, C.; Rein, A. The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: Possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J. Virol. 1999, 73, 4251–4256. [Google Scholar] [CrossRef]
- Wu, W.; Henderson, L.E.; Copeland, T.D.; Gorelick, R.J.; Bosche, W.J.; Rein, A.; Levin, J.G. Human immunodeficiency virus type 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J. Virol. 1996, 70, 7132–7142. [Google Scholar] [CrossRef] [PubMed]
- Tanchou, V.; Gabus, C.; Rogemond, V.; Darlix, J.-L. Formation of stable and functional HIV-1 nucleoprotein complexes in vitro. J. Mol. Biol. 1995, 252, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Darlix, J.-L.; Vincent, C.; Gabus, H.; de Rocquigny, H.; Roques, B. Trans-activation of the 5’ to 3’ viral DNA strand transfer by nucleocapsid protein during reverse transcription of HIV-1. C. R. Acad. Sci. 1993, 316, 763–771. [Google Scholar]
- Allain, B.; Lapadat-Tapolsky, M.; Berlioz, C.; Darlix, J.-L. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 1994, 13, 973–981. [Google Scholar] [CrossRef] [PubMed]
- Peliska, J.A.; Balasubramanian, S.; Giedroc, D.P.; Benkovic, S.J. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalyzed DNA strand transfer reactions and modulates RNaseH activity. Biochemistry 1994, 33, 13817–13823. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Palaniappan, C.; Wu, W.; Fay, P.J.; Bambara, R.A. Evidence for a unique mechanism of strand transfer from the transactivation response region of HIV-1. J. Biol. Chem. 1997, 272, 16769–16777. [Google Scholar] [CrossRef] [PubMed]
- Tsuchihashi, Z.; Brown, P.O. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 1994, 68, 5863–5870. [Google Scholar] [CrossRef] [PubMed]
- Auxilien, S.; Keith, G.; Le Grice, S.F.; Darlix, J.-L. Role of post-transcriptional modifications of primer tRNALys, 3 in the fidelity and efficacy of plus strand DNA transfer during HIV-1 reverse transcription. J. Biol. Chem. 1999, 274, 4412–4420. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Guo, J.; Bess, J.; Henderson, L.E.; Levin, J.G. Molecular requirements for human immunodeficiency virus type 1 plus-strand transfer: Analysis in reconstituted and endogenous reverse transcription systems. J. Virol. 1999, 73, 4794–4805. [Google Scholar] [CrossRef]
- Muthuswami, R.; Chen, J.; Burnett, B.P.; Thimmig, R.L.; Janjic, N.; McHenry, C.S. The HIV plus-strand transfer reaction: Determination of replication-competent intermediates and identification of a novel lentiviral element, the primer over-extension sequence. J. Mol. Biol. 2002, 315, 311–323. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Guo, J.; Wu, T.; Anderson, J.; Kane, B.F.; Johnson, D.G.; Gorelick, R.J.; Henderson, L.E.; Levin, J.G. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J. Virol. 2000, 74, 8980–8988. [Google Scholar] [CrossRef]
- Williams, M.C.; Rouzina, I.; Wenner, J.R.; Gorelick, R.J.; Musier-Forsyth, K.; Bloomfield, V.A. Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc. Natl. Acad. Sci. USA 2001, 98, 6121–6126. [Google Scholar] [CrossRef]
- Johnson, P.E.; Turner, R.B.; Wu, Z.-R.; Hairston, L.; Guo, J.; Levin, J.G.; Summers, M.F. A mechanism for (+) strand transfer enhancement by the HIV-1 nucleocapsid protein during reverse transcription. Biochemistry 2000, 39, 9084–9091. [Google Scholar] [CrossRef]
- Barraud, P.; Gaudin, C.; Dardel, F.; Tisne, C. New insights into the formation of HIV-1 reverse transcription initiation complex. Biochimie 2007, 89, 1204–1210. [Google Scholar] [CrossRef] [PubMed]
- Tisne, C.; Roques, B.P.; Dardel, F. Heteronuclear NMR studies of the interaction of tRNA(Lys)3 with HIV-1 nucleocapsid protein. J. Mol. Biol. 2001, 306, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Bourbigot, S.; Ramalanjaona, N.; Boudier, C.; Salgado, G.F.; Roques, B.P.; Mely, Y.; Bouaziz, S.; Morellet, N. How the HIV-1 nucleocapsid protein binds and destabilises the (-)primer binding site during reverse transcription. J. Mol. Biol. 2008, 383, 1112–1128. [Google Scholar] [CrossRef]
- Bazzi, A.; Zargarian, L.; Chaminade, F.; Boudier, C.; de Rocquigny, H.; Rene, B.; Mely, Y.; Fosse, P.; Mauffret, O. Structural insights into the cTAR DNA recognition by the HIV-1 nucleocapsid protein: Role of sugar deoxyriboses in the binding polarity of NC. Nucleic Acids Res. 2011, 39, 3903–3916. [Google Scholar] [CrossRef]
- Belfetmi, A.; Zargarian, L.; Tisne, C.; Sleiman, D.; Morellet, N.; Lescop, E.; Maskri, O.; Rene, B.; Mely, Y.; Fosse, P.; et al. Insights into the mechanisms of RNA secondary structure destabilization by the HIV-1 nucleocapsid protein. RNA 2016, 22, 506–517. [Google Scholar] [CrossRef] [PubMed]
- Rice, W.G.; Turpin, J.A.; Schaeffer, C.A.; Graham, L.; Clanton, D.; Buckheit, R.W.; Zaharevitz, D.; Summers, M.F.; Wallqvist, A.; Covell, D.G. Evaluation of selected chemotypes in coupled cellular and molecular target-based screens identifies novel HIV-1 zinc finger inhibitors. J. Med. Chem. 1996, 39, 3606–3616. [Google Scholar] [CrossRef]
- Rice, W.G.; Schaeffer, C.A.; Harten, B.; Villinger, F.; South, T.L.; Summers, M.F.; Henderson, L.E.; Bess, J.W., Jr.; Arthur, L.O.; McDougal, J.S.; et al. Inhibition of HIV-1 infectivity by zinc-ejecting aromatic C-nitroso compounds. Nature 1993, 361, 473–475. [Google Scholar] [CrossRef]
- Rice, W.G.; Turpin, J.A.; Huang, M.; Clanton, D.; Buckheit, R.W., Jr.; Covell, D.G.; Wallqvist, A.; McDonnell, N.B.; DeGuzman, R.N.; Summers, M.F.; et al. Azodicarbonamide inhibits HIV-1 replication by targeting the nucleocapsid protein. Nat. Med. 1997, 3, 341–345. [Google Scholar] [CrossRef]
- McDonnell, N.B.; De Guzman, R.N.; Rice, W.G.; Turpin, J.A.; Summers, M.F. Zinc ejection as a new rationale for the use of cystamine and related dislufide-containing antiviral agents in the treatment of AIDS. J. Med. Chem. 1997, 40, 1969–1976. [Google Scholar] [CrossRef]
- Tummino, P.J.; Scholten, J.D.; Harvey, P.J.; Holler, T.P.; Maloney, L.; Gogliotti, R.; Domagala, J.; Hupe, D. The in vitro ejection of zinc from human immunodeficiency virus (HIV) type 1 nucleocapsid protein by disulfide benzamides with cellular anti-HIV activity. Proc. Natl. Acad. Sci. USA 1996, 93, 969–973. [Google Scholar] [CrossRef]
- Deshmukh, L.; Tugarinov, V.; Appella, D.H.; Clore, G.M. Targeting a dark excited state of HIV-1 nucleocapsid by antiretroviral thioesters revealed by NMR spectroscopy. Angew. Chem. Int. Ed. 2018, 57, 2687–2691. [Google Scholar] [CrossRef] [PubMed]
- Goudreau, N.; Hucke, O.; Faucher, A.M.; Grand-Maitre, C.; Lepage, O.; Bonneau, P.R.; Mason, S.W.; Titolo, S. Discovery and structural characterization of a new inhibitor series of HIV-1 nucleocapsid function: NMR solution structure determination of a ternary complex involving a 2:1 inhibitor/NC stoichiometry. J. Mol. Biol. 2013, 425, 1982–1998. [Google Scholar] [CrossRef] [PubMed]
- Le Grice, S.F. Targeting the HIV RNA genome: High-hanging fruit only needs a longer ladder. Curr. Top. Microbiol. Immunol. 2015, 389, 147–169. [Google Scholar] [PubMed]
- Warui, D.M.; Baranger, A.M. Identification of small molecule inhibitors of the HIV-1 nucleocapsid-stem-loop 3 RNA complex. J. Med. Chem. 2012, 55, 4132–4141. [Google Scholar] [CrossRef]
- Bell, N.M.; L’Hernault, A.; Murat, P.; Richards, J.E.; Lever, A.M.; Balasubramanian, S. Targeting RNA-protein interactions within the human immunodeficiency virus type 1 lifecycle. Biochemistry 2013, 52, 9269–9274. [Google Scholar] [CrossRef]
- Ingemarsdotter, C.K.; Zeng, J.; Long, Z.; Lever, A.M.L.; Kenyon, J.C. An RNA-binding compound that stabilizes the HIV-1 gRNA packaging signal structure and specifically blocks HIV-1 RNA encapsidation. Retrovirology 2018, 15, 25. [Google Scholar] [CrossRef]
- Dietz, J.; Koch, J.; Kaur, A.; Raja, C.; Stein, S.; Grez, M.; Pustowka, A.; Mensch, S.; Ferner, J.; Moller, L.; et al. Inhibition of HIV-1 by a peptide ligand of the genomic RNA packaging signal Psi. ChemMedChem 2008, 3, 749–755. [Google Scholar] [CrossRef]
- Chung, J.; Ulyanov, N.B.; Guilbert, C.; Mujeeb, A.; James, T.L. Binding characteristics of small molecules that mimic nucleocapsid protein-induced maturation of stem-loop 1 of HIV-1 RNA. Biochemistry 2010, 49, 6341–6351. [Google Scholar] [CrossRef][Green Version]
- Tria, G.; Mertens, H.D.; Kachala, M.; Svergun, D.I. Advanced ensemble modelling of flexible macromolecules using X-ray solution scattering. IUCrJ 2015, 2, 207–217. [Google Scholar] [CrossRef]
- Fang, X.; Stagno, J.R.; Bhandari, Y.R.; Zuo, X.; Wang, Y.X. Small-angle X-ray scattering: A bridge between RNA secondary structures and three-dimensional topological structures. Curr. Opin. Struct. Biol. 2015, 30, 147–160. [Google Scholar] [CrossRef]
- Grishaev, A.; Ying, J.; Canny, M.D.; Pardi, A.; Bax, A. Solution structure of tRNAVal from refinement of homology model against residual dipolar coupling and SAXS data. J. Biomol. NMR 2008, 42, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zuo, X.; Yu, P.; Xu, H.; Starich, M.R.; Tiede, D.M.; Shapiro, B.A.; Schwieters, C.D.; Wang, Y.X. A method for helical RNA global structure determination in solution using small-angle x-ray scattering and NMR measurements. J. Mol. Biol. 2009, 393, 717–734. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.X.; Zuo, X.; Wang, J.; Yu, P.; Butcher, S.E. Rapid global structure determination of large RNA and RNA complexes using NMR and small-angle X-ray scattering. Methods 2010, 52, 180–191. [Google Scholar] [CrossRef] [PubMed]
- Jain, N.; Morgan, C.E.; Rife, B.D.; Salemi, M.; Tolbert, B.S. Solution structure of the HIV-1 Intron splicing silencer and its interactions with the UP1 domain of heterogeneous nuclear Ribonucleoprotein (hnRNP) A1. J. Biol. Chem. 2016, 291, 2331–2344. [Google Scholar] [CrossRef]
- Fang, X.; Wang, J.; O’Carroll, I.P.; Mitchell, M.; Zuo, X.; Wang, Y.; Yu, P.; Liu, Y.; Rausch, J.W.; Dyba, M.A.; et al. An unusual topological structure of the HIV-1 Rev response element. Cell 2013, 155, 594–605. [Google Scholar] [CrossRef]
- Zuo, X.; Wang, J.; Yu, P.; Eyler, D.; Xu, H.; Starich, M.R.; Tiede, D.M.; Simon, A.E.; Kasprzak, W.; Schwieters, C.D.; et al. Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3’ UTR of turnip crinkle virus. Proc. Natl. Acad. Sci. USA 2010, 107, 1385–1390. [Google Scholar] [CrossRef]
- Burke, J.E.; Sashital, D.G.; Zuo, X.; Wang, Y.X.; Butcher, S.E. Structure of the yeast U2/U6 snRNA complex. RNA 2012, 18, 673–683. [Google Scholar] [CrossRef][Green Version]
- Cornilescu, G.; Didychuk, A.L.; Rodgers, M.L.; Michael, L.A.; Burke, J.E.; Montemayor, E.J.; Hoskins, A.A.; Butcher, S.E. Structural Analysis of multi-helical RNAs by NMR-SAXS/WAXS: Application to the U4/U6 di-snRNA. J. Mol. Biol. 2016, 428, 777–789. [Google Scholar] [CrossRef]
- Imai, S.; Kumar, P.; Hellen, C.U.; D’Souza, V.M.; Wagner, G. An accurately preorganized IRES RNA structure enables eIF4G capture for initiation of viral translation. Nat. Struct. Mol. Biol. 2016, 23, 859–864. [Google Scholar] [CrossRef]
- Luque, D.; Caston, J.R. Cryo-electron microscopy for the study of virus assembly. Nat. Chem. Biol. 2020, 16, 231–239. [Google Scholar] [CrossRef]
- Merk, A.; Bartesaghi, A.; Banerjee, S.; Falconieri, V.; Rao, P.; Davis, M.I.; Pragani, R.; Boxer, M.B.; Earl, L.A.; Milne, J.L.S.; et al. Breaking Cryo-EM resolution barriers to facilitate drug discovery. Cell 2016, 165, 1698–1707. [Google Scholar] [CrossRef] [PubMed]
- Geraets, J.A.; Pothula, K.R.; Schroder, G.F. Integrating cryo-EM and NMR data. Curr. Opin. Struct. Biol. 2020, 61, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Levin, J.G.; Rosenak, M.J. Synthesis of murine leukemia virus proteins associated with virions assembled in actinomycin D-treated cells: Evidence for persistence of viral messenger RNA. Proc. Natl. Acad. Sci. USA 1976, 73, 1154–1158. [Google Scholar] [CrossRef] [PubMed]
- Mougel, M.; Akkawi, C.; Chamontin, C.; Feuillard, J.; Pessel-Vivares, L.; Socol, M.; Laine, S. NXF1 and CRM1 nuclear export pathways orchestrate nuclear export, translation and packaging of murine leukaemia retrovirus unspliced RNA. RNA Biol. 2020, 17, 528–538. [Google Scholar] [CrossRef]
- Malim, M.H.; Hauber, J.; Le, S.Y.; Maizel, J.V.; Cullen, B.R. The HIV-1 rev transactivator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 1989, 338, 254–257. [Google Scholar] [CrossRef]
- Zhang, H.; Keane, S.C. Advances that facilitate the study of large RNA structure and dynamics by nuclear magnetic resonance spectroscopy. Wiley Interdiscip. Rev. RNA 2019, 10, e1541. [Google Scholar] [CrossRef]
- Liu, Y.; Holmstrom, E.; Zhang, J.; Yu, P.; Wang, J.; Dyba, M.A.; Chen, D.; Ying, J.; Lockett, S.; Nesbitt, D.J.; et al. Synthesis and applications of RNAs with position-selective labelling and mosaic composition. Nature 2015, 522, 368–372. [Google Scholar] [CrossRef]
- Marchant, J.; Bax, A.; Summers, M.F. Accurate measurement of residual dipolar couplings in large RNAs by Variable flip angle NMR. J. Am. Chem. Soc. 2018, 140, 6978–6983. [Google Scholar] [CrossRef]
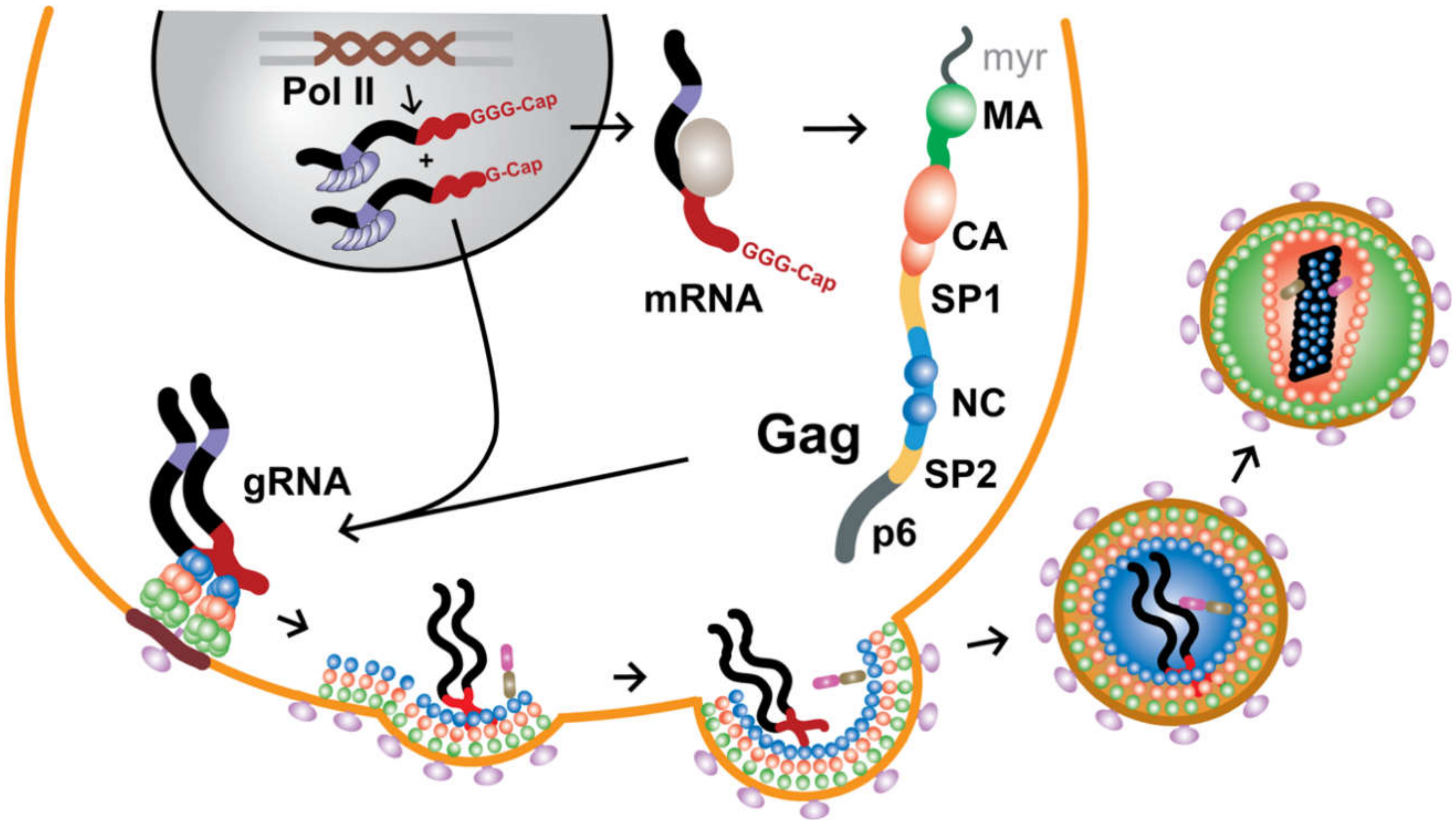
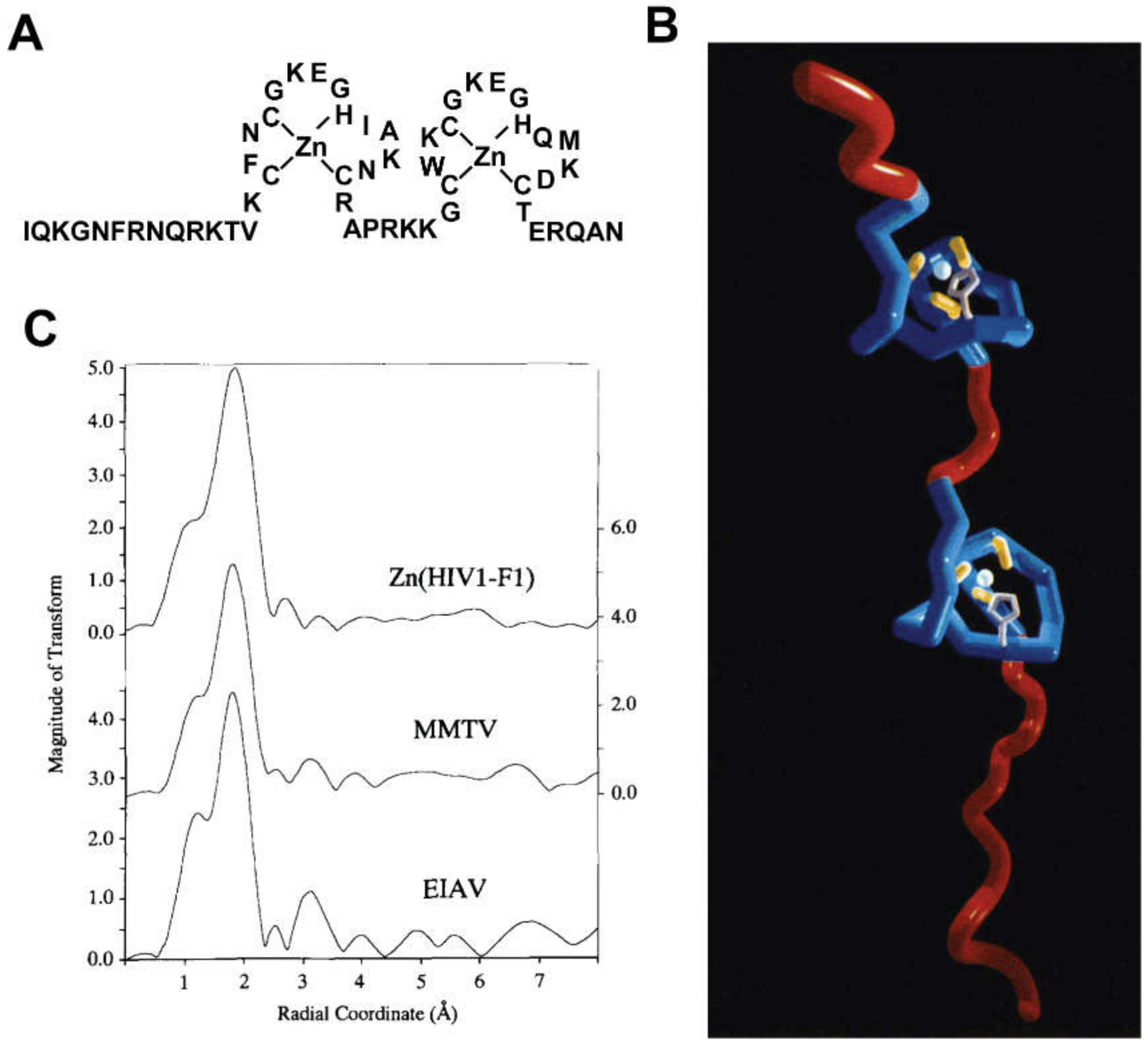
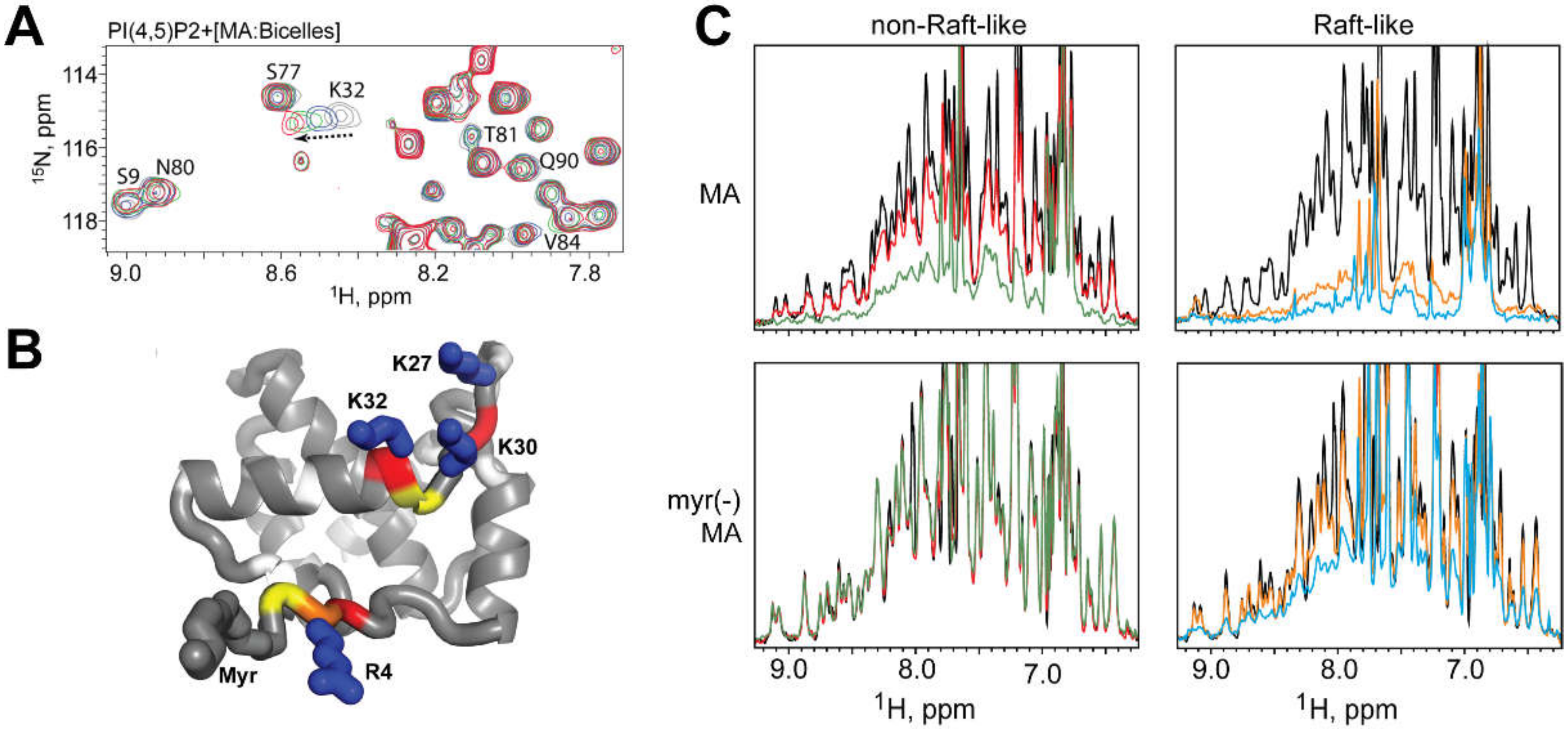
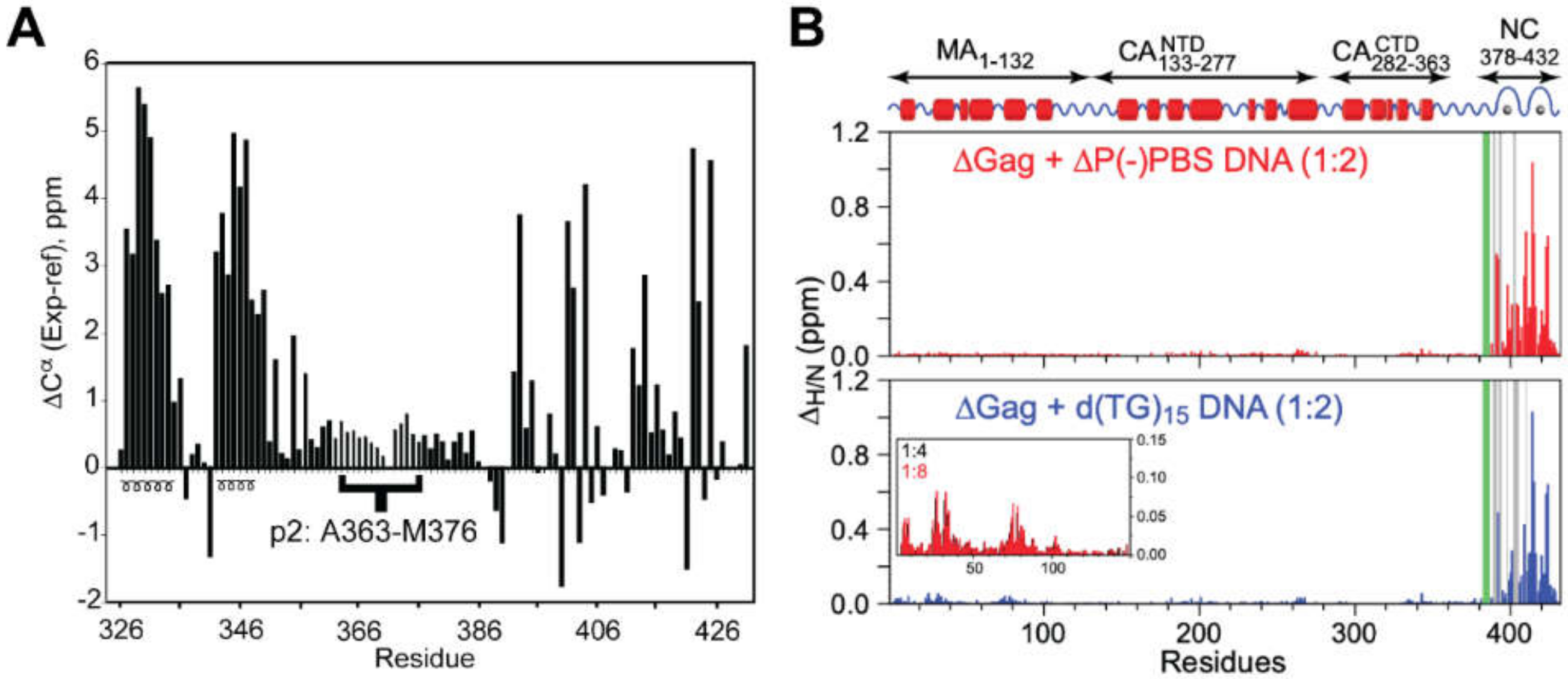
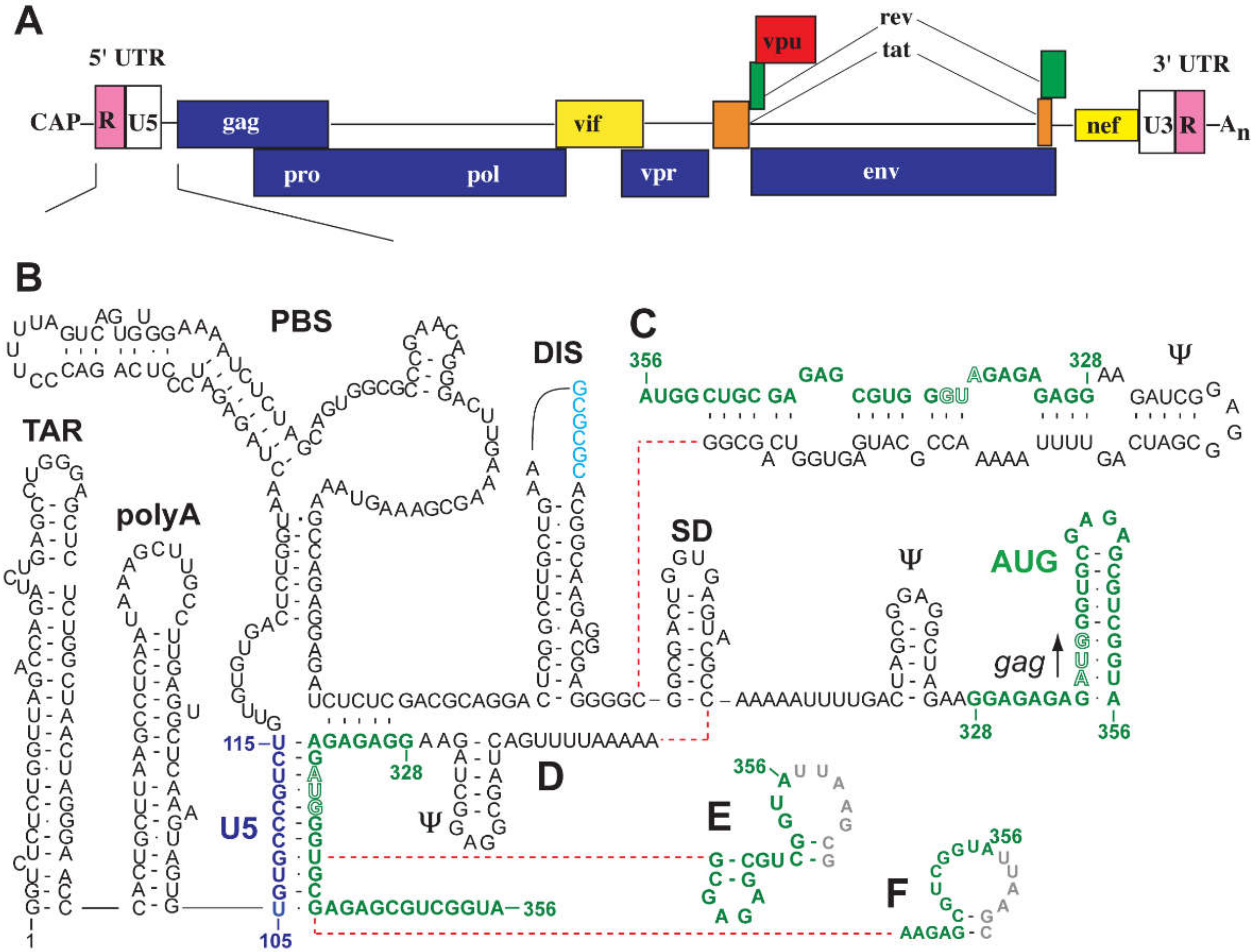
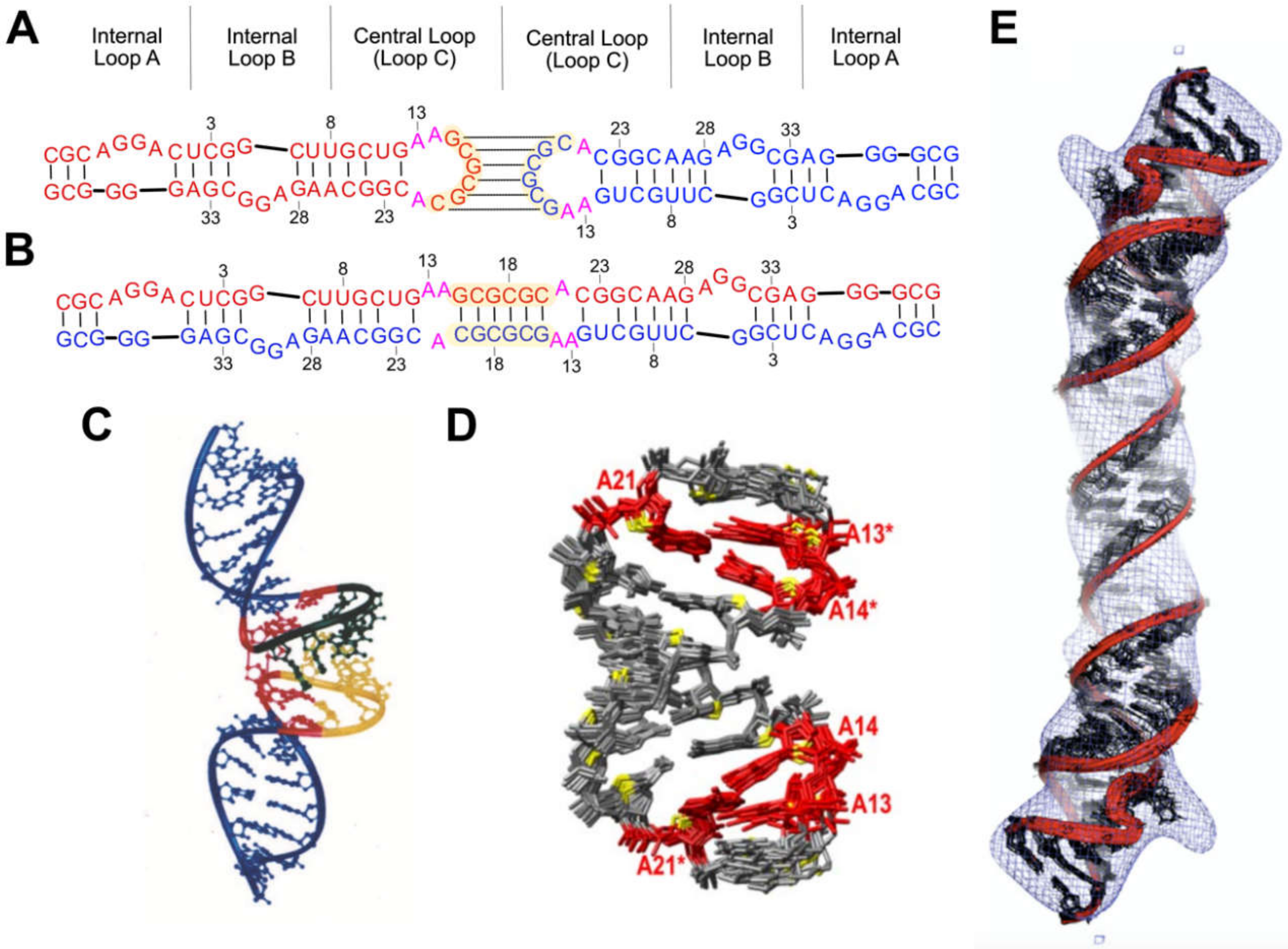
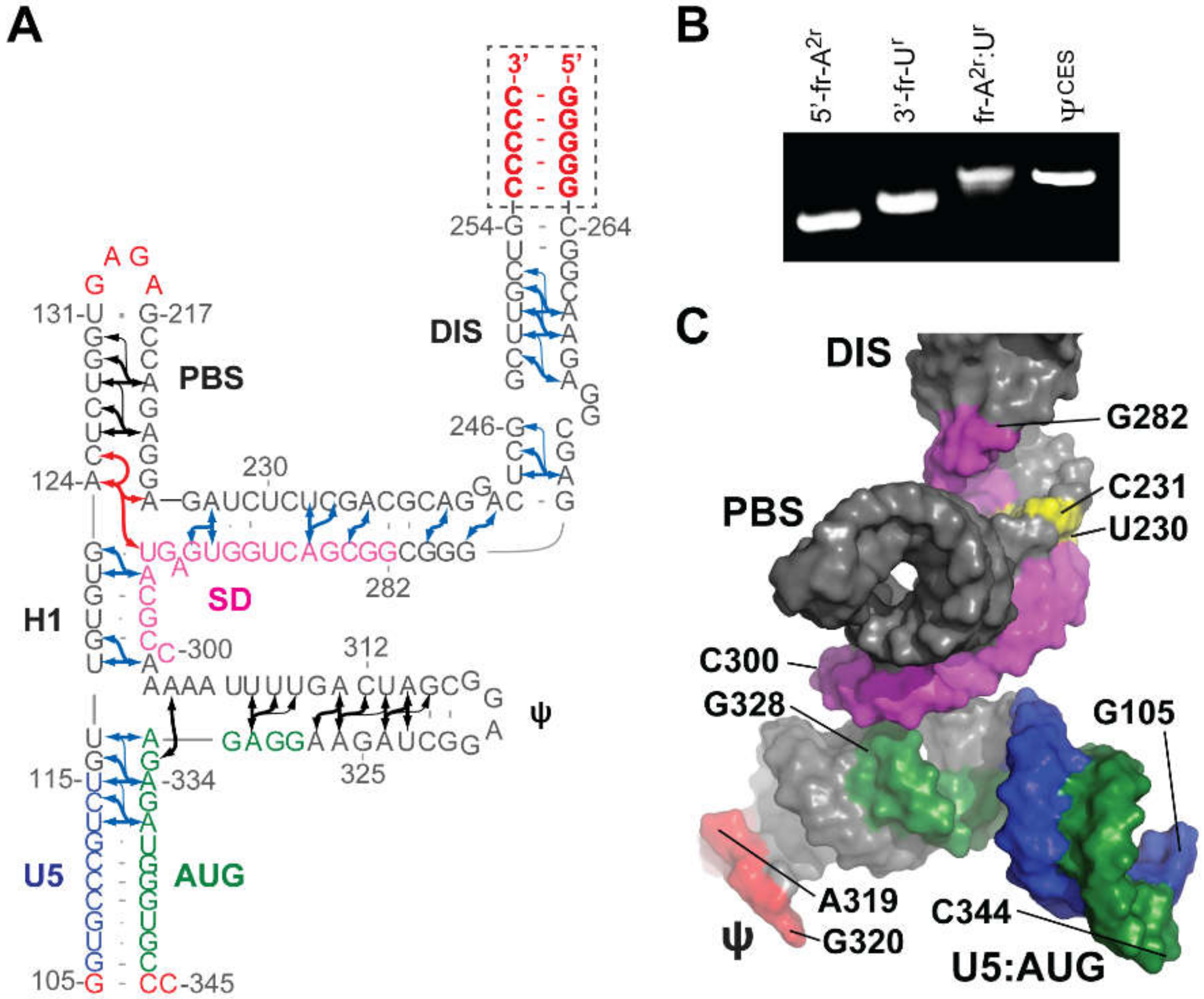
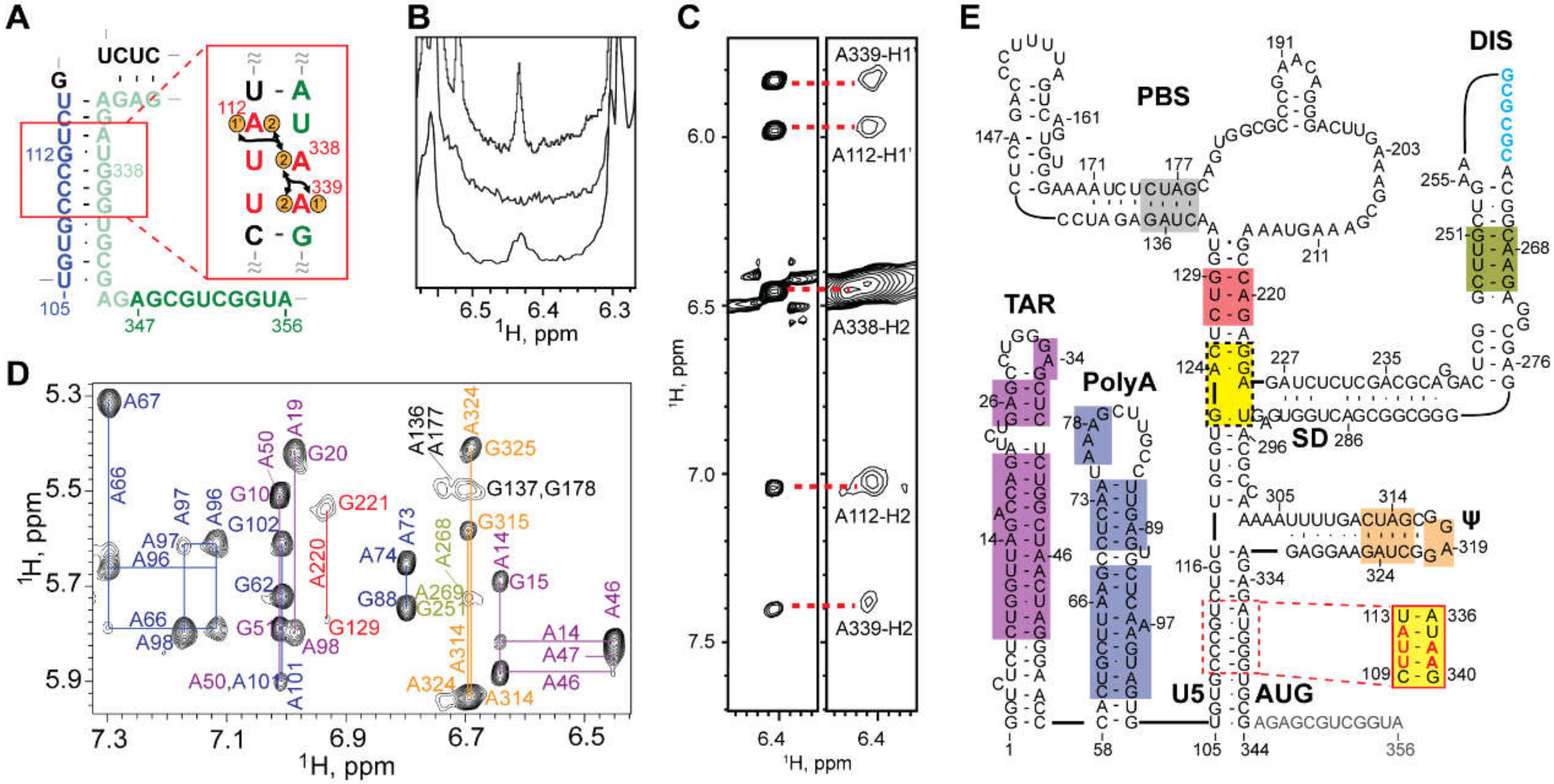
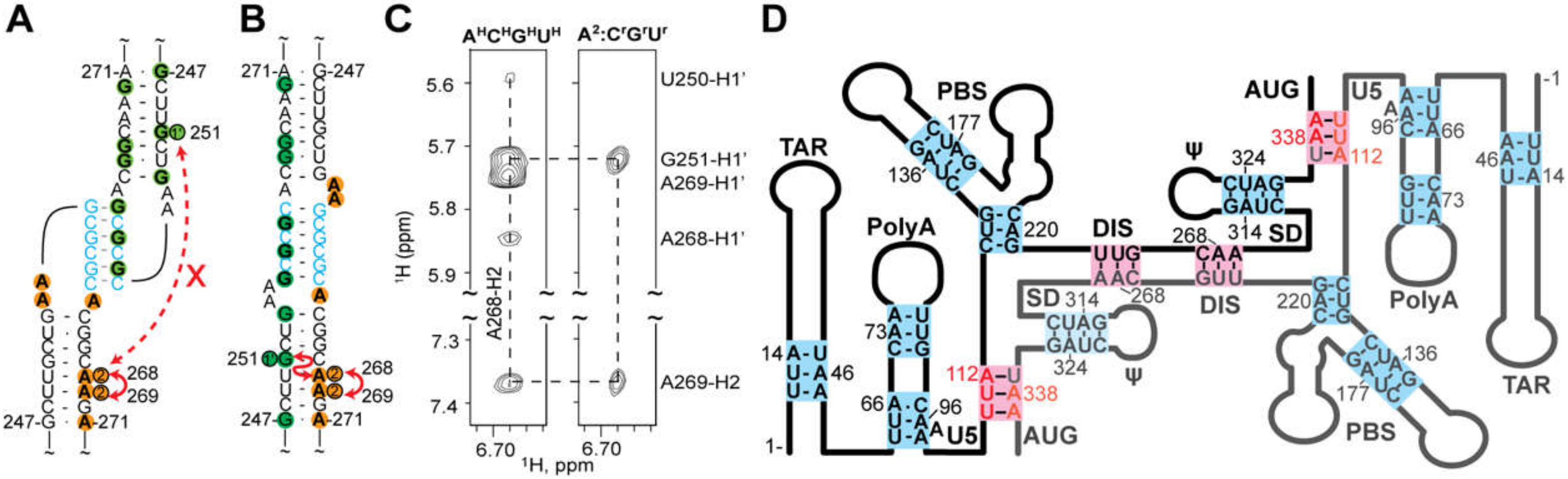
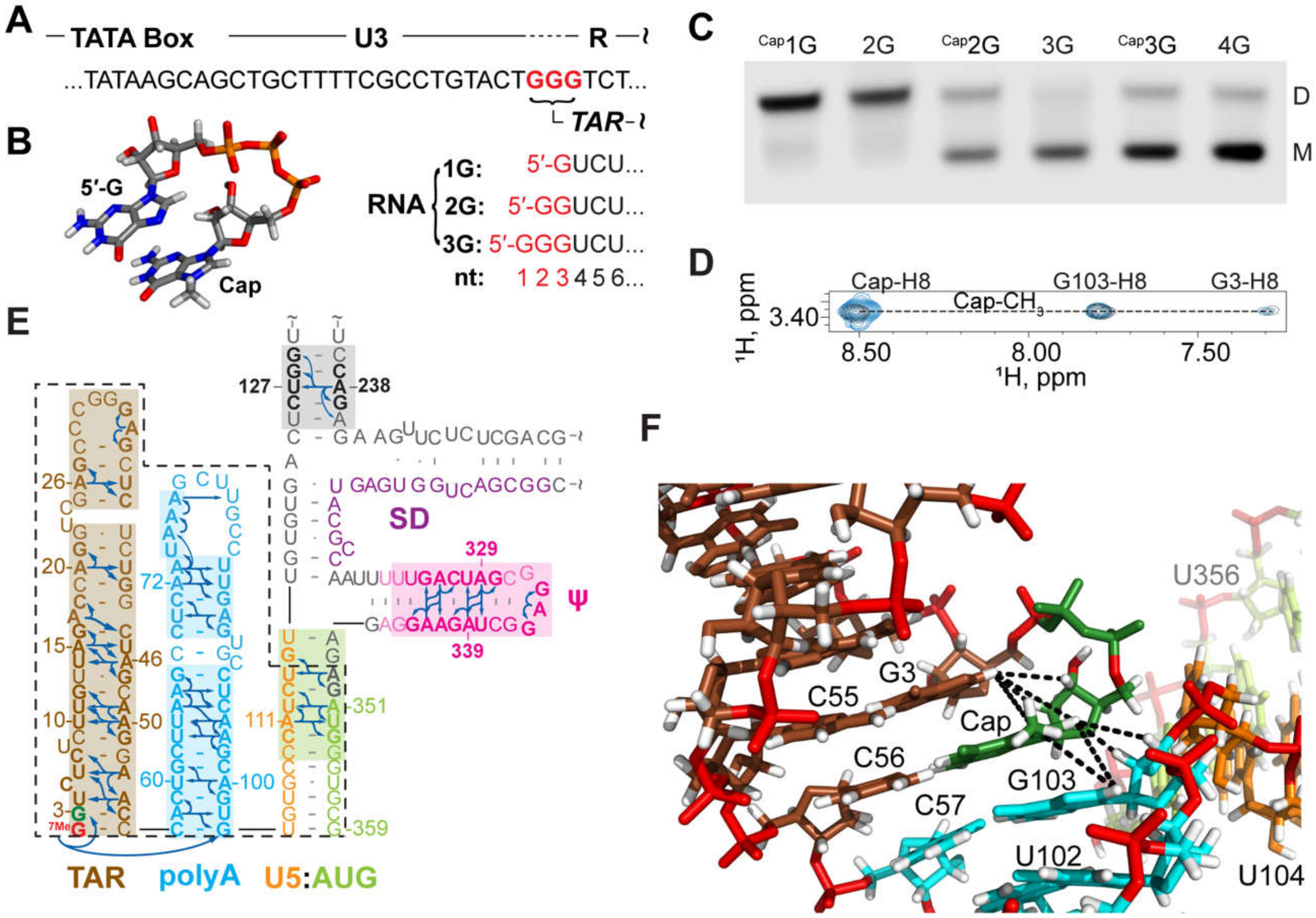
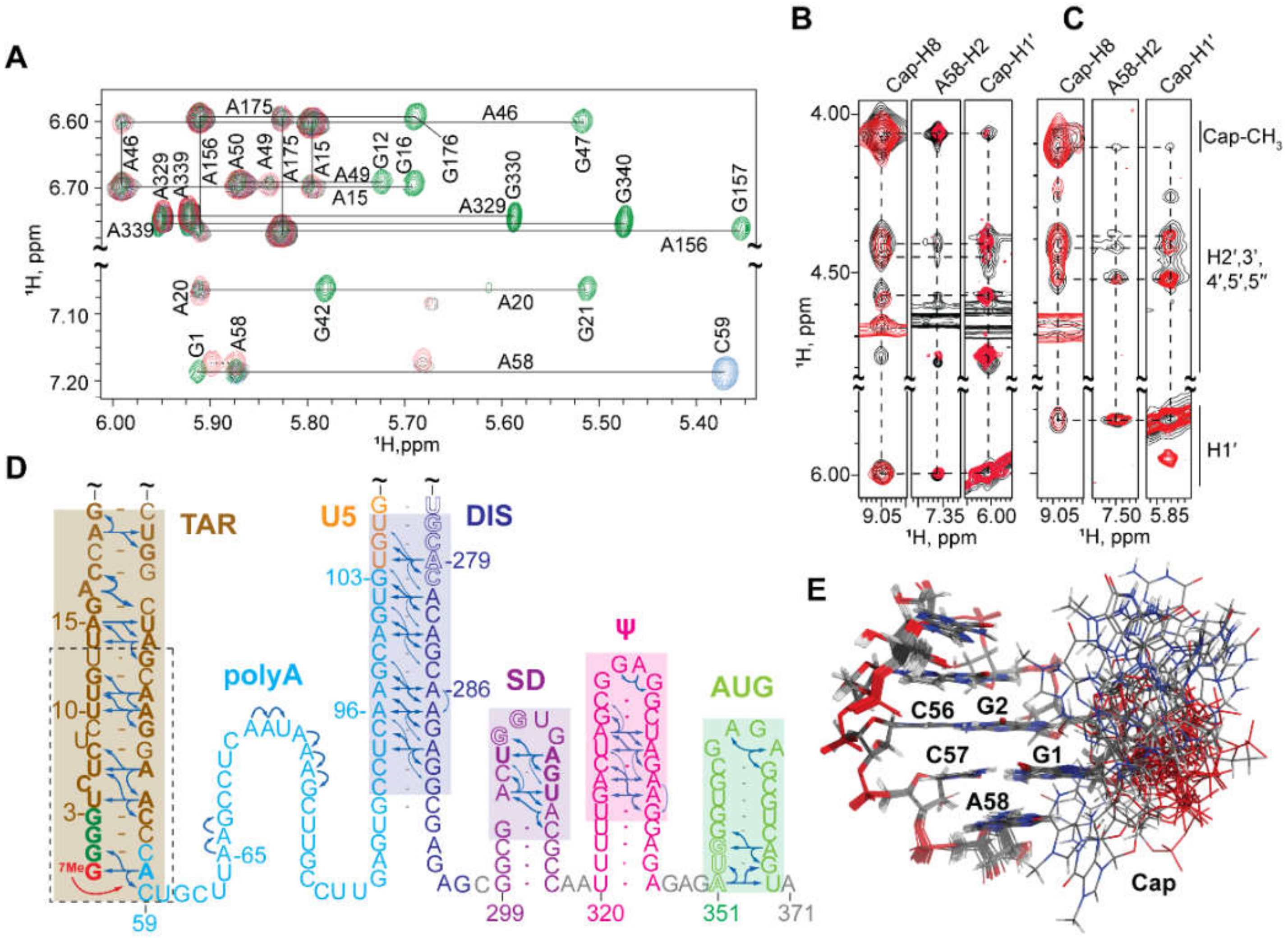
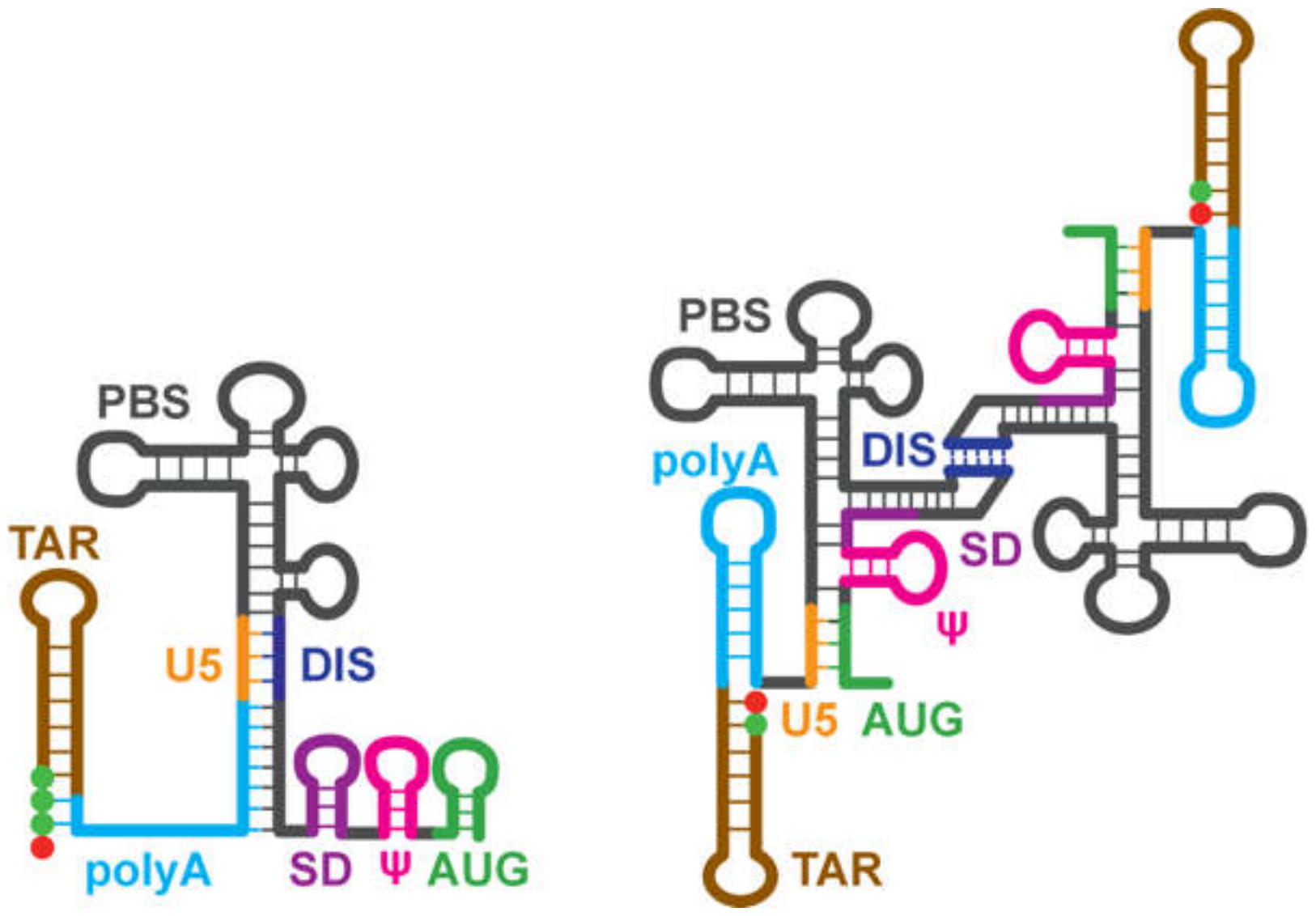
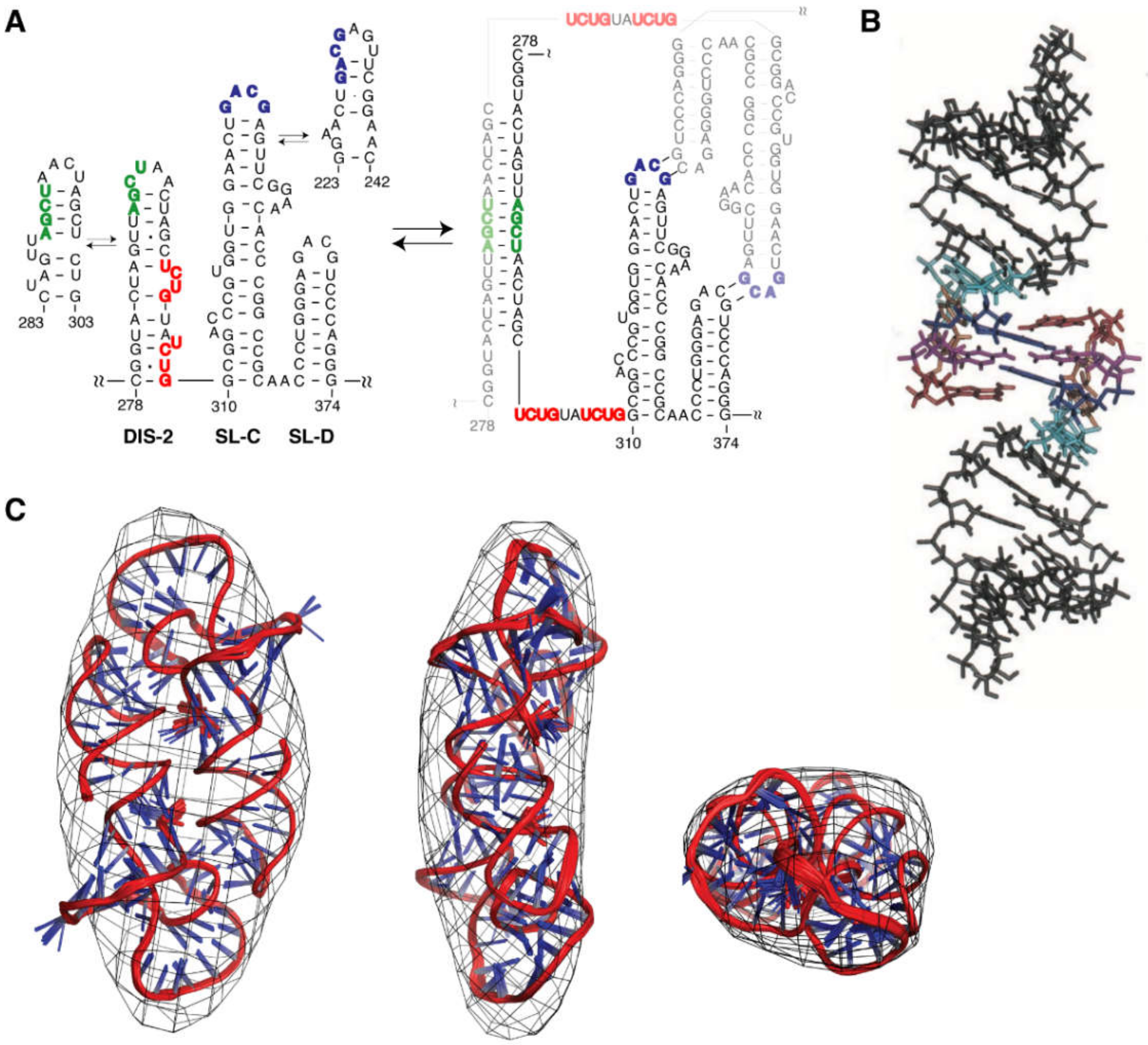

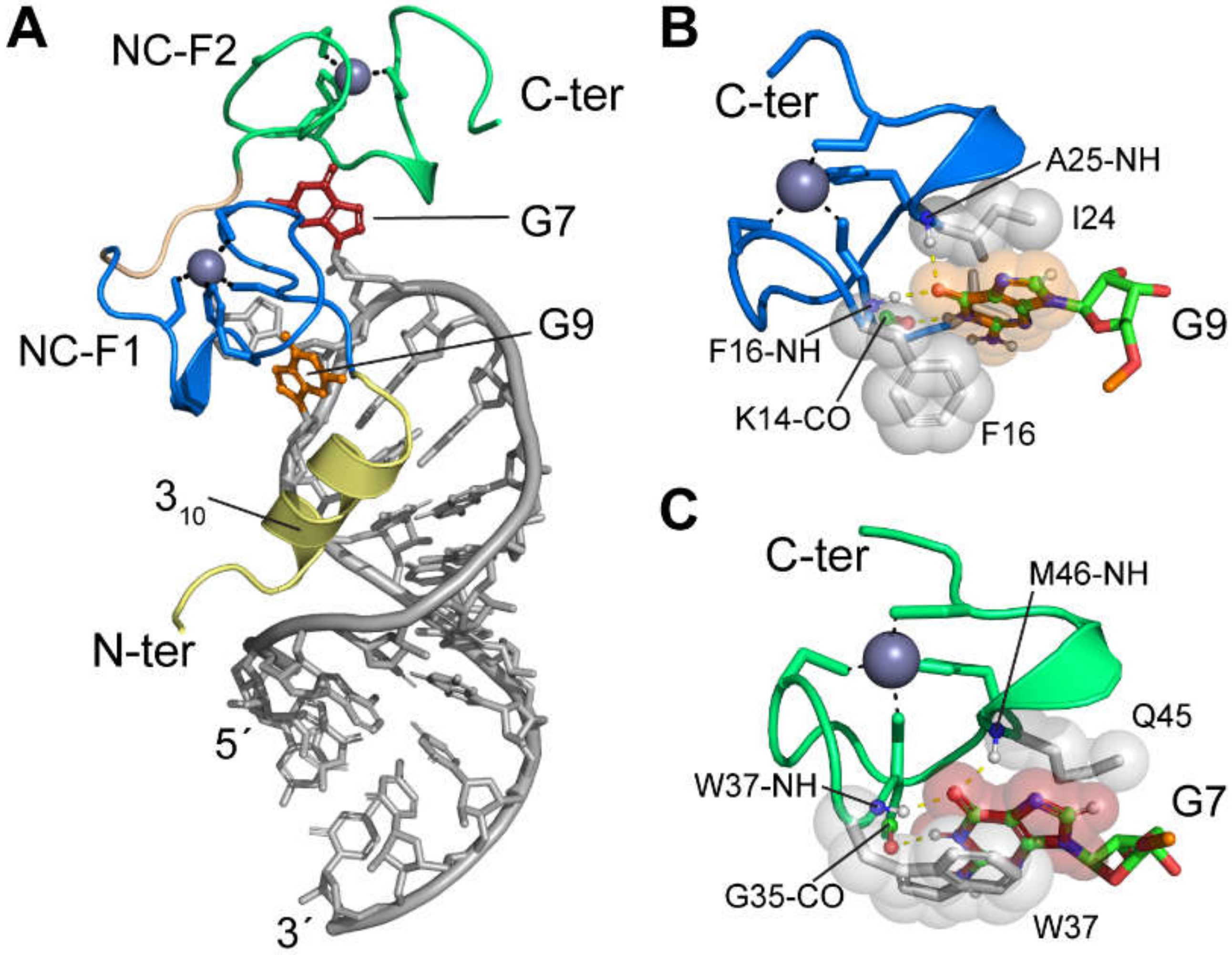
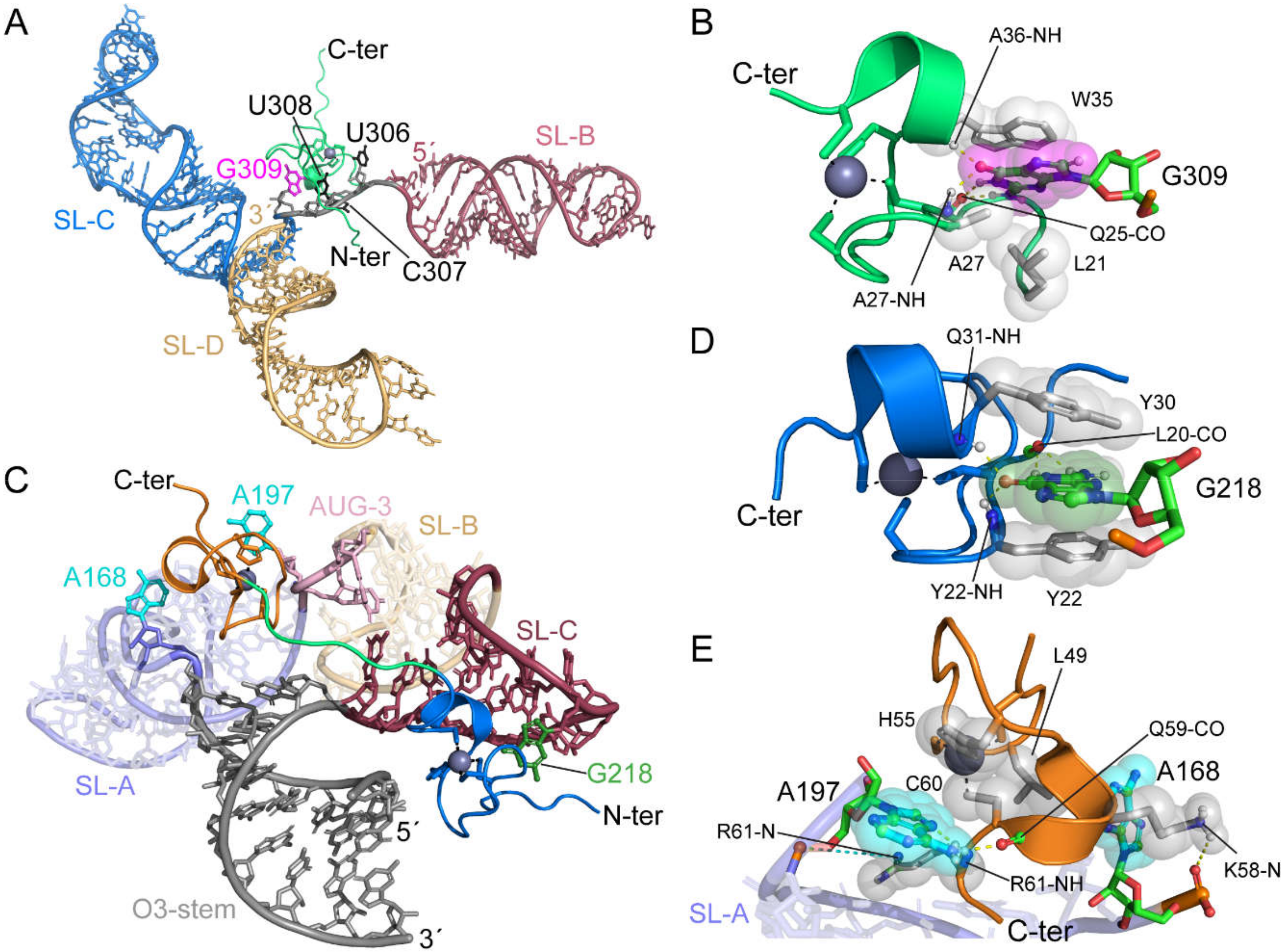
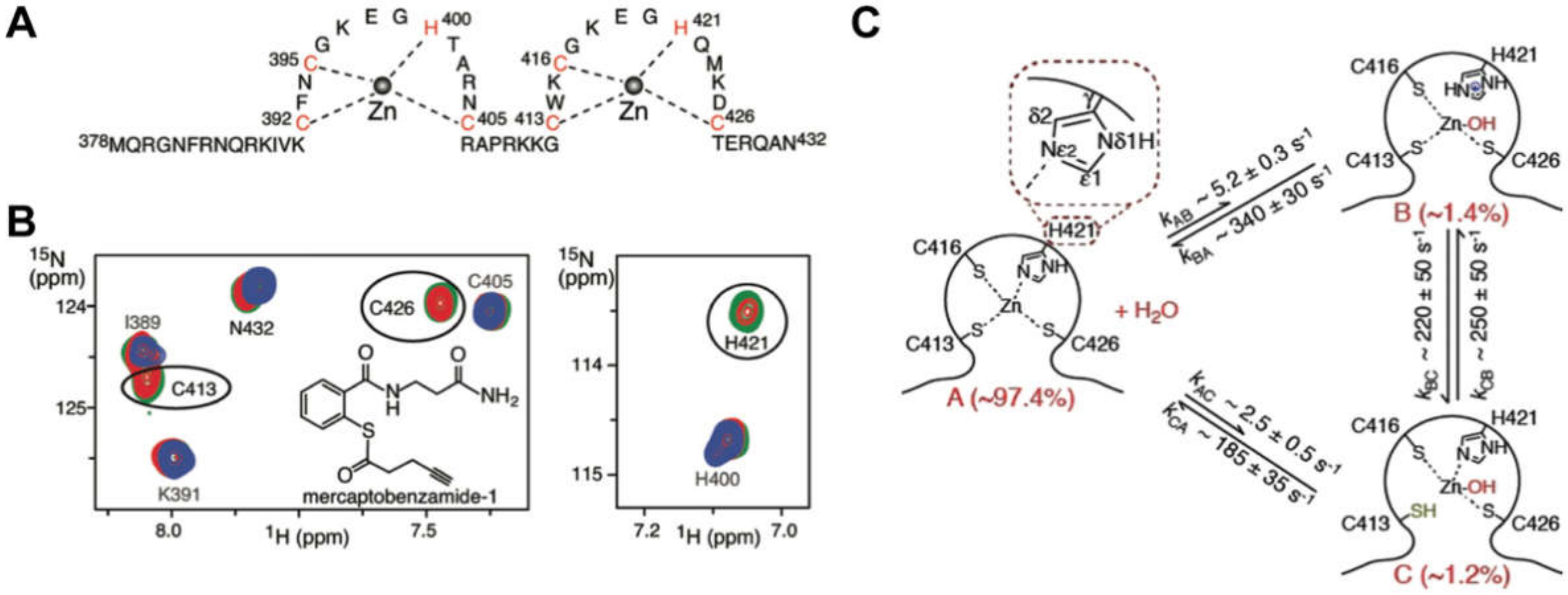
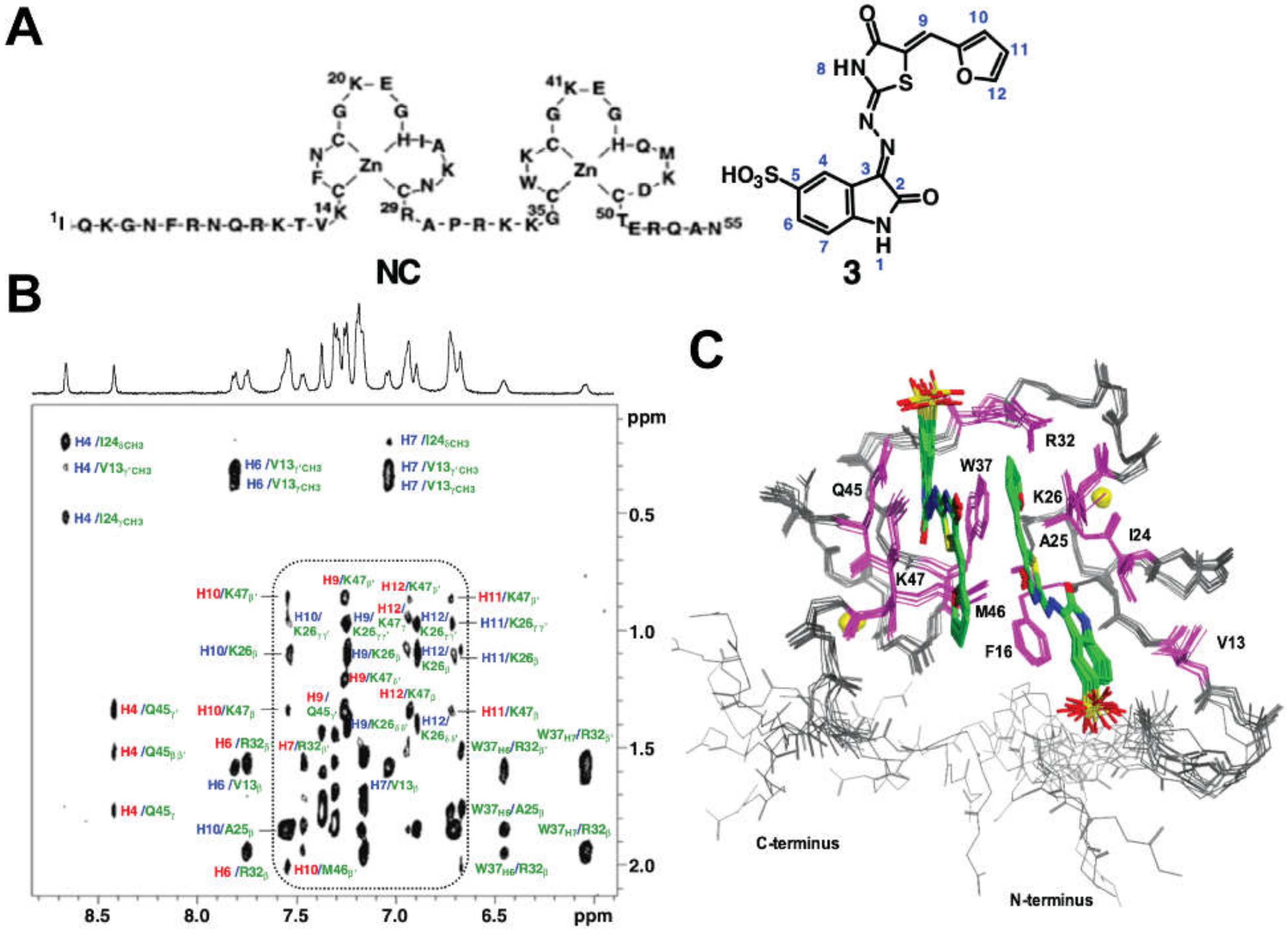
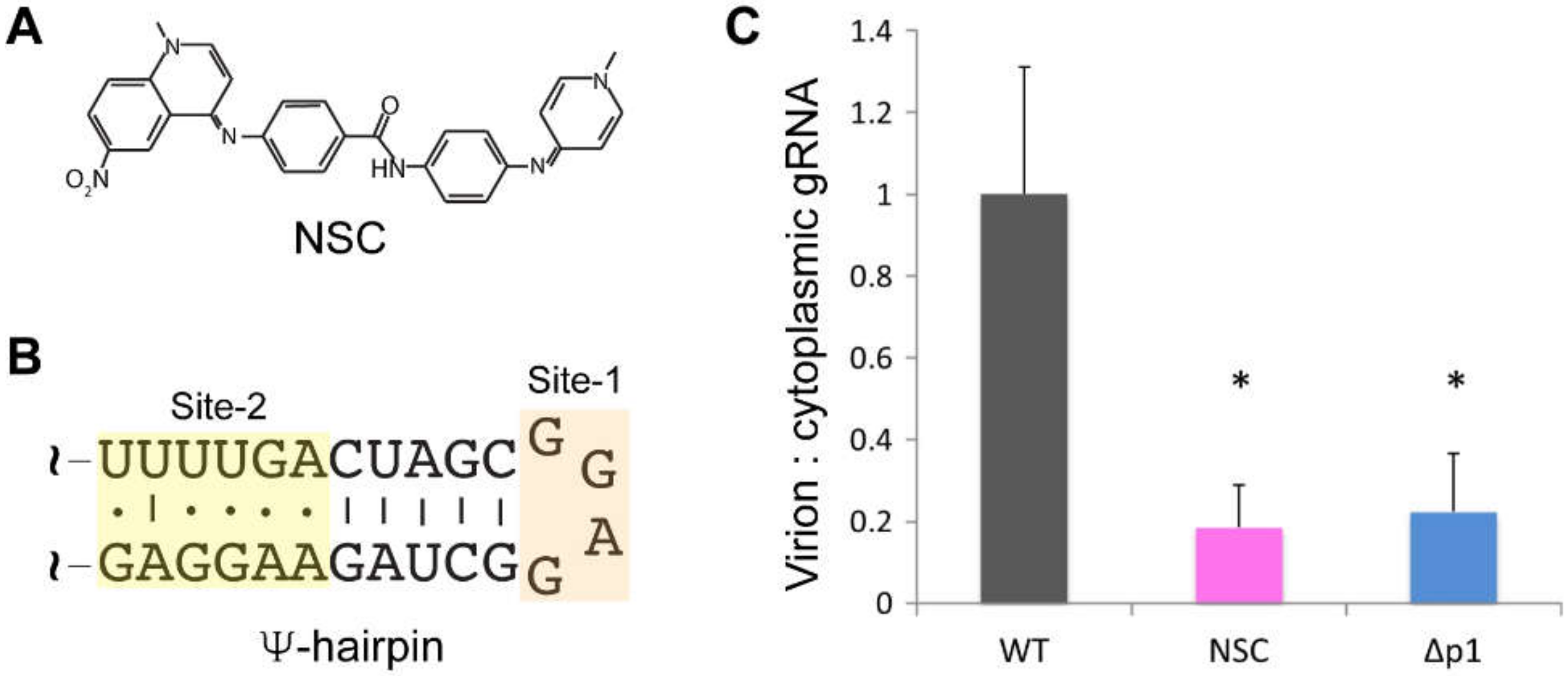
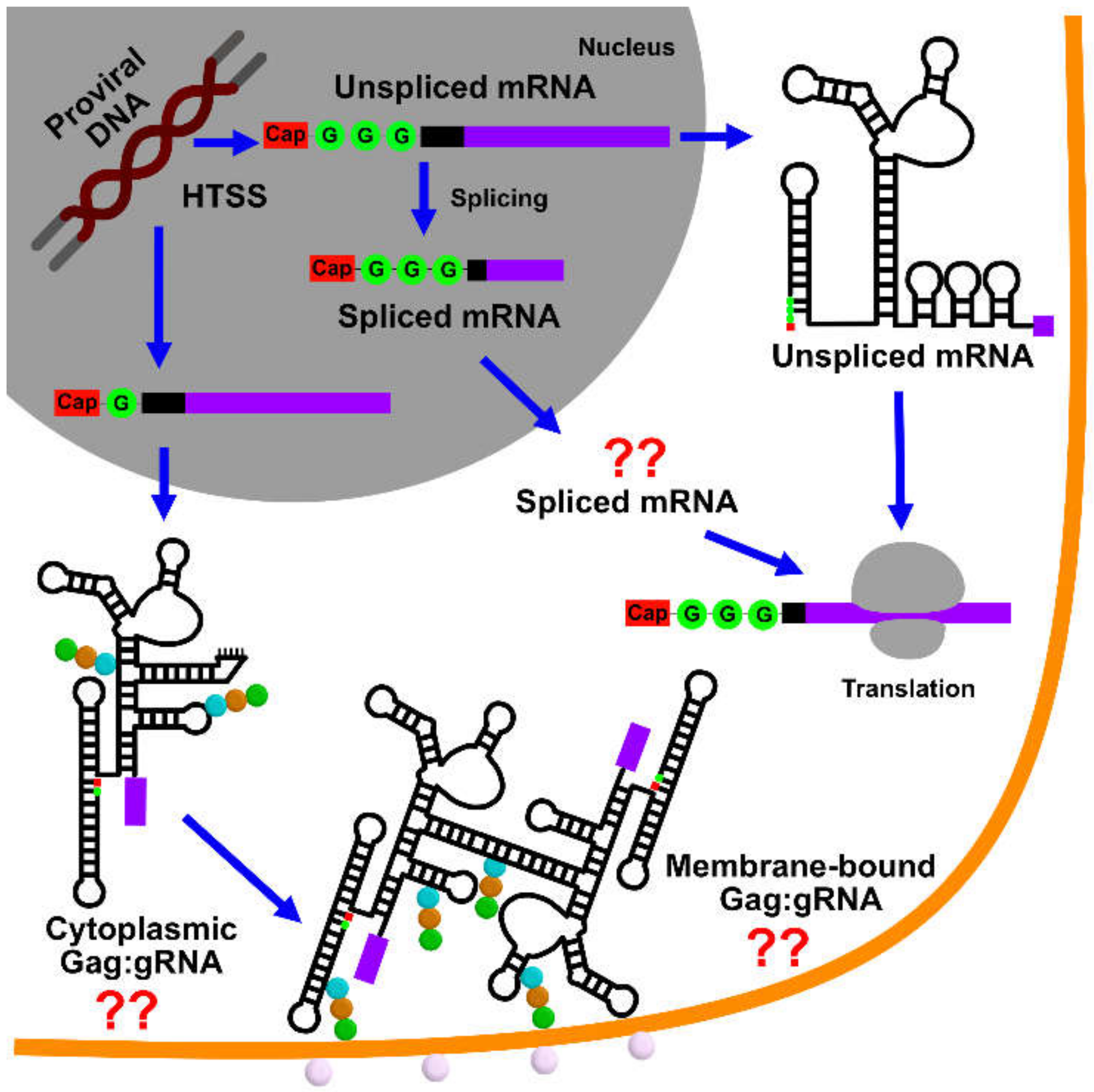
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boyd, P.S.; Brown, J.B.; Brown, J.D.; Catazaro, J.; Chaudry, I.; Ding, P.; Dong, X.; Marchant, J.; O’Hern, C.T.; Singh, K.; et al. NMR Studies of Retroviral Genome Packaging. Viruses 2020, 12, 1115. https://doi.org/10.3390/v12101115
Boyd PS, Brown JB, Brown JD, Catazaro J, Chaudry I, Ding P, Dong X, Marchant J, O’Hern CT, Singh K, et al. NMR Studies of Retroviral Genome Packaging. Viruses. 2020; 12(10):1115. https://doi.org/10.3390/v12101115
Chicago/Turabian StyleBoyd, Patricia S., Janae B. Brown, Joshua D. Brown, Jonathan Catazaro, Issac Chaudry, Pengfei Ding, Xinmei Dong, Jan Marchant, Colin T. O’Hern, Karndeep Singh, and et al. 2020. "NMR Studies of Retroviral Genome Packaging" Viruses 12, no. 10: 1115. https://doi.org/10.3390/v12101115
APA StyleBoyd, P. S., Brown, J. B., Brown, J. D., Catazaro, J., Chaudry, I., Ding, P., Dong, X., Marchant, J., O’Hern, C. T., Singh, K., Swanson, C., Summers, M. F., & Yasin, S. (2020). NMR Studies of Retroviral Genome Packaging. Viruses, 12(10), 1115. https://doi.org/10.3390/v12101115






