Host Gene Regulation by Transposable Elements: The New, the Old and the Ugly
Abstract
1. Introduction
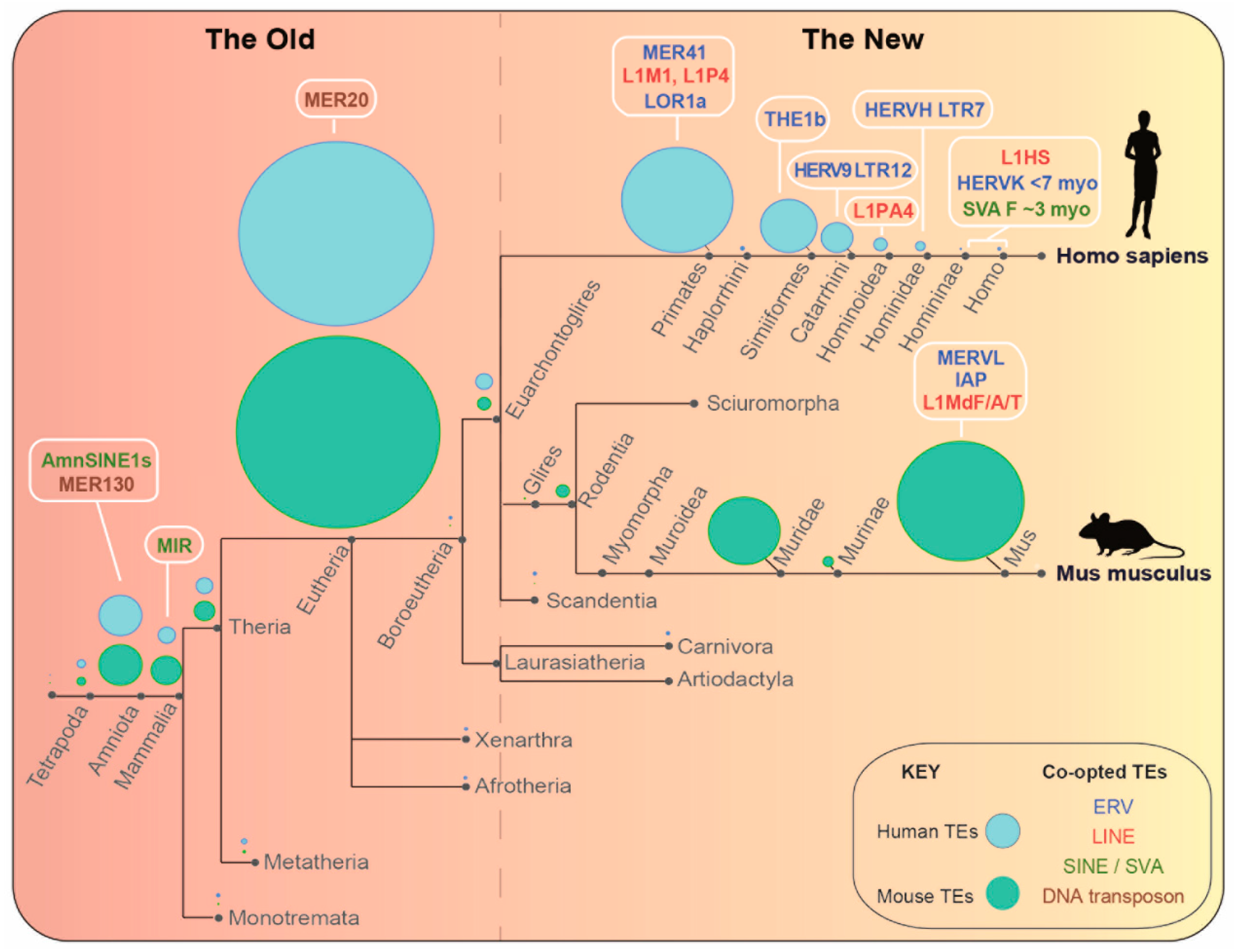
2. Gene Regulation by Transposable Elements: The New
2.1. New Transposable Elements in Mouse
2.1.1. Co-Option of TEs to Regulate Gene Networks
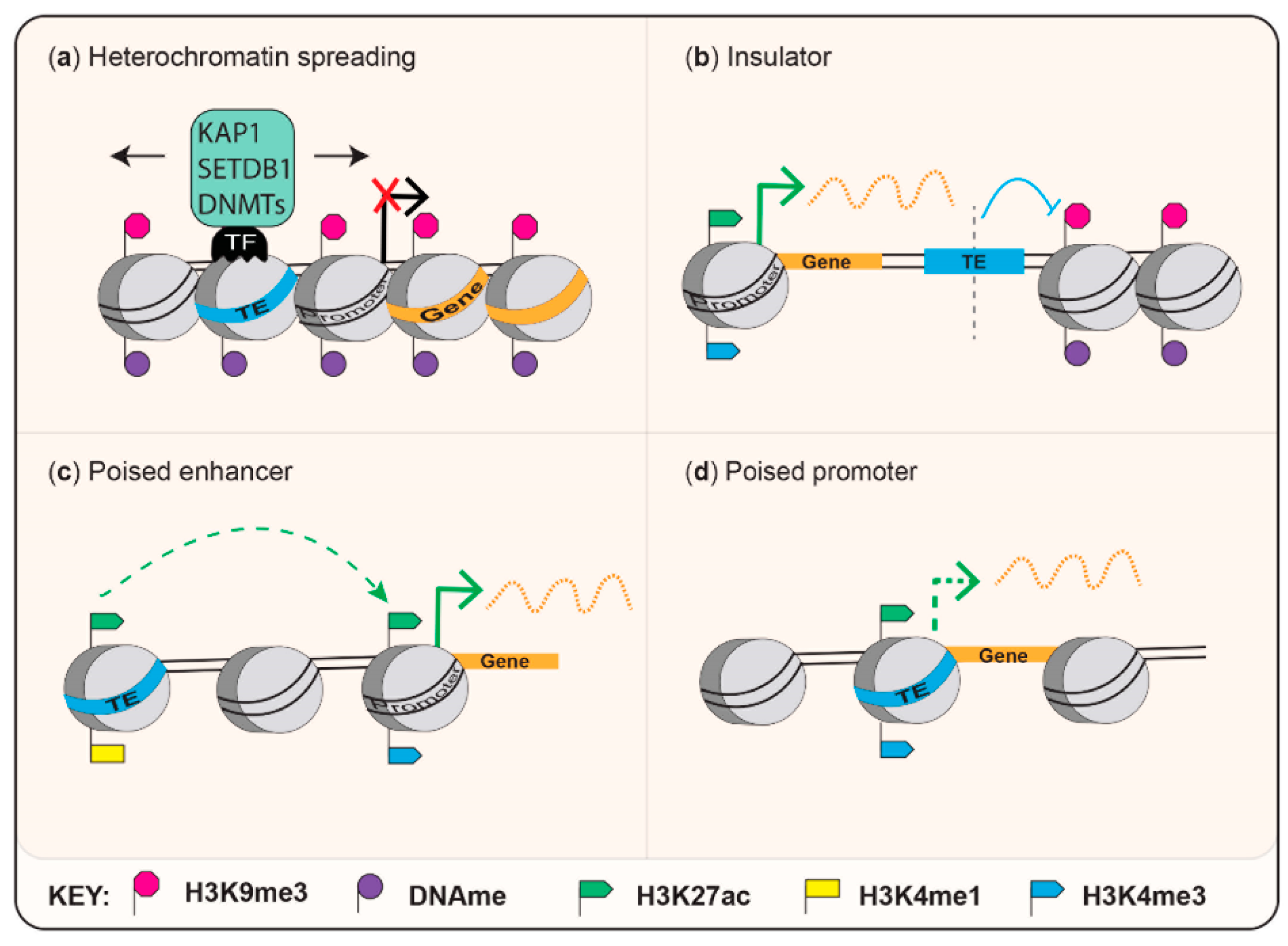
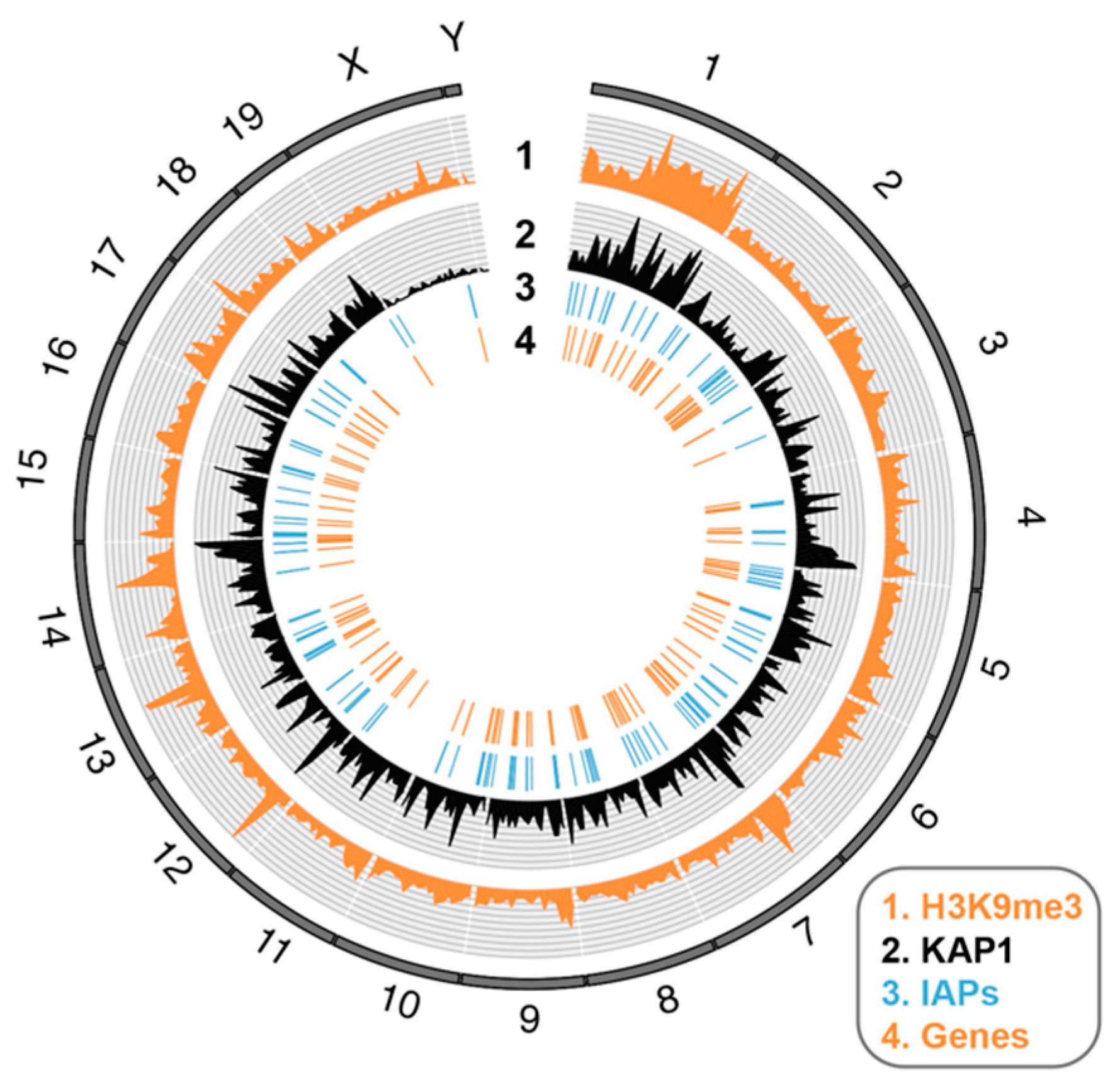
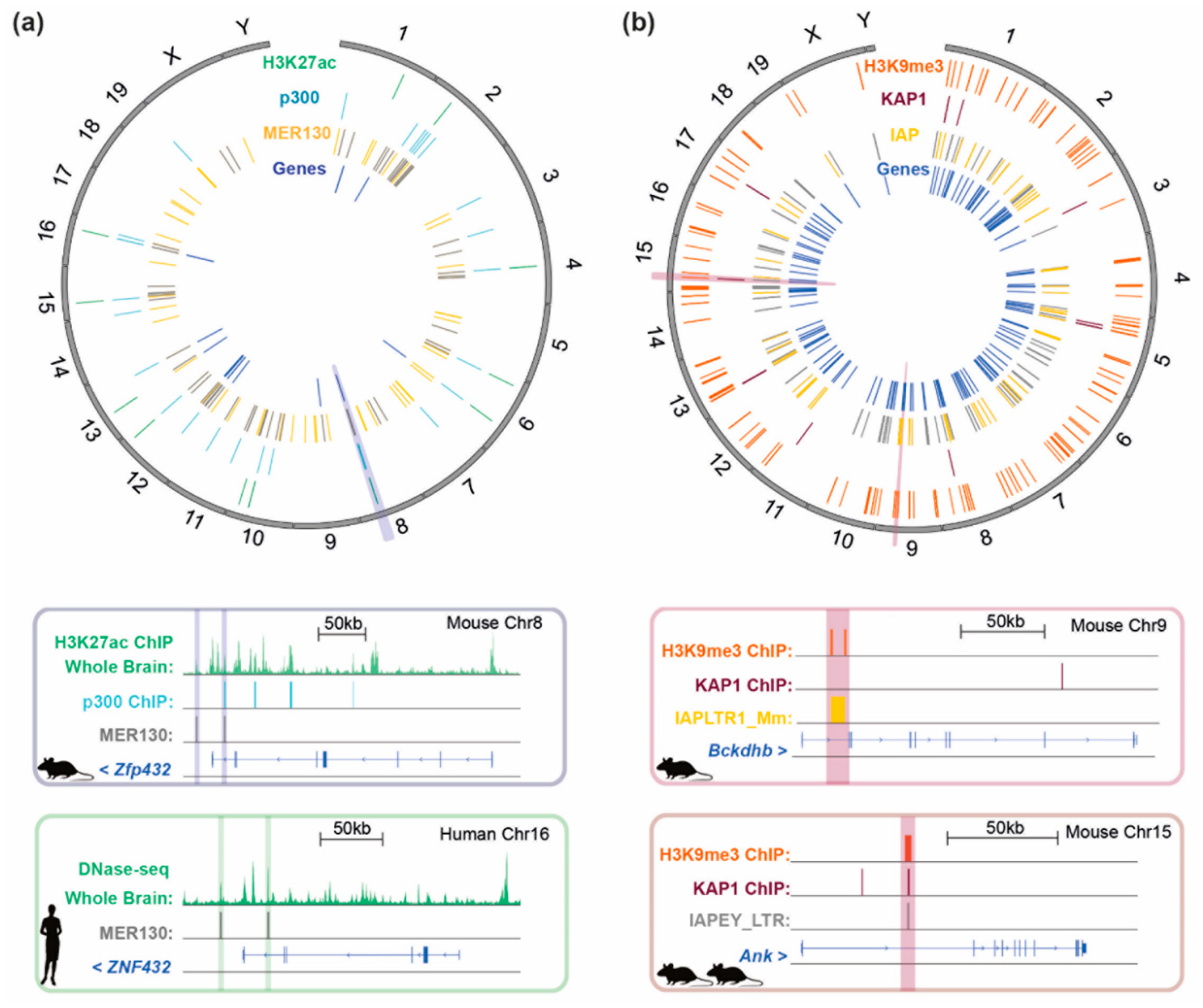
2.1.2. Sequence-Specific Epigenetic Silencing
2.1.3. Metastable Epialleles
2.1.4. Position-Effect Variegation
2.2. New TEs in Humans and Convergent Co-Option
2.2.1. The HUSH Complex
2.2.2. A TE Origin to Genomic Imprinting
2.2.3. Fighting Fire with Fire: TEs as Effectors of Immunity
3. Gene Regulation by Transposable Elements: The Old
3.1. When X-Chromosome Inactivation Is on the LINE
3.2. Ancient TEs Shaping the Brain of Mammals
3.3. Ancient Mammalian-Conserved TEs as Insulators
4. Gene Regulation by Transposable Elements: The Ugly
4.1. Gene Dysregulation by TEs in Cancer
4.2. Gene Dysregulation by TEs in Autoimmune Disease
5. Conclusions
6. Methods
6.1. Expression Analysis in Mouse Pre-Implantation Development
6.2. Genome-Wide Visualization of Features
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Theunissen, T.W.; Jaenisch, R. Mechanisms of gene regulation in human embryos and pluripotent stem cells. Development 2017, 144, 4496–4509. [Google Scholar] [CrossRef] [PubMed]
- de Koning, A.P.; Gu, W.; Castoe, T.A.; Batzer, M.A.; Pollock, D.D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011, 7, e1002384. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Babaian, A.; Mager, D.L. Endogenous retroviral promoter exaptation in human cancer. Mob. DNA 2016, 7, 24. [Google Scholar] [CrossRef]
- Friedli, M.; Trono, D. The developmental control of transposable elements and the evolution of higher species. Annu. Rev. Cell Dev. Biol. 2015, 31, 429–451. [Google Scholar] [CrossRef]
- Percharde, M.; Sultana, T.; Ramalho-Santos, M. What Doesn’t Kill You Makes You Stronger: Transposons as Dual Players in Chromatin Regulation and Genomic Variation. BioEssays News Rev. Mol. Cell. Dev. Biol. 2020, 42, e1900232. [Google Scholar] [CrossRef]
- Robbez-Masson, L.; Rowe, H.M. Retrotransposons shape species-specific embryonic stem cell gene expression. Retrovirology 2015, 12, 45. [Google Scholar] [CrossRef]
- Thompson, P.J.; Macfarlan, T.S.; Lorincz, M.C. Long Terminal Repeats: From Parasitic Elements to Building Blocks of the Transcriptional Regulatory Repertoire. Mol. Cell 2016, 62, 766–776. [Google Scholar] [CrossRef]
- Bruno, M.; Mahgoub, M.; Macfarlan, T.S. The Arms Race Between KRAB-Zinc Finger Proteins and Endogenous Retroelements and Its Impact on Mammals. Annu. Rev. Genet. 2019, 53, 393–416. [Google Scholar] [CrossRef]
- Deniz, O.; Frost, J.M.; Branco, M.R. Regulation of transposable elements by DNA modifications. Nat. Rev. Genet. 2019, 20, 417–431. [Google Scholar] [CrossRef]
- Fukuda, K.; Okuda, A.; Yusa, K.; Shinkai, Y. A CRISPR knockout screen identifies SETDB1-target retroelement silencing factors in embryonic stem cells. Genome Res. 2018, 28, 846–858. [Google Scholar] [CrossRef] [PubMed]
- Magiorkinis, G.; Blanco-Melo, D.; Belshaw, R. The decline of human endogenous retroviruses: Extinction and survival. Retrovirology 2015, 12, 8. [Google Scholar] [CrossRef]
- Brouha, B.; Schustak, J.; Badge, R.M.; Lutz-Prigge, S.; Farley, A.H.; Moran, J.V.; Kazazian, H.H., Jr. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. USA 2003, 100, 5280–5285. [Google Scholar] [CrossRef] [PubMed]
- Gianfrancesco, O.; Geary, B.; Savage, A.L.; Billingsley, K.J.; Bubb, V.J.; Quinn, J.P. The Role of SINE-VNTR-Alu (SVA) Retrotransposons in Shaping the Human Genome. Int. J. Mol. Sci. 2019, 20, 5977. [Google Scholar] [CrossRef] [PubMed]
- Campos-Sanchez, R.; Cremona, M.A.; Pini, A.; Chiaromonte, F.; Makova, K.D. Integration and Fixation Preferences of Human and Mouse Endogenous Retroviruses Uncovered with Functional Data Analysis. PLoS Comput. Biol. 2016, 12, e1004956. [Google Scholar] [CrossRef] [PubMed]
- Goodier, J.L.; Ostertag, E.M.; Du, K.; Kazazian, H.H., Jr. A novel active L1 retrotransposon subfamily in the mouse. Genome Res. 2001, 11, 1677–1685. [Google Scholar] [CrossRef] [PubMed]
- Ribet, D.; Harper, F.; Dupressoir, A.; Dewannieux, M.; Pierron, G.; Heidmann, T. An infectious progenitor for the murine IAP retrotransposon: Emergence of an intracellular genetic parasite from an ancient retrovirus. Genome Res. 2008, 18, 597–609. [Google Scholar] [CrossRef]
- Kazachenka, A.; Bertozzi, T.M.; Sjoberg-Herrera, M.K.; Walker, N.; Gardner, J.; Gunning, R.; Pahita, E.; Adams, S.; Adams, D.; Ferguson-Smith, A.C. Identification, Characterization, and Heritability of Murine Metastable Epialleles: Implications for Non-genetic Inheritance. Cell 2018, 175, 1717. [Google Scholar] [CrossRef]
- Lilue, J.; Doran, A.G.; Fiddes, I.T.; Abrudan, M.; Armstrong, J.; Bennett, R.; Chow, W.; Collins, J.; Collins, S.; Czechanski, A.; et al. Sixteen diverse laboratory mouse reference genomes define strain-specific haplotypes and novel functional loci. Nat. Genet. 2018, 50, 1574–1583. [Google Scholar] [CrossRef]
- Rebollo, R.; Karimi, M.M.; Bilenky, M.; Gagnier, L.; Miceli-Royer, K.; Zhang, Y.; Goyal, P.; Keane, T.M.; Jones, S.; Hirst, M.; et al. Retrotransposon-induced heterochromatin spreading in the mouse revealed by insertional polymorphisms. PLoS Genet. 2011, 7, e1002301. [Google Scholar] [CrossRef]
- Rowe, H.M.; Kapopoulou, A.; Corsinotti, A.; Fasching, L.; Macfarlan, T.S.; Tarabay, Y.; Viville, S.; Jakobsson, J.; Pfaff, S.L.; Trono, D. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res. 2013, 23, 452–461. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Vicente-Garcia, C.; Seruggia, D.; Molto, E.; Fernandez-Minan, A.; Neto, A.; Lee, E.; Gomez-Skarmeta, J.L.; Montoliu, L.; Lunyak, V.V.; et al. MIR retrotransposon sequences provide insulators to the human genome. Proc. Natl. Acad. Sci. USA 2015, 112, E4428–E4437. [Google Scholar] [CrossRef]
- Macfarlan, T.S.; Gifford, W.D.; Driscoll, S.; Lettieri, K.; Rowe, H.M.; Bonanomi, D.; Firth, A.; Singer, O.; Trono, D.; Pfaff, S.L. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 2012, 487, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 2016, 351, 1083–1087. [Google Scholar] [CrossRef]
- Eckersley-Maslin, M.A.; Alda-Catalinas, C.; Reik, W. Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat. Rev. Mol. Cell Biol. 2018, 19, 436–450. [Google Scholar] [CrossRef] [PubMed]
- Torres-Padilla, M.E. On transposons and totipotency. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 20190339. [Google Scholar] [CrossRef]
- Izsvak, Z.; Wang, J.; Singh, M.; Mager, D.L.; Hurst, L.D. Pluripotency and the endogenous retrovirus HERVH: Conflict or serendipity? BioEssays News Rev. Mol. Cell. Dev. Biol. 2016, 38, 109–117. [Google Scholar] [CrossRef]
- Peaston, A.E.; Evsikov, A.V.; Graber, J.H.; de Vries, W.N.; Holbrook, A.E.; Solter, D.; Knowles, B.B. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell 2004, 7, 597–606. [Google Scholar] [CrossRef]
- Ishiuchi, T.; Enriquez-Gasca, R.; Mizutani, E.; Boskovic, A.; Ziegler-Birling, C.; Rodriguez-Terrones, D.; Wakayama, T.; Vaquerizas, J.M.; Torres-Padilla, M.E. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat. Struct. Mol. Biol. 2015, 22, 662–671. [Google Scholar] [CrossRef]
- Eckersley-Maslin, M.A.; Svensson, V.; Krueger, C.; Stubbs, T.M.; Giehr, P.; Krueger, F.; Miragaia, R.J.; Kyriakopoulos, C.; Berrens, R.V.; Milagre, I.; et al. MERVL/Zscan4 Network Activation Results in Transient Genome-wide DNA Demethylation of mESCs. Cell Rep. 2016, 17, 179–192. [Google Scholar] [CrossRef]
- Alda-Catalinas, C.; Bredikhin, D.; Hernando-Herraez, I.; Santos, F.; Kubinyecz, O.; Eckersley-Maslin, M.A.; Stegle, O.; Reik, W. A Single-Cell Transcriptomics CRISPR-Activation Screen Identifies Epigenetic Regulators of the Zygotic Genome Activation Program. Cell Syst. 2020. [Google Scholar] [CrossRef] [PubMed]
- De Iaco, A.; Coudray, A.; Duc, J.; Trono, D. DPPA2 and DPPA4 are necessary to establish a 2C-like state in mouse embryonic stem cells. EMBO Rep. 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- De Iaco, A.; Planet, E.; Coluccio, A.; Verp, S.; Duc, J.; Trono, D. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 2017, 49, 941–945. [Google Scholar] [CrossRef]
- Eckersley-Maslin, M.; Alda-Catalinas, C.; Blotenburg, M.; Kreibich, E.; Krueger, C.; Reik, W. Dppa2 and Dppa4 directly regulate the Dux-driven zygotic transcriptional program. Genes Dev. 2019, 33, 194–208. [Google Scholar] [CrossRef] [PubMed]
- Hendrickson, P.G.; Dorais, J.A.; Grow, E.J.; Whiddon, J.L.; Lim, J.W.; Wike, C.L.; Weaver, B.D.; Pflueger, C.; Emery, B.R.; Wilcox, A.L.; et al. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 2017, 49, 925–934. [Google Scholar] [CrossRef]
- Young, J.M.; Whiddon, J.L.; Yao, Z.; Kasinathan, B.; Snider, L.; Geng, L.N.; Balog, J.; Tawil, R.; van der Maarel, S.M.; Tapscott, S.J. DUX4 binding to retroelements creates promoters that are active in FSHD muscle and testis. PLoS Genet. 2013, 9, e1003947. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, F.; Chen, R.; Xie, D.; Yang, J.; Zhao, X.; Guo, R.; Zhang, Y.; Shen, Y.; Goke, J.; et al. Zscan4c activates endogenous retrovirus MERVL and cleavage embryo genes. Nucleic Acids Res. 2019, 47, 8485–8501. [Google Scholar] [CrossRef]
- Srinivasan, R.; Nady, N.; Arora, N.; Hsieh, L.J.; Swigut, T.; Narlikar, G.J.; Wossidlo, M.; Wysocka, J. Zscan4 binds nucleosomal microsatellite DNA and protects mouse two-cell embryos from DNA damage. Sci. Adv. 2020, 6, eaaz9115. [Google Scholar] [CrossRef]
- Wu, J.; Huang, B.; Chen, H.; Yin, Q.; Liu, Y.; Xiang, Y.; Zhang, B.; Liu, B.; Wang, Q.; Xia, W.; et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 2016, 534, 652–657. [Google Scholar] [CrossRef]
- Jachowicz, J.W.; Bing, X.; Pontabry, J.; Boskovic, A.; Rando, O.J.; Torres-Padilla, M.E. LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat. Genet. 2017, 49, 1502–1510. [Google Scholar] [CrossRef]
- Percharde, M.; Lin, C.J.; Yin, Y.; Guan, J.; Peixoto, G.A.; Bulut-Karslioglu, A.; Biechele, S.; Huang, B.; Shen, X.; Ramalho-Santos, M. A LINE1-Nucleolin Partnership Regulates Early Development and ESC Identity. Cell 2018, 174, 391–405.e319. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Leng, L.; Liu, C.; Lu, C.; Yuan, Y.; Wu, L.; Gong, F.; Zhang, S.; Wei, X.; Wang, M.; et al. An integrated chromatin accessibility and transcriptome landscape of human pre-implantation embryos. Nat. Commun. 2019, 10, 364. [Google Scholar] [CrossRef] [PubMed]
- Wolf, G.; de Iaco, A.; Sun, M.A.; Bruno, M.; Tinkham, M.; Hoang, D.; Mitra, A.; Ralls, S.; Trono, D.; Macfarlan, T.S. KRAB-zinc finger protein gene expansion in response to active retrotransposons in the murine lineage. eLife 2020, 9. [Google Scholar] [CrossRef]
- Karimi, M.M.; Goyal, P.; Maksakova, I.A.; Bilenky, M.; Leung, D.; Tang, J.X.; Shinkai, Y.; Mager, D.L.; Jones, S.; Hirst, M.; et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 2011, 8, 676–687. [Google Scholar] [CrossRef]
- Matsui, T.; Leung, D.; Miyashita, H.; Maksakova, I.A.; Miyachi, H.; Kimura, H.; Tachibana, M.; Lorincz, M.C.; Shinkai, Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 2010, 464, 927–931. [Google Scholar] [CrossRef] [PubMed]
- Rowe, H.M.; Friedli, M.; Offner, S.; Verp, S.; Mesnard, D.; Marquis, J.; Aktas, T.; Trono, D. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development 2013, 140, 519–529. [Google Scholar] [CrossRef]
- Rowe, H.M.; Jakobsson, J.; Mesnard, D.; Rougemont, J.; Reynard, S.; Aktas, T.; Maillard, P.V.; Layard-Liesching, H.; Verp, S.; Marquis, J.; et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 2010, 463, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Ecco, G.; Cassano, M.; Kauzlaric, A.; Duc, J.; Coluccio, A.; Offner, S.; Imbeault, M.; Rowe, H.M.; Turelli, P.; Trono, D. Transposable Elements and Their KRAB-ZFP Controllers Regulate Gene Expression in Adult Tissues. Dev. Cell 2016, 36, 611–623. [Google Scholar] [CrossRef]
- Fasching, L.; Kapopoulou, A.; Sachdeva, R.; Petri, R.; Jonsson, M.E.; Manne, C.; Turelli, P.; Jern, P.; Cammas, F.; Trono, D.; et al. TRIM28 Represses Transcription of Endogenous Retroviruses in Neural Progenitor Cells. Cell Rep. 2015, 10, 20–28. [Google Scholar] [CrossRef]
- Hummel, B.; Hansen, E.C.; Yoveva, A.; Aprile-Garcia, F.; Hussong, R.; Sawarkar, R. The evolutionary capacitor HSP90 buffers the regulatory effects of mammalian endogenous retroviruses. Nat. Struct. Mol. Biol. 2017, 24, 234–242. [Google Scholar] [CrossRef]
- Seah, M.K.Y.; Wang, Y.; Goy, P.A.; Loh, H.M.; Peh, W.J.; Low, D.H.P.; Han, B.Y.; Wong, E.; Leong, E.L.; Wolf, G.; et al. The KRAB-zinc-finger protein ZFP708 mediates epigenetic repression at RMER19B retrotransposons. Development 2019, 146. [Google Scholar] [CrossRef] [PubMed]
- Duhl, D.M.; Vrieling, H.; Miller, K.A.; Wolff, G.L.; Barsh, G.S. Neomorphic agouti mutations in obese yellow mice. Nat. Genet. 1994, 8, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Morgan, H.D.; Sutherland, H.G.; Martin, D.I.; Whitelaw, E. Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet. 1999, 23, 314–318. [Google Scholar] [CrossRef] [PubMed]
- Rakyan, V.K.; Chong, S.; Champ, M.E.; Cuthbert, P.C.; Morgan, H.D.; Luu, K.V.; Whitelaw, E. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc. Natl. Acad. Sci. USA 2003, 100, 2538–2543. [Google Scholar] [CrossRef]
- Elmer, J.L.; Ferguson-Smith, A.C. Strain-Specific Epigenetic Regulation of Endogenous Retroviruses: The Role of Trans-Acting Modifiers. Viruses 2020, 12, 810. [Google Scholar] [CrossRef]
- Rebollo, R.; Galvao-Ferrarini, M.; Gagnier, L.; Zhang, Y.; Ferraj, A.; Beck, C.R.; Lorincz, M.C.; Mager, D.L. Inter-Strain Epigenomic Profiling Reveals a Candidate IAP Master Copy in C3H Mice. Viruses 2020, 12, 783. [Google Scholar] [CrossRef]
- Sanchez-Luque, F.J.; Kempen, M.H.C.; Gerdes, P.; Vargas-Landin, D.B.; Richardson, S.R.; Troskie, R.L.; Jesuadian, J.S.; Cheetham, S.W.; Carreira, P.E.; Salvador-Palomeque, C.; et al. LINE-1 Evasion of Epigenetic Repression in Humans. Mol. Cell 2019, 75, 590–604.e512. [Google Scholar] [CrossRef]
- Tchasovnikarova, I.A.; Timms, R.T.; Matheson, N.J.; Wals, K.; Antrobus, R.; Gottgens, B.; Dougan, G.; Dawson, M.A.; Lehner, P.J. GENE SILENCING. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science 2015, 348, 1481–1485. [Google Scholar] [CrossRef]
- Maeda-Smithies, N.; Hiller, S.; Dong, S.; Kim, H.S.; Bennett, B.J.; Kayashima, Y. Ectopic expression of the Stabilin2 gene triggered by an intracisternal A particle (IAP) element in DBA/2J strain of mice. Mamm. Genome 2020, 31, 2–16. [Google Scholar] [CrossRef]
- Jonsson, M.E.; Ludvik Brattas, P.; Gustafsson, C.; Petri, R.; Yudovich, D.; Pircs, K.; Verschuere, S.; Madsen, S.; Hansson, J.; Larsson, J.; et al. Activation of neuronal genes via LINE-1 elements upon global DNA demethylation in human neural progenitors. Nat. Commun. 2019, 10, 3182. [Google Scholar] [CrossRef]
- Denli, A.M.; Narvaiza, I.; Kerman, B.E.; Pena, M.; Benner, C.; Marchetto, M.C.; Diedrich, J.K.; Aslanian, A.; Ma, J.; Moresco, J.J.; et al. Primate-specific ORF0 contributes to retrotransposon-mediated diversity. Cell 2015, 163, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, F.M.; Greenberg, D.; Nguyen, N.; Haeussler, M.; Ewing, A.D.; Katzman, S.; Paten, B.; Salama, S.R.; Haussler, D. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 2014, 516, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Turelli, P.; Castro-Diaz, N.; Marzetta, F.; Kapopoulou, A.; Raclot, C.; Duc, J.; Tieng, V.; Quenneville, S.; Trono, D. Interplay of TRIM28 and DNA methylation in controlling human endogenous retroelements. Genome Res. 2014, 24, 1260–1270. [Google Scholar] [CrossRef] [PubMed]
- Friedli, M.; Turelli, P.; Kapopoulou, A.; Rauwel, B.; Castro-Diaz, N.; Rowe, H.M.; Ecco, G.; Unzu, C.; Planet, E.; Lombardo, A.; et al. Loss of transcriptional control over endogenous retroelements during reprogramming to pluripotency. Genome Res. 2014, 24, 1251–1259. [Google Scholar] [CrossRef] [PubMed]
- Wolf, D.; Goff, S.P. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 2007, 131, 46–57. [Google Scholar] [CrossRef]
- Wolf, D.; Goff, S.P. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 2009, 458, 1201–1204. [Google Scholar] [CrossRef]
- Wolf, G.; Yang, P.; Fuchtbauer, A.C.; Fuchtbauer, E.M.; Silva, A.M.; Park, C.; Wu, W.; Nielsen, A.L.; Pedersen, F.S.; Macfarlan, T.S. The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes Dev. 2015, 29, 538–554. [Google Scholar] [CrossRef]
- Tie, C.H.; Fernandes, L.; Conde, L.; Robbez-Masson, L.; Sumner, R.P.; Peacock, T.; Rodriguez-Plata, M.T.; Mickute, G.; Gifford, R.; Towers, G.J.; et al. KAP1 regulates endogenous retroviruses in adult human cells and contributes to innate immune control. EMBO Rep. 2018, 19. [Google Scholar] [CrossRef]
- Wolf, D.; Hug, K.; Goff, S.P. TRIM28 mediates primer binding site-targeted silencing of Lys1,2 tRNA-utilizing retroviruses in embryonic cells. Proc. Natl. Acad. Sci. USA 2008, 105, 12521–12526. [Google Scholar] [CrossRef]
- Wang, J.; Xie, G.; Singh, M.; Ghanbarian, A.T.; Rasko, T.; Szvetnik, A.; Cai, H.; Besser, D.; Prigione, A.; Fuchs, N.V.; et al. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature 2014, 516, 405–409. [Google Scholar] [CrossRef]
- Romer, C.; Singh, M.; Hurst, L.D.; Izsvak, Z. How to tame an endogenous retrovirus: HERVH and the evolution of human pluripotency. Curr. Opin. Virol. 2017, 25, 49–58. [Google Scholar] [CrossRef]
- Grow, E.J.; Flynn, R.A.; Chavez, S.L.; Bayless, N.L.; Wossidlo, M.; Wesche, D.J.; Martin, L.; Ware, C.B.; Blish, C.A.; Chang, H.Y.; et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature 2015, 522, 221–225. [Google Scholar] [CrossRef]
- Collins, P.L.; Kyle, K.E.; Egawa, T.; Shinkai, Y.; Oltz, E.M. The histone methyltransferase SETDB1 represses endogenous and exogenous retroviruses in B lymphocytes. Proc. Natl. Acad. Sci. USA 2015, 112, 8367–8372. [Google Scholar] [CrossRef] [PubMed]
- Faulkner, G.J.; Kimura, Y.; Daub, C.O.; Wani, S.; Plessy, C.; Irvine, K.M.; Schroder, K.; Cloonan, N.; Steptoe, A.L.; Lassmann, T.; et al. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 2009, 41, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Takemoto, K.; Shinkai, Y. A somatic role for the histone methyltransferase Setdb1 in endogenous retrovirus silencing. Nat. Commun. 2018, 9, 1683. [Google Scholar] [CrossRef] [PubMed]
- Pehrsson, E.C.; Choudhary, M.N.K.; Sundaram, V.; Wang, T. The epigenomic landscape of transposable elements across normal human development and anatomy. Nat. Commun. 2019, 10, 5640. [Google Scholar] [CrossRef] [PubMed]
- Trizzino, M.; Park, Y.; Holsbach-Beltrame, M.; Aracena, K.; Mika, K.; Caliskan, M.; Perry, G.H.; Lynch, V.J.; Brown, C.D. Transposable elements are the primary source of novelty in primate gene regulation. Genome Res. 2017, 27, 1623–1633. [Google Scholar] [CrossRef]
- Villar, D.; Berthelot, C.; Aldridge, S.; Rayner, T.F.; Lukk, M.; Pignatelli, M.; Park, T.J.; Deaville, R.; Erichsen, J.T.; Jasinska, A.J.; et al. Enhancer Evolution across 20 Mammalian Species. Cell 2015, 160, 554–566. [Google Scholar] [CrossRef]
- Kaneko, S.; Bonasio, R.; Saldana-Meyer, R.; Yoshida, T.; Son, J.; Nishino, K.; Umezawa, A.; Reinberg, D. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol. Cell 2014, 53, 290–300. [Google Scholar] [CrossRef]
- Tchasovnikarova, I.A.; Timms, R.T.; Douse, C.H.; Roberts, R.C.; Dougan, G.; Kingston, R.E.; Modis, Y.; Lehner, P.J. Hyperactivation of HUSH complex function by Charcot-Marie-Tooth disease mutation in MORC2. Nat. Genet. 2017, 49, 1035–1044. [Google Scholar] [CrossRef]
- Douse, C.H.; Bloor, S.; Liu, Y.; Shamin, M.; Tchasovnikarova, I.A.; Timms, R.T.; Lehner, P.J.; Modis, Y. Neuropathic MORC2 mutations perturb GHKL ATPase dimerization dynamics and epigenetic silencing by multiple structural mechanisms. Nat. Commun. 2018, 9, 651. [Google Scholar] [CrossRef]
- Liu, N.; Lee, C.H.; Swigut, T.; Grow, E.; Gu, B.; Bassik, M.C.; Wysocka, J. Selective silencing of euchromatic L1s revealed by genome-wide screens for L1 regulators. Nature 2018, 553, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, G.Z.; Cingoz, O.; Goff, S.P. NP220 mediates silencing of unintegrated retroviral DNA. Nature 2018, 564, 278–282. [Google Scholar] [CrossRef] [PubMed]
- Harten, S.K.; Bruxner, T.J.; Bharti, V.; Blewitt, M.; Nguyen, T.M.; Whitelaw, E.; Epp, T. The first mouse mutants of D14Abb1e (Fam208a) show that it is critical for early development. Mamm. Genome 2014, 25, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Robbez-Masson, L.; Tie, C.H.C.; Conde, L.; Tunbak, H.; Husovsky, C.; Tchasovnikarova, I.A.; Timms, R.T.; Herrero, J.; Lehner, P.J.; Rowe, H.M. The HUSH complex cooperates with TRIM28 to repress young retrotransposons and new genes. Genome Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Ardeljan, D.; Steranka, J.P.; Liu, C.; Li, Z.; Taylor, M.S.; Payer, L.M.; Gorbounov, M.; Sarnecki, J.S.; Deshpande, V.; Hruban, R.H.; et al. Cell fitness screens reveal a conflict between LINE-1 retrotransposition and DNA replication. Nat. Struct. Mol. Biol. 2020, 27, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Douse, C.H.; Tchasovnikarova, I.A.; Timms, R.T.; Protasio, A.V.; Seczynska, M.; Prigozhin, D.M.; Albecka, A.; Wagstaff, J.; Williamson, J.C.; Freund, S.M.V.; et al. TASOR Is a pseudo-PARP that Directs HUSH Complex Assembly and Epigenetic Transposon Control. bioRxiv. 2020. Available online: https://www.biorxiv.org/content/10.1101/2020.03.09.974832v1 (accessed on 3 August 2020).
- Ferguson-Smith, A.C. Genomic imprinting: The emergence of an epigenetic paradigm. Nat. Rev. Genet. 2011, 12, 565–575. [Google Scholar] [CrossRef]
- Reik, W.; Walter, J. Genomic imprinting: Parental influence on the genome. Nat. Rev. Genet. 2001, 2, 21–32. [Google Scholar] [CrossRef]
- Barlow, D.P. Methylation and imprinting: From host defense to gene regulation? Science 1993, 260, 309–310. [Google Scholar] [CrossRef]
- Ondicova, M.; Oakey, R.J.; Walsh, C.P. Is imprinting the result of “friendly fire” by the host defense system? PLoS Genet. 2020, 16, e1008599. [Google Scholar] [CrossRef]
- Wood, A.J.; Bourc’his, D.; Bestor, T.H.; Oakey, R.J. Allele-specific demethylation at an imprinted mammalian promoter. Nucleic Acids Res. 2007, 35, 7031–7039. [Google Scholar] [CrossRef]
- Wood, A.J.; Roberts, R.G.; Monk, D.; Moore, G.E.; Schulz, R.; Oakey, R.J. A screen for retrotransposed imprinted genes reveals an association between X chromosome homology and maternal germ-line methylation. PLoS Genet. 2007, 3, e20. [Google Scholar] [CrossRef]
- Youngson, N.A.; Kocialkowski, S.; Peel, N.; Ferguson-Smith, A.C. A small family of sushi-class retrotransposon-derived genes in mammals and their relation to genomic imprinting. J. Mol. Evol. 2005, 61, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Bogutz, A.B.; Brind’Amour, J.; Kobayashi, H.; Jensen, K.N.; Nakabayashi, K.; Imai, H.; Lorincz, M.C.; Lefebvre, L. Evolution of imprinting via lineage-specific insertion of retroviral promoters. Nat. Commun. 2019, 10, 5674. [Google Scholar] [CrossRef]
- Hanna, C.W.; Perez-Palacios, R.; Gahurova, L.; Schubert, M.; Krueger, F.; Biggins, L.; Andrews, S.; Colome-Tatche, M.; Bourc’his, D.; Dean, W.; et al. Endogenous retroviral insertions drive non-canonical imprinting in extra-embryonic tissues. Genome Biol. 2019, 20, 225. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Coluccio, A.; Thorball, C.W.; Planet, E.; Shi, H.; Offner, S.; Turelli, P.; Imbeault, M.; Ferguson-Smith, A.C.; Trono, D. ZNF445 is a primary regulator of genomic imprinting. Genes Dev. 2019, 33, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Imbeault, M.; Helleboid, P.Y.; Trono, D. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature 2017, 543, 550–554. [Google Scholar] [CrossRef] [PubMed]
- Strogantsev, R.; Krueger, F.; Yamazawa, K.; Shi, H.; Gould, P.; Goldman-Roberts, M.; McEwen, K.; Sun, B.; Pedersen, R.; Ferguson-Smith, A.C. Allele-specific binding of ZFP57 in the epigenetic regulation of imprinted and non-imprinted monoallelic expression. Genome Biol. 2015, 16, 112. [Google Scholar] [CrossRef]
- Swanzey, E.; McNamara, T.F.; Apostolou, E.; Tahiliani, M.; Stadtfeld, M. A Susceptibility Locus on Chromosome 13 Profoundly Impacts the Stability of Genomic Imprinting in Mouse Pluripotent Stem Cells. Cell Rep. 2020, 30, 3597–3604.e3593. [Google Scholar] [CrossRef]
- Krebs, C.J.; Larkins, L.K.; Khan, S.M.; Robins, D.M. Expansion and diversification of KRAB zinc-finger genes within a cluster including Regulator of sex-limitation 1 and 2. Genomics 2005, 85, 752–761. [Google Scholar] [CrossRef]
- Barreiro, L.B.; Marioni, J.C.; Blekhman, R.; Stephens, M.; Gilad, Y. Functional comparison of innate immune signaling pathways in primates. PLoS Genet. 2010, 6, e1001249. [Google Scholar] [CrossRef] [PubMed]
- Platanias, L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005, 5, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Schroder, K.; Irvine, K.M.; Taylor, M.S.; Bokil, N.J.; Le Cao, K.A.; Masterman, K.A.; Labzin, L.I.; Semple, C.A.; Kapetanovic, R.; Fairbairn, L.; et al. Conservation and divergence in Toll-like receptor 4-regulated gene expression in primary human versus mouse macrophages. Proc. Natl. Acad. Sci. USA 2012, 109, E944–E953. [Google Scholar] [CrossRef] [PubMed]
- Bao, W.; Kojima, K.K.; Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 2015, 6, 11. [Google Scholar] [CrossRef]
- Silva, J.C.; Shabalina, S.A.; Harris, D.G.; Spouge, J.L.; Kondrashovi, A.S. Conserved fragments of transposable elements in intergenic regions: Evidence for widespread recruitment of MIR- and L2-derived sequences within the mouse and human genomes. Genet. Res. 2003, 82, 1–18. [Google Scholar] [CrossRef]
- Brockdorff, N.; Ashworth, A.; Kay, G.F.; Cooper, P.; Smith, S.; McCabe, V.M.; Norris, D.P.; Penny, G.D.; Patel, D.; Rastan, S. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature 1991, 351, 329–331. [Google Scholar] [CrossRef] [PubMed]
- Loda, A.; Heard, E. Xist RNA in action: Past, present, and future. PLoS Genet. 2019, 15, e1008333. [Google Scholar] [CrossRef] [PubMed]
- Lyon, M.F. X-chromosome inactivation: A repeat hypothesis. Cytogenet. Cell Genet. 1998, 80, 133–137. [Google Scholar] [CrossRef]
- Bailey, J.A.; Carrel, L.; Chakravarti, A.; Eichler, E.E. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: The Lyon repeat hypothesis. Proc. Natl. Acad. Sci. USA 2000, 97, 6634–6639. [Google Scholar] [CrossRef] [PubMed]
- Chow, J.C.; Ciaudo, C.; Fazzari, M.J.; Mise, N.; Servant, N.; Glass, J.L.; Attreed, M.; Avner, P.; Wutz, A.; Barillot, E.; et al. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell 2010, 141, 956–969. [Google Scholar] [CrossRef] [PubMed]
- Larson, A.G.; Elnatan, D.; Keenen, M.M.; Trnka, M.J.; Johnston, J.B.; Burlingame, A.L.; Agard, D.A.; Redding, S.; Narlikar, G.J. Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature 2017, 547, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Strom, A.R.; Emelyanov, A.V.; Mir, M.; Fyodorov, D.V.; Darzacq, X.; Karpen, G.H. Phase separation drives heterochromatin domain formation. Nature 2017, 547, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Britten, R.J.; Davidson, E.H. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q. Rev. Biol. 1971, 46, 111–138. [Google Scholar] [CrossRef]
- Tashiro, K.; Teissier, A.; Kobayashi, N.; Nakanishi, A.; Sasaki, T.; Yan, K.; Tarabykin, V.; Vigier, L.; Sumiyama, K.; Hirakawa, M.; et al. A mammalian conserved element derived from SINE displays enhancer properties recapitulating Satb2 expression in early-born callosal projection neurons. PLoS ONE 2011, 6, e28497. [Google Scholar] [CrossRef] [PubMed]
- Nishihara, H.; Smit, A.F.; Okada, N. Functional noncoding sequences derived from SINEs in the mammalian genome. Genome Res. 2006, 16, 864–874. [Google Scholar] [CrossRef]
- Alcamo, E.A.; Chirivella, L.; Dautzenberg, M.; Dobreva, G.; Farinas, I.; Grosschedl, R.; McConnell, S.K. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron 2008, 57, 364–377. [Google Scholar] [CrossRef] [PubMed]
- Flower, W.H. On the Commissures of the Cerebral Hemispheres of the Marsupialia and Monotremata, as Compared with Those of the Placental Mammals. Proc. R. Soc. Lond. 1865, 14, 71–74. [Google Scholar]
- Notwell, J.H.; Chung, T.; Heavner, W.; Bejerano, G. A family of transposable elements co-opted into developmental enhancers in the mouse neocortex. Nat. Commun. 2015, 6, 6644. [Google Scholar] [CrossRef] [PubMed]
- Jjingo, D.; Huda, A.; Gundapuneni, M.; Marino-Ramirez, L.; Jordan, I.K. Effect of the transposable element environment of human genes on gene length and expression. Genome Biol. Evol. 2011, 3, 259–271. [Google Scholar] [CrossRef]
- Samstein, R.M.; Josefowicz, S.Z.; Arvey, A.; Treuting, P.M.; Rudensky, A.Y. Extrathymic generation of regulatory T cells in placental mammals mitigates maternal-fetal conflict. Cell 2012, 150, 29–38. [Google Scholar] [CrossRef]
- Lynch, V.J.; Leclerc, R.D.; May, G.; Wagner, G.P. Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat. Genet. 2011, 43, 1154–1159. [Google Scholar] [CrossRef] [PubMed]
- Lamprecht, B.; Walter, K.; Kreher, S.; Kumar, R.; Hummel, M.; Lenze, D.; Kochert, K.; Bouhlel, M.A.; Richter, J.; Soler, E.; et al. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat. Med. 2010, 16, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Kreher, S.; Bouhlel, M.A.; Cauchy, P.; Lamprecht, B.; Li, S.; Grau, M.; Hummel, F.; Kochert, K.; Anagnostopoulos, I.; Johrens, K.; et al. Mapping of transcription factor motifs in active chromatin identifies IRF5 as key regulator in classical Hodgkin lymphoma. Proc. Natl. Acad. Sci. USA 2014, 111, E4513–E4522. [Google Scholar] [CrossRef]
- Babaian, A.; Romanish, M.T.; Gagnier, L.; Kuo, L.Y.; Karimi, M.M.; Steidl, C.; Mager, D.L. Onco-exaptation of an endogenous retroviral LTR drives IRF5 expression in Hodgkin lymphoma. Oncogene 2016, 35, 2542–2546. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.; Trac, C.; Kato, H.; Costello, K.R.; Chen, Z.; Natarajan, R.; Schones, D.E. LTRs activated by Epstein-Barr virus-induced transformation of B cells alter the transcriptome. Genome Res. 2018, 28, 1791–1798. [Google Scholar] [CrossRef]
- Deniz, O.; Ahmed, M.; Todd, C.D.; Rio-Machin, A.; Dawson, M.A.; Branco, M.R. Endogenous retroviruses are a source of enhancers with oncogenic potential in acute myeloid leukaemia. Nat. Commun. 2020, 11, 3506. [Google Scholar] [CrossRef]
- Ng, K.W.; Attig, J.; Young, G.R.; Ottina, E.; Papamichos, S.I.; Kotsianidis, I.; Kassiotis, G. Soluble PD-L1 generated by endogenous retroelement exaptation is a receptor antagonist. eLife 2019, 8. [Google Scholar] [CrossRef]
- Groger, V.; Cynis, H. Human Endogenous Retroviruses and Their Putative Role in the Development of Autoimmune Disorders such as Multiple Sclerosis. Front. Microbiol. 2018, 9, 265. [Google Scholar] [CrossRef]
- Rolland, A.; Jouvin-Marche, E.; Viret, C.; Faure, M.; Perron, H.; Marche, P.N. The envelope protein of a human endogenous retrovirus-W family activates innate immunity through CD14/TLR4 and promotes Th1-like responses. J. Immunol. 2006, 176, 7636–7644. [Google Scholar] [CrossRef]
- Wu, Z.; Mei, X.; Zhao, D.; Sun, Y.; Song, J.; Pan, W.; Shi, W. DNA methylation modulates HERV-E expression in CD4+ T cells from systemic lupus erythematosus patients. J. Dermatol. Sci. 2015, 77, 110–116. [Google Scholar] [CrossRef]
- Sukapan, P.; Promnarate, P.; Avihingsanon, Y.; Mutirangura, A.; Hirankarn, N. Types of DNA methylation status of the interspersed repetitive sequences for LINE-1, Alu, HERV-E and HERV-K in the neutrophils from systemic lupus erythematosus patients and healthy controls. J. Hum. Genet. 2014, 59, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Treger, R.S.; Pope, S.D.; Kong, Y.; Tokuyama, M.; Taura, M.; Iwasaki, A. The Lupus Susceptibility Locus Sgp3 Encodes the Suppressor of Endogenous Retrovirus Expression SNERV. Immunity 2019, 50, 334–347.e9. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, M.E.; Garza, R.; Johansson, P.A.; Jakobsson, J. Transposable Elements: A Common Feature of Neurodevelopmental and Neurodegenerative Disorders. Trends Genet. 2020, 36, 610–623. [Google Scholar] [CrossRef] [PubMed]
- Tam, O.H.; Ostrow, L.W.; Gale Hammell, M. Diseases of the nERVous system: Retrotransposon activity in neurodegenerative disease. Mob. DNA 2019, 10, 32. [Google Scholar] [CrossRef]
- Benoit, M.; Drost, H.G.; Catoni, M.; Gouil, Q.; Lopez-Gomollon, S.; Baulcombe, D.; Paszkowski, J. Environmental and epigenetic regulation of Rider retrotransposons in tomato. PLoS Genet. 2019, 15, e1008370. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Jin, Y.; Tam, O.H.; Paniagua, E.; Hammell, M. TEtranscripts: A package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics 2015, 31, 3593–3599. [Google Scholar] [CrossRef]
- Nellaker, C.; Keane, T.M.; Yalcin, B.; Wong, K.; Agam, A.; Belgard, T.G.; Flint, J.; Adams, D.J.; Frankel, W.N.; Ponting, C.P. The genomic landscape shaped by selection on transposable elements across 18 mouse strains. Genome Biol. 2012, 13, R45. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Wenger, A.M.; Clarke, S.L.; Notwell, J.H.; Chung, T.; Tuteja, G.; Guturu, H.; Schaar, B.T.; Bejerano, G. The enhancer landscape during early neocortical development reveals patterns of dense regulation and co-option. PLoS Genet. 2013, 9, e1003728. [Google Scholar] [CrossRef]
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Pollard, K.S.; Hubisz, M.J.; Rosenbloom, K.R.; Siepel, A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010, 20, 110–121. [Google Scholar] [CrossRef]
- Karolchik, D.; Hinrichs, A.S.; Furey, T.S.; Roskin, K.M.; Sugnet, C.W.; Haussler, D.; Kent, W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004, 32, D493–D496. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Enriquez-Gasca, R.; Gould, P.A.; Rowe, H.M. Host Gene Regulation by Transposable Elements: The New, the Old and the Ugly. Viruses 2020, 12, 1089. https://doi.org/10.3390/v12101089
Enriquez-Gasca R, Gould PA, Rowe HM. Host Gene Regulation by Transposable Elements: The New, the Old and the Ugly. Viruses. 2020; 12(10):1089. https://doi.org/10.3390/v12101089
Chicago/Turabian StyleEnriquez-Gasca, Rocio, Poppy A. Gould, and Helen M. Rowe. 2020. "Host Gene Regulation by Transposable Elements: The New, the Old and the Ugly" Viruses 12, no. 10: 1089. https://doi.org/10.3390/v12101089
APA StyleEnriquez-Gasca, R., Gould, P. A., & Rowe, H. M. (2020). Host Gene Regulation by Transposable Elements: The New, the Old and the Ugly. Viruses, 12(10), 1089. https://doi.org/10.3390/v12101089




