Distinct MCM10 Proteasomal Degradation Profiles by Primate Lentiviruses Vpr Proteins
Abstract
1. Introduction
2. Materials and Methods
2.1. Phylogenetic Analysis and Vpr Alleles Preparation
2.2. Plasmid Construction
2.3. Cell Culture, Transfection, and Drug Treatment
2.4. Co-Immunoprecipitation Assay
2.5. Western Blotting
2.6. Immunofluorescence Staining
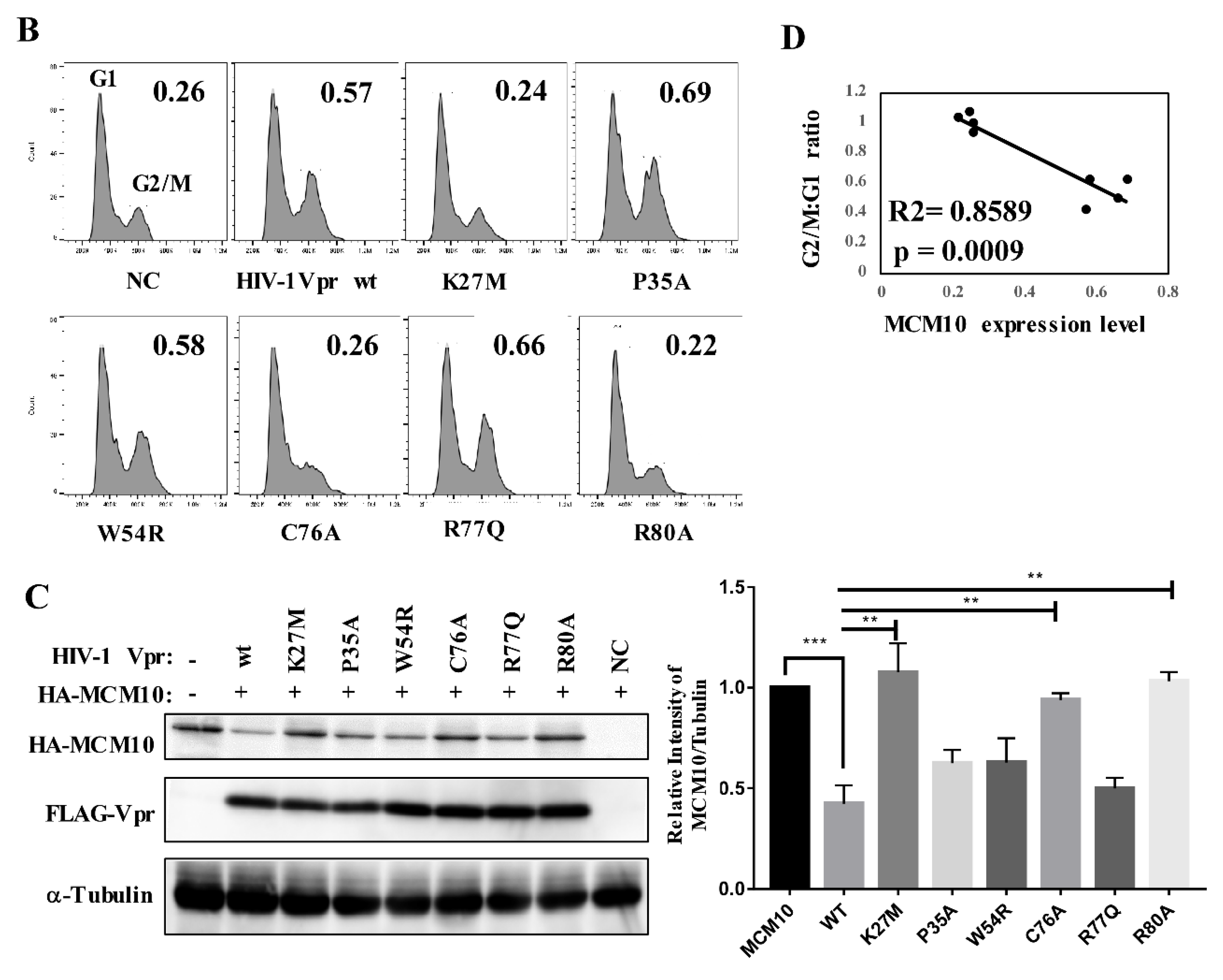
2.7. Cell Cycle Analysis
2.8. Real-Time qRT-PCR Analysis of Human MCM10 mRNA Expression
2.9. Statistical Analysis
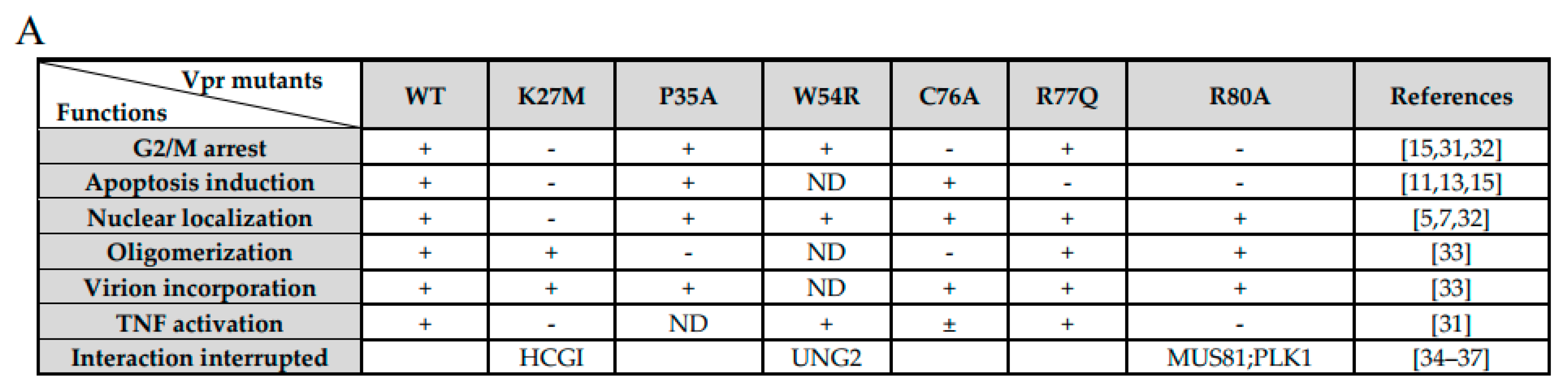
3. Results
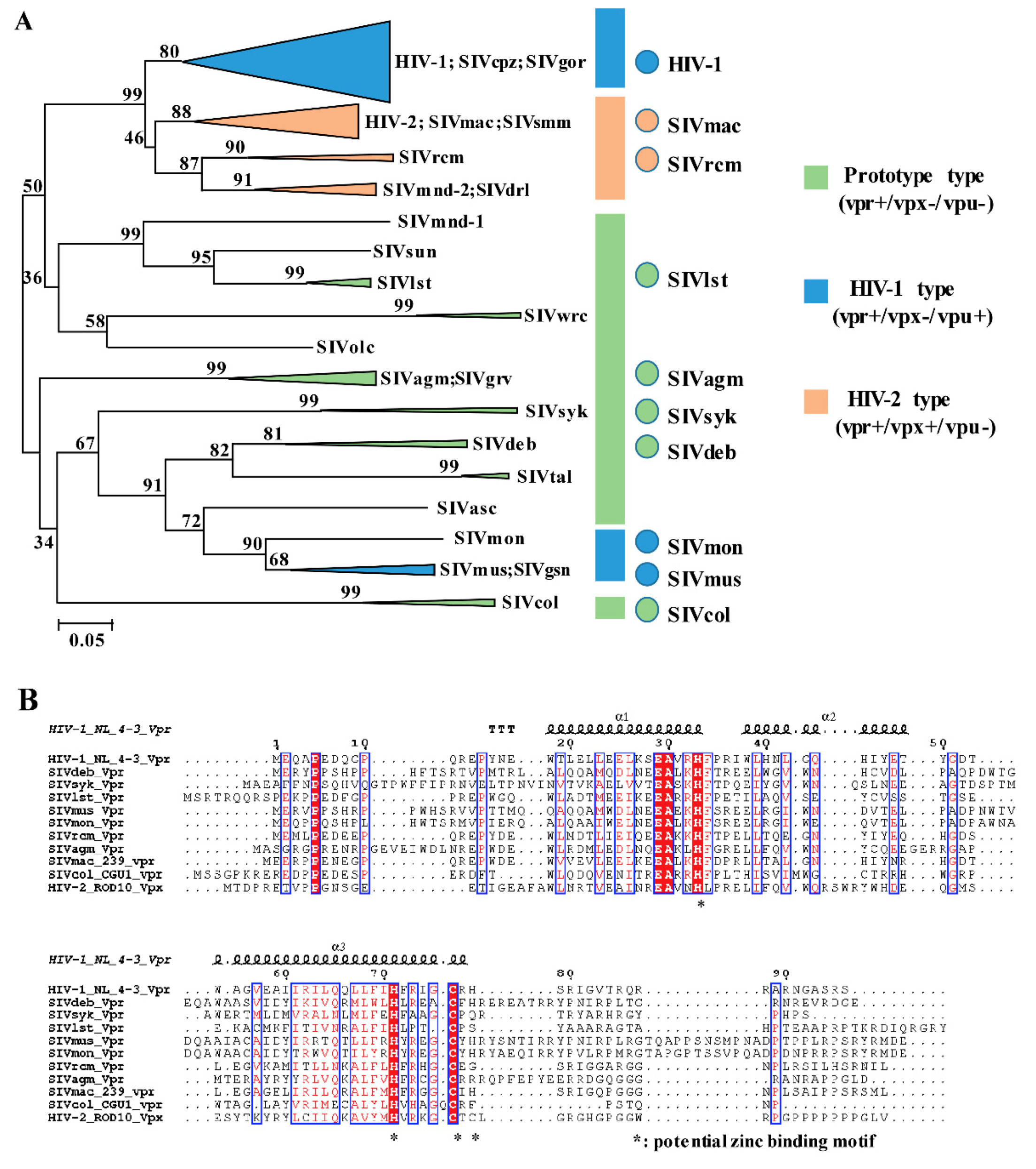
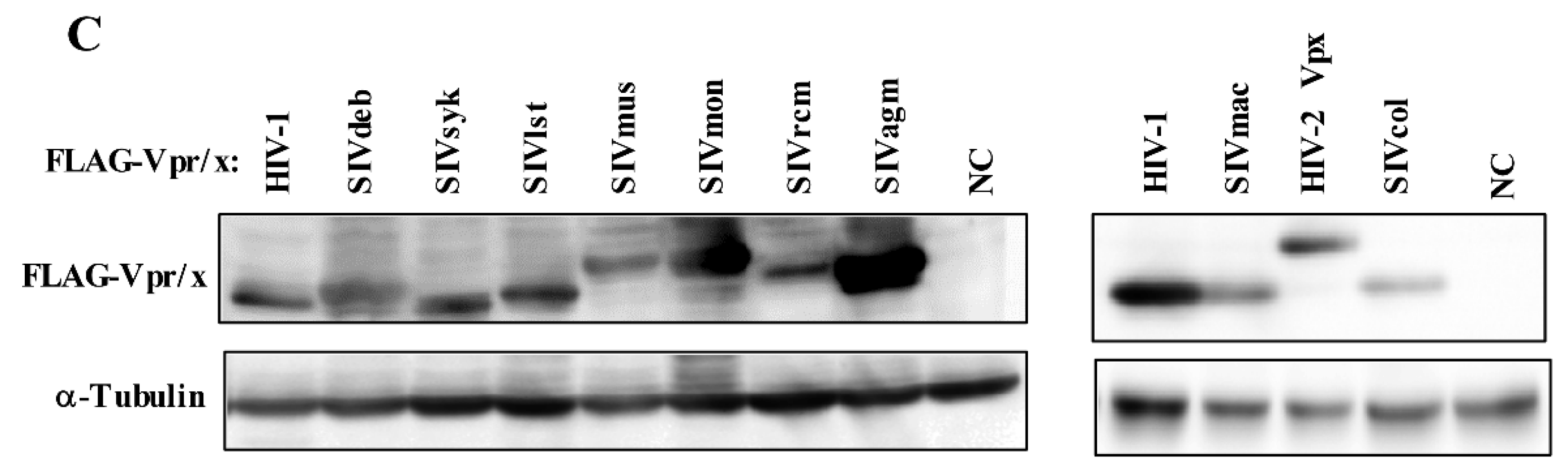
3.1. Phylogeny, Multiple Alignments, and Expression of Vpr/x from Representative Strains
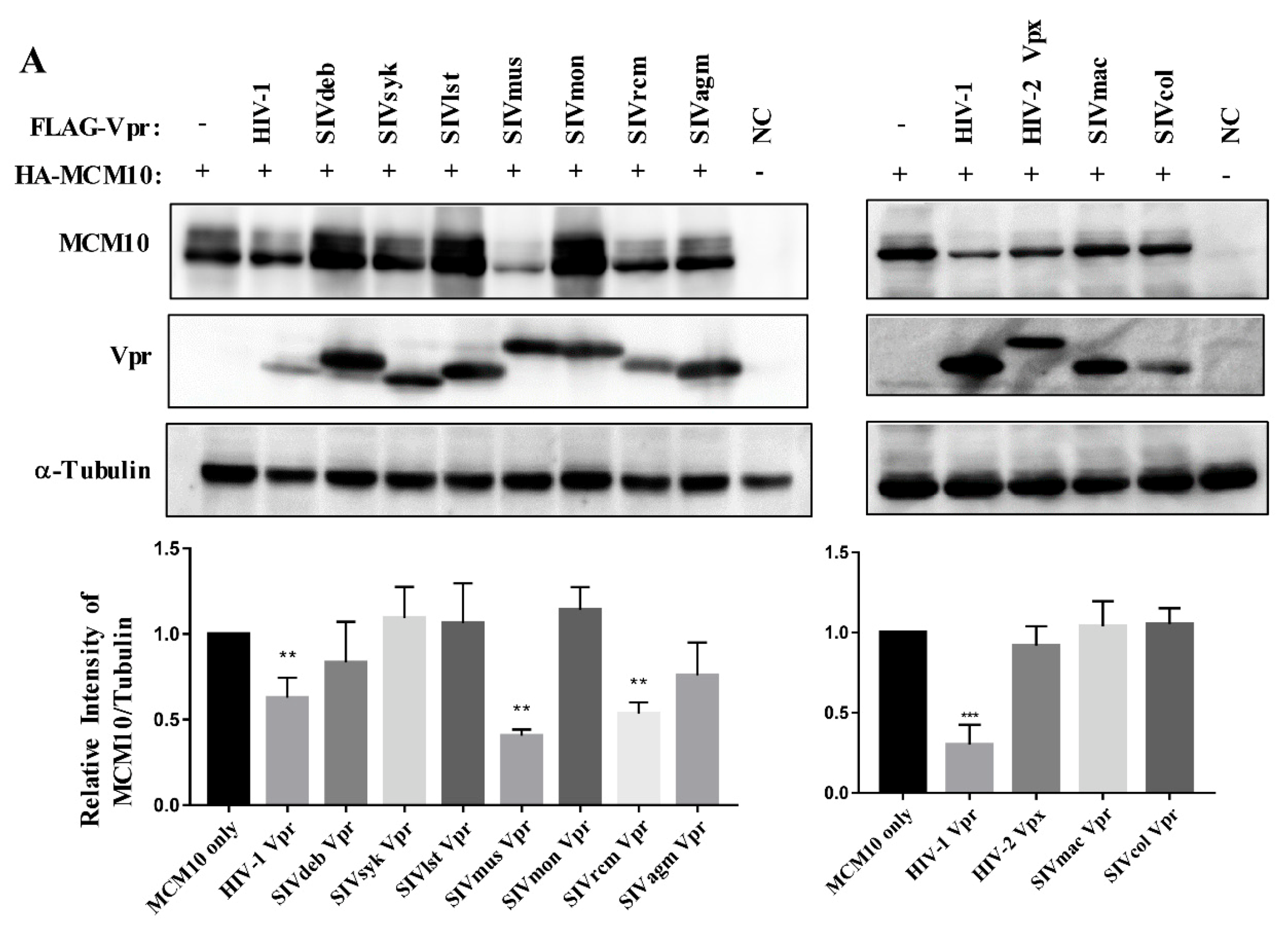
3.2. MCM10 down-Regulation by Primate Lentiviruses Vpr/x Proteins
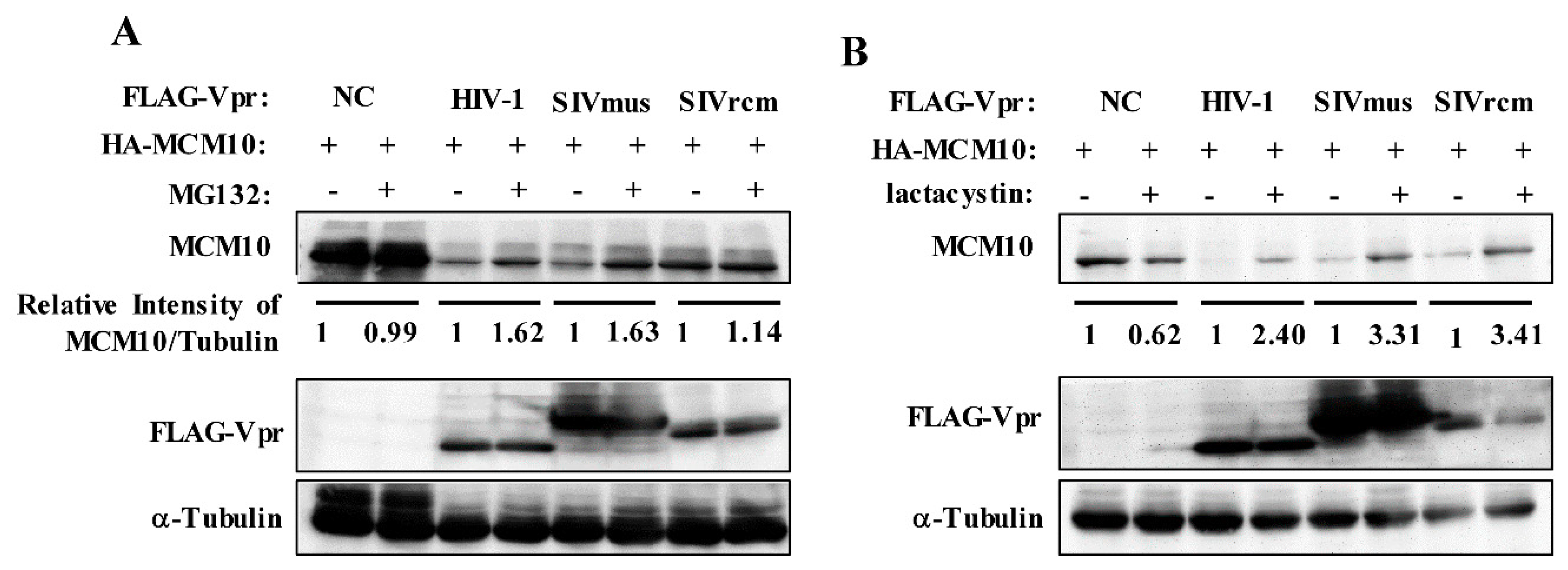
3.3. MCM10 Degradation via Proteasome Dependent Pathway
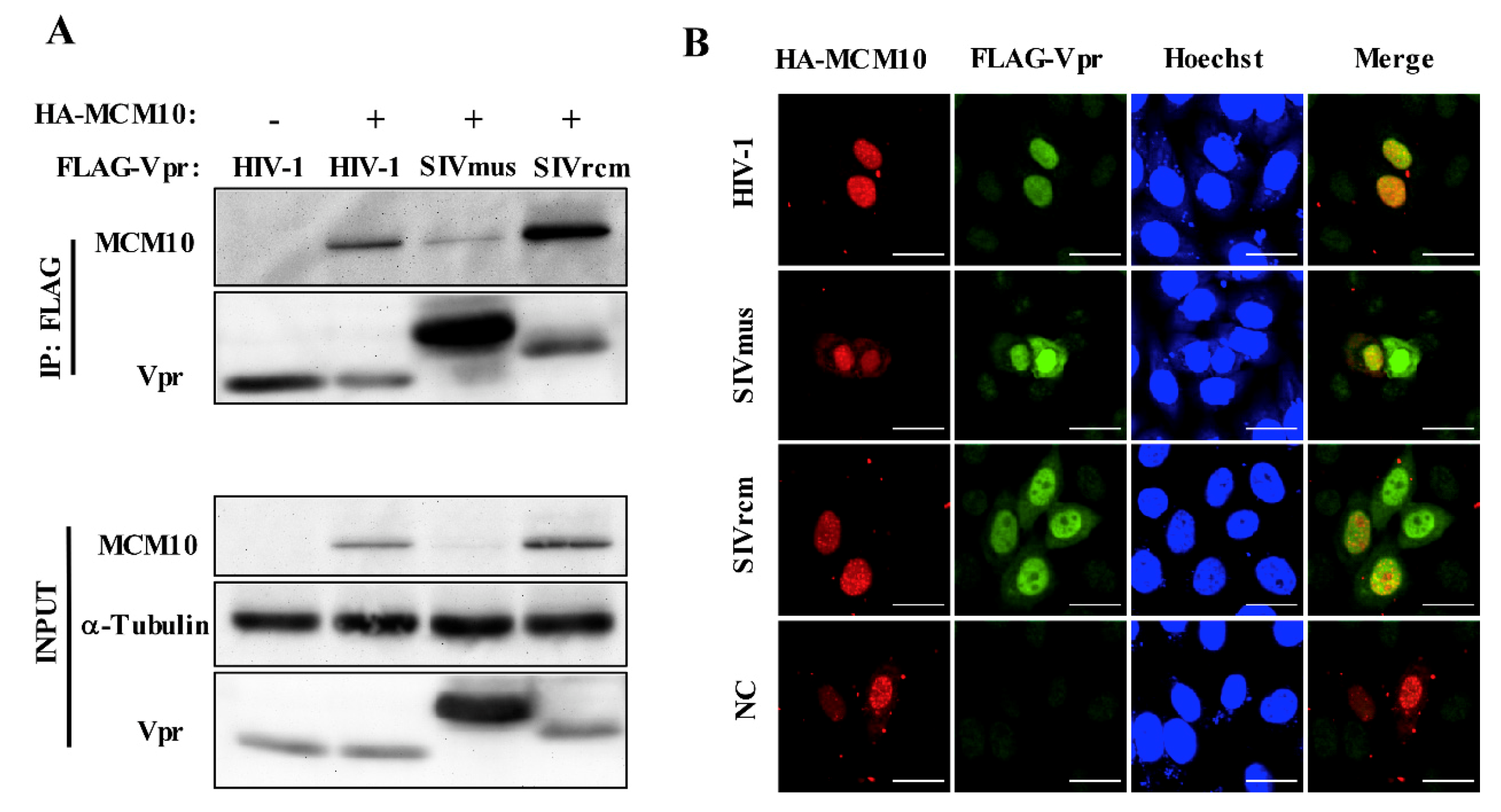
3.4. Interaction between MCM10 and HIV-1, SIVmus, and SIVrcm Vprs
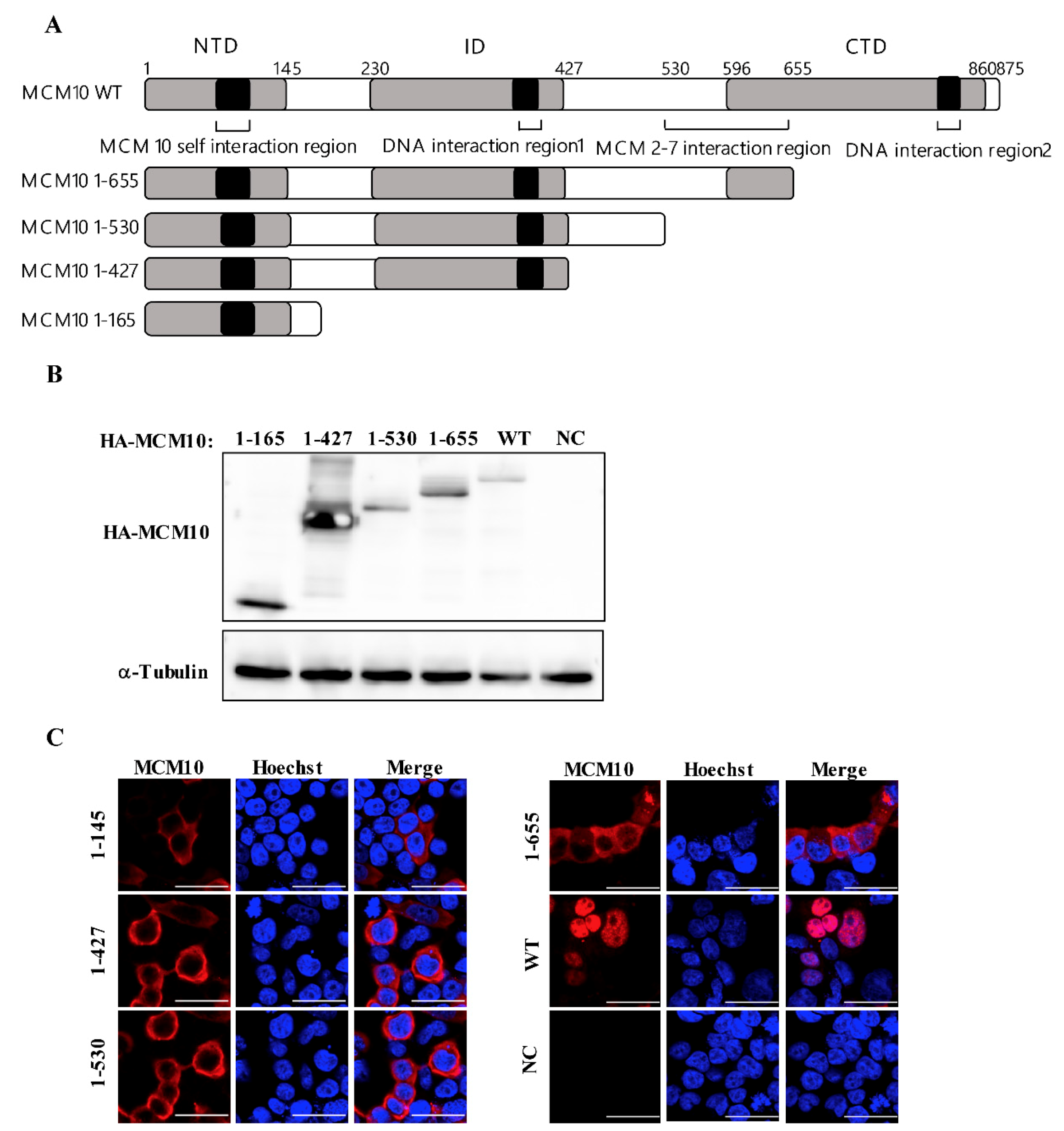
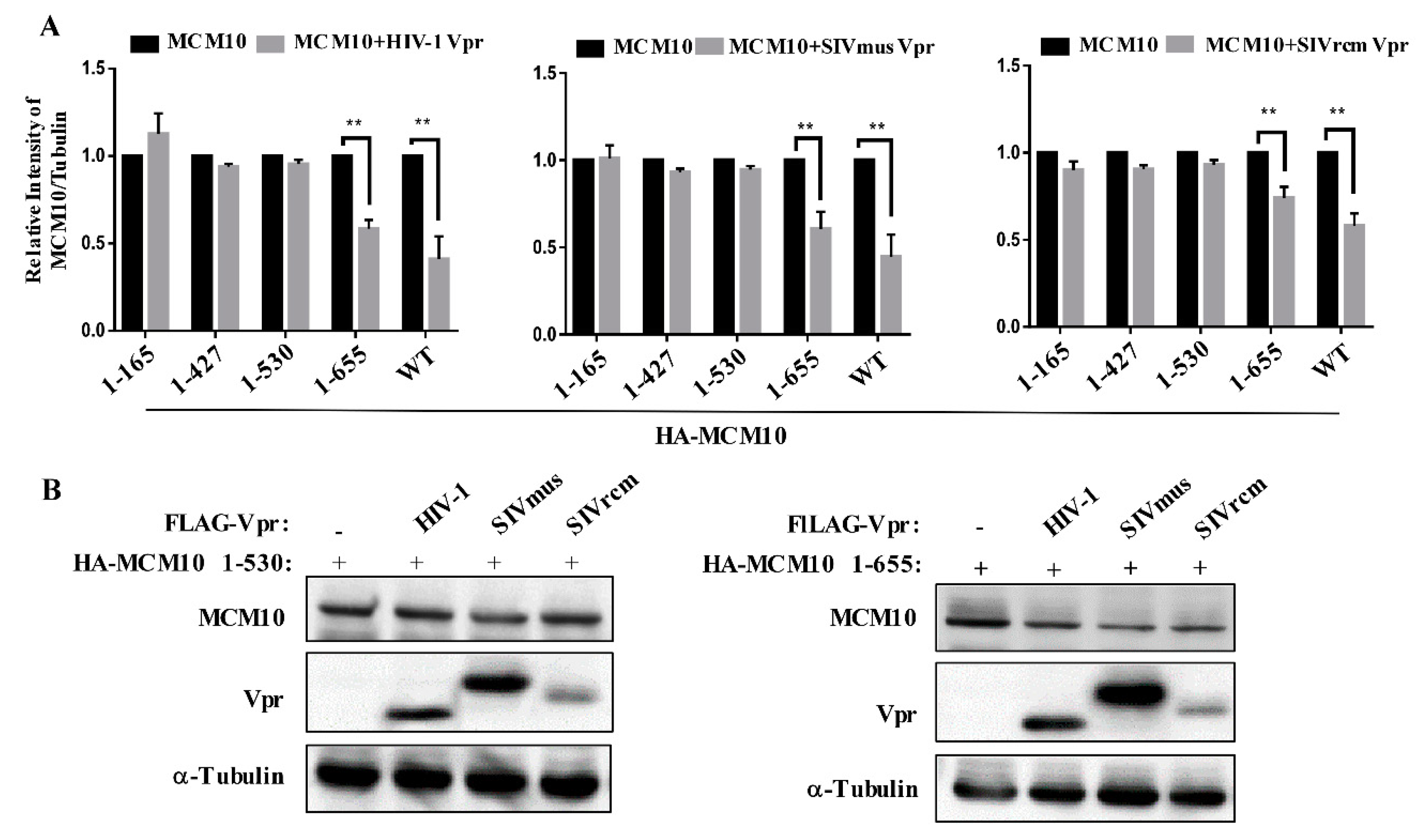
3.5. MCM 2-7 Interaction Region of MCM10 Susceptible to Degradation by Vprs
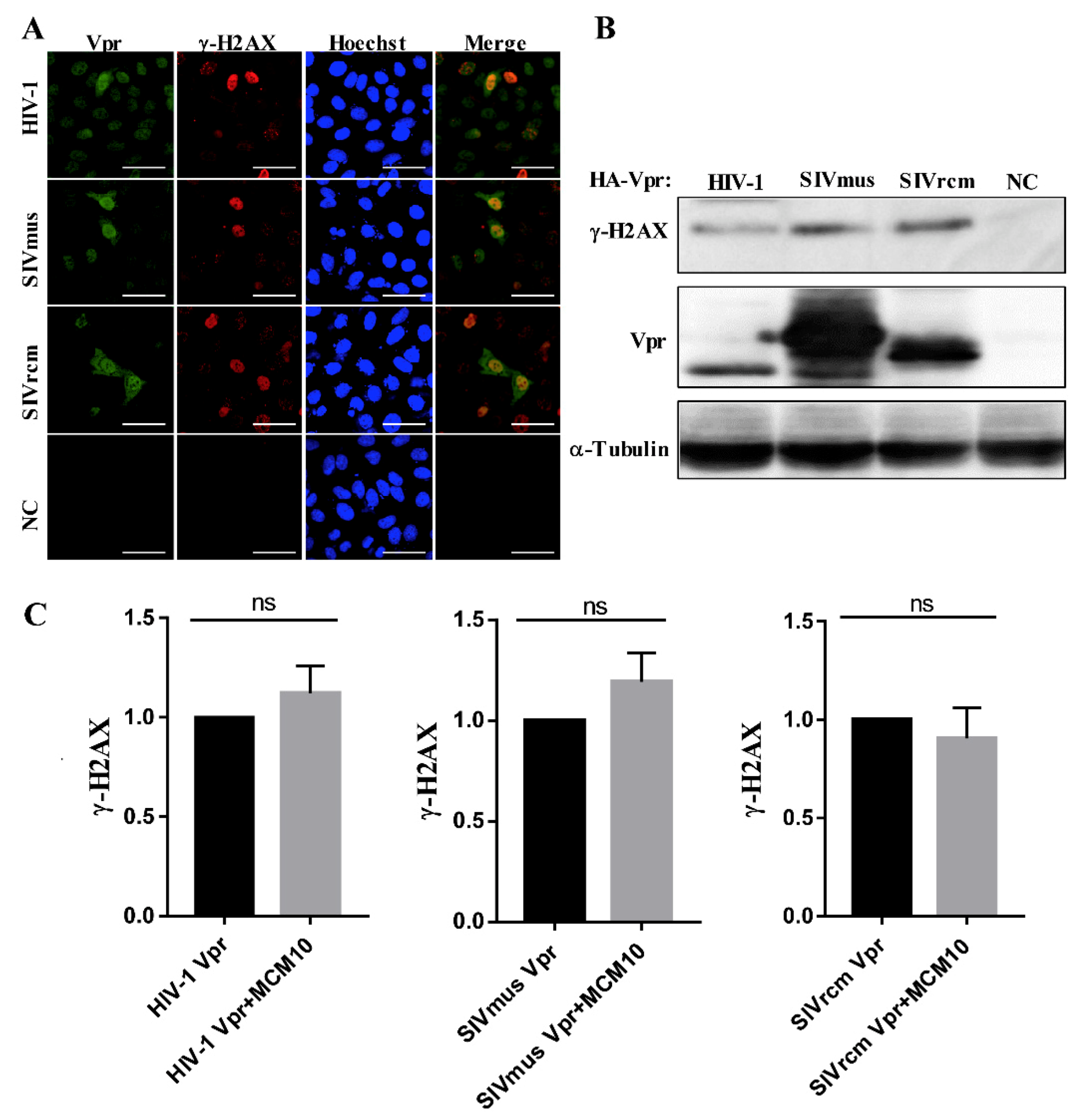
3.6. MCM10 Failure to Alleviate DDR Inducted by Primate Lentiviruses Vprs
3.7. Correlation of MCM10 Degradation with HIV-1 Vpr G2/M Arrest
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Malim, M.H.; Emerman, M. HIV-1 accessory proteins—Ensuring viral survival in a hostile environment. Cell Host Microbe 2008, 3, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Balliet, J.W.; Kolson, D.L.; Eiger, G.; Kim, F.M.; Mcgann, K.A.; Srinivasan, A.; Collman, R. Distinct Effects in Primary Macrophages and Lymphocytes of the Human-Immunodeficiency-Virus Type-1 Accessory Genes Vpr, Vpu, and Nef-Mutational Analysis of a Primary Hiv-1 Isolate. Virology 1994, 200, 623–631. [Google Scholar] [CrossRef]
- Mansky, L.M. The mutation rate of human immunodeficiency virus type 1 is influenced by the vpr gene. Virology 1996, 222, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Stark, L.A.; Hay, R.T. Human immunodeficiency virus type 1 HIV-1 viral protein R Vpr interacts with Lys-tRNA synthetase: Implications for priming of HIV-1 reverse transcription. J. Virol. 1998, 72, 3037–3044. [Google Scholar] [CrossRef] [PubMed]
- Heinzinger, N.K.; Bukrinsky, M.I.; Haggerty, S.A.; Ragland, A.M.; Kewalramani, V.; Lee, M.A.; Gendelman, H.E.; Ratner, L.; Stevenson, M.; Emerman, M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc. Natl. Acad. Sci. USA 1994, 91, 7311–7315. [Google Scholar] [CrossRef]
- Mahalingam, S.; Collman, R.G.; Patel, M.; Monken, C.E.; Srinivasan, A. Functional analysis of HIV-1 Vpr: Identification of determinants essential for subcellular localization. Virology 1995, 212, 331–339. [Google Scholar] [CrossRef]
- Nitahara-Kasahara, Y.; Kamata, M.; Yamamoto, T.; Zhang, X.; Miyamoto, Y.; Muneta, K.; Iijima, S.; Yoneda, Y.; Tsunetsugu-Yokota, Y.; Aida, Y. Novel nuclear import of Vpr promoted by importin alpha is crucial for human immunodeficiency virus type 1 replication in macrophages. J. Virol. 2007, 81, 5284–5293. [Google Scholar] [CrossRef]
- Felzien, L.K.; Woffendin, C.; Hottiger, M.O.; Subbramanian, R.A.; Cohen, E.A.; Nabel, G.J. HIV transcriptional activation by the accessory protein, VPR, is mediated by the p300 co-activator. Proc. Natl. Acad. Sci. USA 1998, 95, 5281–5286. [Google Scholar] [CrossRef]
- Kuramitsu, M.; Hashizume, C.; Yamamoto, N.; Azuma, A.; Kamata, M.; Yamamoto, N.; Tanaka, Y.; Aida, Y. A novel role for Vpr of human immunodeficiency virus type 1 as a regulator of the splicing of cellular pre-mRNA. Microbes Infect. 2005, 7, 1150–1160. [Google Scholar] [CrossRef]
- Hashizume, C.; Kuramitsu, M.; Zhang, X.F.; Kurosawa, T.; Kamata, M.; Aida, Y. Human immunodeficiency virus type 1 Vpr interacts with spliceosomal protein SAP145 to mediate cellular pre-mRNA splicing inhibition. Microbes Infect. 2007, 9, 490–497. [Google Scholar] [CrossRef]
- Rogel, M.E.; Wu, L.I.; Emerman, M. The human immunodeficiency virus type 1 vpr gene prevents cell proliferation during chronic infection. J. Virol. 1995, 69, 882–888. [Google Scholar] [CrossRef]
- Stewart, S.A.; Poon, B.; Jowett, J.B.; Chen, I.S. Human immunodeficiency virus type 1 Vpr induces apoptosis following cell cycle arrest. J. Virol. 1997, 71, 5579–5592. [Google Scholar] [CrossRef]
- Nishizawa, M.; Kamata, M.; Mojin, T.; Nakai, Y.; Aida, Y. Induction of apoptosis by the Vpr protein of human immunodeficiency virus type 1 occurs independently of G2 arrest of the cell cycle. Virology 2000, 276, 16–26. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fregoso, O.I.; Emerman, M. Activation of the DNA Damage Response Is a Conserved Function of HIV-1 and HIV-2 Vpr That Is Independent of SLX4 Recruitment. MBio 2016, 7, e01433-16. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.L.; DeHart, J.L.; Zimmerman, E.S.; Ardon, O.; Kim, B.; Jacquot, G.; Benichou, S.; Planelles, V. HIV-1 Vpr-induced apoptosis is cell cycle dependent and requires Bax but not ANT. PLoS Pathog. 2006, 2, e127. [Google Scholar] [CrossRef] [PubMed]
- Casey, L.; Wen, X.Y.; de Noronha, C.M.C. The functions of the HIV1 protein Vpr and its action through the DCAF1.DDB1. Cullin4 ubiquitin ligase. Cytokine 2010, 51, 1–9. [Google Scholar] [CrossRef]
- Laguette, N.; Bregnard, C.; Hue, P.; Basbous, J.; Yatim, A.; Larroque, M.; Kirchhoff, F.; Constantinou, A.; Sobhian, B.; Benkirane, M. Premature Activation of the SLX4 Complex by Vpr Promotes G2/M Arrest and Escape from Innate Immune Sensing. Cell 2014, 156, 134–145. [Google Scholar] [CrossRef]
- Romani, B.; Baygloo, N.S.; Aghasadeghi, M.R.; Allahbakhshi, E. HIV-1 Vpr Protein Enhances Proteasomal Degradation of MCM10 DNA Replication Factor through the Cul4-DDB1[VprBP] E3 Ubiquitin Ligase to Induce G2/M Cell Cycle Arrest. J. Biol. Chem. 2015, 290, 17380–17389. [Google Scholar] [CrossRef]
- Kaur, M.; Khan, M.M.; Kar, A.; Sharma, A.; Saxena, S. CRL4-DDB1-VPRBP ubiquitin ligase mediates the stress triggered proteolysis of Mcm10. Nucleic Acids Res. 2012, 40, 7332–7346. [Google Scholar] [CrossRef]
- Thu, Y.M.; Bielinsky, A.K. MCM10: One tool for all-Integrity, maintenance and damage control. Semin. Cell Dev. Biol. 2014, 30, 121–130. [Google Scholar] [CrossRef]
- Baxley, R.M.; Bielinsky, A.K. Mcm10: A Dynamic Scaffold at Eukaryotic Replication Forks. Genes 2017, 8, 73. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Murakami, T.; Aida, Y. Visualizing Vpr-Induced G2 Arrest and Apoptosis. PLoS ONE 2014, 9, e86840. [Google Scholar] [CrossRef] [PubMed]
- Chutiwitoonchai, N.; Aida, Y. NXT1, a Novel Influenza A NP Binding Protein, Promotes the Nuclear Export of NP via a CRM1-Dependent Pathway. Viruses 2016, 8, 209. [Google Scholar] [CrossRef]
- Morellet, N.; Bouaziz, S.; Petitjean, P.; Roques, B.P. NMR structure of the HIV-1 regulatory protein VPR. J. Mol. Biol. 2003, 327, 215–227. [Google Scholar] [CrossRef]
- Miyatake, H.; Sanjoh, A.; Murakami, T.; Murakami, H.; Matsuda, G.; Hagiwara, K.; Yokoyama, M.; Sato, H.; Miyamoto, Y.; Dohmae, N.; et al. Molecular Mechanism of HIV-1 Vpr for Binding to Importin-alpha. J. Mol. Biol. 2016, 428, 2744–2757. [Google Scholar] [CrossRef]
- Wu, Y.; Zhou, X.H.; Barnes, C.O.; DeLucia, M.; Cohen, A.E.; Gronenborn, A.M.; Ahn, J.; Calero, G. The DDB1-DCAF1-Vpr-UNG2 crystal structure reveals how HIV-1 Vpr steers human UNG2 toward destruction. Nat. Struct. Mol. Biol. 2016, 23, 933–940. [Google Scholar] [CrossRef]
- Wang, H.; Guo, H.R.; Su, J.M.; Rui, Y.J.; Zheng, W.W.; Gao, W.Y.; Zhang, W.Y.; Li, Z.L.; Liu, G.C.; Markham, R.B.; et al. Inhibition of Vpx-Mediated SAMHD1 and Vpr-Mediated Host Helicase Transcription Factor Degradation by Selective Disruption of Viral CRL4 DCAF1 E3 Ubiquitin Ligase Assembly. J. Virol. 2017, 91, e00225-17. [Google Scholar] [CrossRef]
- Chang, H.; Siarot, L.; Murakami, T.; Aida, Y. Viral Infectious Diseases Unit, RIKEN, Wako, Saitama, Japan. Subcellular Distribution of Primate Lentiviruses 11 Vpr/x Proteins. Unpublished work. 2020. [Google Scholar]
- Chang, H.; Aida, Y. Viral Infectious Diseases Unit, RIKEN, Wako, Saitama, Japan. MCM10 Down-Regulation by Primate Lentiviruses 11 Vpr/x via Dose-Dependent Assay. Unpublished work. 2020. [Google Scholar]
- Roesch, F.; Richard, L.; Rua, R.; Porrot, F.; Casartelli, N.; Schwartz, O. Vpr Enhances Tumor Necrosis Factor Production by HIV-1-Infected T Cells. J. Virol. 2015, 89, 12118–12130. [Google Scholar] [CrossRef]
- Jacquot, G.; Le Rouzic, E.; David, A.; Mazzolini, J.; Bouchet, J.; Bouaziz, S.; Niedergang, F.; Pancino, G.; Benichou, S. Localization of HIV-1 Vpr to the nuclear envelope: Impact on Vpr functions and virus replication in macrophages. Retrovirology 2007, 4, 84. [Google Scholar] [CrossRef]
- Venkatachari, N.J.; Walker, L.A.; Tastan, O.; Le, T.; Dempsey, T.M.; Li, Y.; Yanamala, N.; Srinivasan, A.; Klein-Seetharaman, J.; Montelaro, R.C.; et al. Human immunodeficiency virus type 1 Vpr: Oligomerization is an essential feature for its incorporation into virus particles. Virol. J. 2010, 7, 119. [Google Scholar] [CrossRef] [PubMed]
- Mansky, L.M.; Preveral, S.; Selig, L.; Benarous, R.; Benichou, S. The interaction of Vpr with uracil DNA glycosylase modulates the human immunodeficiency virus type 1 in vivo mutation rate. J. Virol. 2000, 74, 7039–7047. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.H.; DeLucia, M.; Ahn, J. SLX4-SLX1 Protein-independent Down-regulation of MUS81-EME1 Protein by HIV-1 Viral Protein R Vpr. J. Biol. Chem. 2016, 291, 16936–16947. [Google Scholar] [CrossRef] [PubMed]
- Lahouassa, H.; Blondot, M.L.; Chauveau, L.; Chougui, G.; Morel, M.; Leduc, M.; Guillonneau, F.; Ramirez, B.C.; Schwartz, O.; Margottin-Goguet, F. HIV-1 Vpr degrades the HLTF DNA translocase in T cells and macrophages. Proc. Natl. Acad. Sci. USA 2016, 113, 5311–5316. [Google Scholar] [CrossRef]
- Lv, L.; Wang, Q.; Xu, Y.P.; Tsao, L.C.; Nakagawa, T.; Guo, H.T.; Su, L.S.; Xiong, Y. Vpr Targets TET2 for Degradation by CRL4VprBP E3 Ligase to Sustain IL-6 Expression and Enhance HIV-1 Replication. Mol. Cell 2018, 70, 961–970. [Google Scholar] [CrossRef]
- Tellinghuisen, T.L.; Marcotrigiano, J.; Rice, C.M. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 2005, 435, 374–379. [Google Scholar] [CrossRef]
- Mehle, A.; Thomas, E.R.; Rajendran, K.S.; Gabuzda, D. A zinc-binding region in vif binds cul5 and determines cullin selection. J. Biol. Chem. 2006, 281, 17259–17265. [Google Scholar] [CrossRef]
- Guo, Y.; Dong, L.; Qiu, X.; Wang, Y.; Zhang, B.; Liu, H.; Yu, Y.; Zang, Y.; Yang, M.; Huang, Z. Structural basis for hijacking CBF-beta and CUL5 E3 ligase complex by HIV-1 Vif. Nature 2014, 505, 229–233. [Google Scholar] [CrossRef]
- Dannull, J.; Surovoy, A.; Jung, G.; Moelling, K. Specific Binding of Hiv-1 Nucleocapsid Protein to Psi-Rna in-Vitro Requires N-Terminal Zinc-Finger and Flanking Basic-Amino-Acid Residues. EMBO J. 1994, 13, 1525–1533. [Google Scholar] [CrossRef]
- Rein, A. RNA Packaging in HIV. Trends Microbiol. 2019, 27, 715–723. [Google Scholar] [CrossRef]
- Izumi, M.; Mizuno, T.; Yanagi, K.; Sugimura, K.; Okumura, K.; Imamoto, N.; Abe, T.; Hanaoka, F. The Mcm2-7-interacting domain of human mini-chromosome maintenance 10 Mcm10 protein is important for stable chromatin association and origin firing. J. Biol. Chem. 2017, 292, 13008–13021. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.E.; Dyson, H.J. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999, 293, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Warren, E.M.; Huang, H.; Fanning, E.; Chazin, W.J.; Eichman, B.F. Physical Interactions between Mcm10, DNA, and DNA Polymerase alpha. J. Biol. Chem. 2009, 284, 24662–24672. [Google Scholar] [CrossRef] [PubMed]
- Tompa, P. Intrinsically disordered proteins: A 10-year recap. Trends Biochem. Sci. 2012, 37, 509–516. [Google Scholar] [CrossRef]
- Romani, B.; Baygloo, N.S.; Hamidi-Fard, M.; Aghasadeghi, M.R.; Allahbakhshi, E. HIV-1 Vpr Protein Induces Proteasomal Degradation of Chromatin-associated Class I HDACs to Overcome Latent Infection of Macrophages. J. Biol. Chem. 2016, 291, 2696–2711. [Google Scholar] [CrossRef]
- Yan, J.P.; Shun, M.C.; Hao, C.L.; Zhang, Y.; Qian, J.; Hrecka, K.; DeLucia, M.; Monnie, C.; Ahn, J.; Skowronski, J. HIV-1 Vpr Reprograms CLR4DCAF1 E3 Ubiquitin Ligase to Antagonize Exonuclease 1-Mediated Restriction of HIV-1 Infection. MBio 2018, 9, e01732-18. [Google Scholar] [CrossRef]
- Tomimatsu, N.; Mukherjee, B.; Harris, J.L.; Boffo, F.L.; Hardebeck, M.C.; Potts, P.R.; Khanna, K.K.; Burma, S. DNA-damage-induced degradation of EXO1 exonuclease limits DNA end resection to ensure accurate DNA repair. J. Biol. Chem. 2017, 292, 10779–10790. [Google Scholar] [CrossRef]
- Chattopadhyay, S.; Bielinsky, A.K. Human Mcm10 regulates the catalytic subunit of DNA polymerase-alpha and prevents DNA damage during replication. Mol. Biol. Cell 2007, 18, 4085–4095. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, H.; Siarot, L.; Matsuura, R.; Lo, C.-W.; Sato, H.; Otsuki, H.; Aida, Y. Distinct MCM10 Proteasomal Degradation Profiles by Primate Lentiviruses Vpr Proteins. Viruses 2020, 12, 98. https://doi.org/10.3390/v12010098
Chang H, Siarot L, Matsuura R, Lo C-W, Sato H, Otsuki H, Aida Y. Distinct MCM10 Proteasomal Degradation Profiles by Primate Lentiviruses Vpr Proteins. Viruses. 2020; 12(1):98. https://doi.org/10.3390/v12010098
Chicago/Turabian StyleChang, Hao, Lowela Siarot, Ryosuke Matsuura, Chieh-Wen Lo, Hirotaka Sato, Hiroyuki Otsuki, and Yoko Aida. 2020. "Distinct MCM10 Proteasomal Degradation Profiles by Primate Lentiviruses Vpr Proteins" Viruses 12, no. 1: 98. https://doi.org/10.3390/v12010098
APA StyleChang, H., Siarot, L., Matsuura, R., Lo, C.-W., Sato, H., Otsuki, H., & Aida, Y. (2020). Distinct MCM10 Proteasomal Degradation Profiles by Primate Lentiviruses Vpr Proteins. Viruses, 12(1), 98. https://doi.org/10.3390/v12010098




