Fetal Calf Serum Exerts an Inhibitory Effect on Replication of Duck Hepatitis A Virus Genotype 1 in Duck Embryo Fibroblast Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells, Viruses, and Antiserum
2.2. Virus Propagation
2.3. RT-qPCR Assay
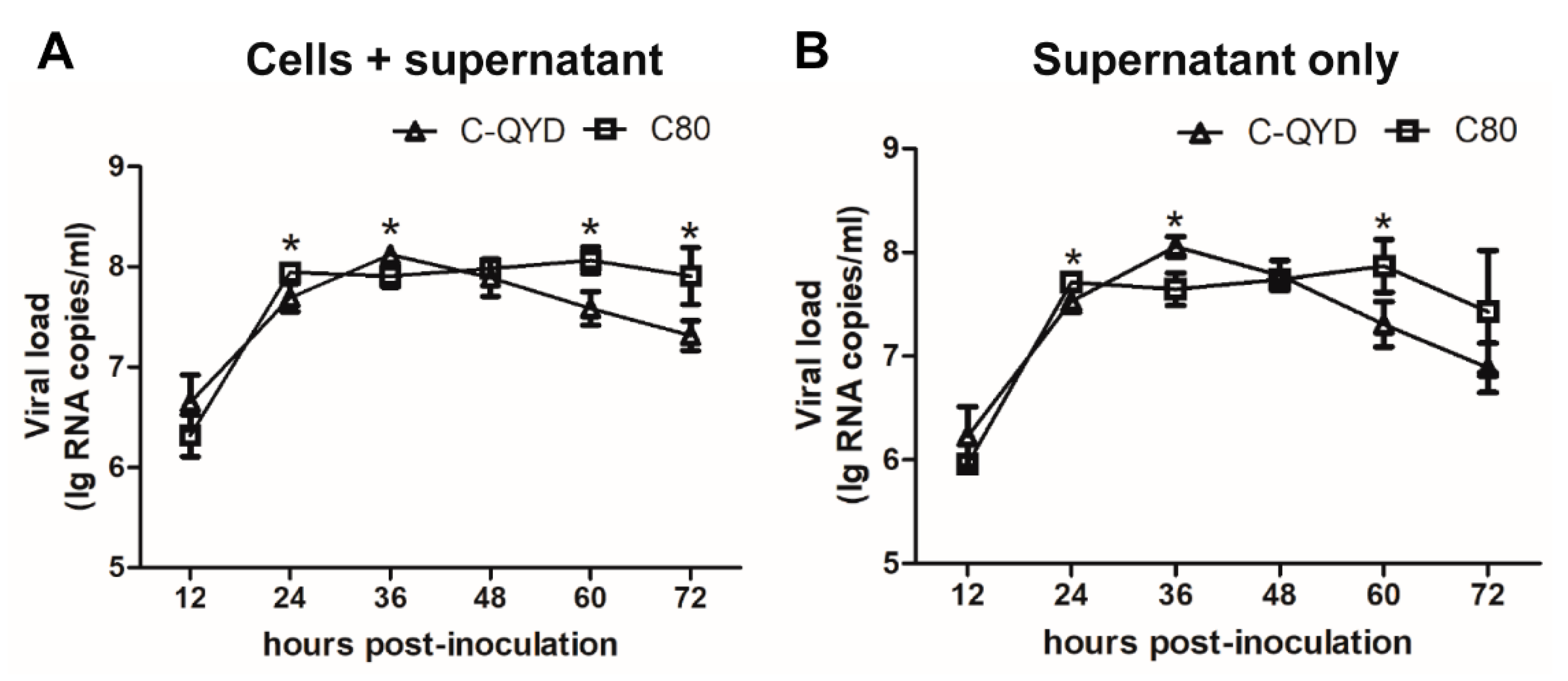
2.4. Determination of Viral Growth Kinetics
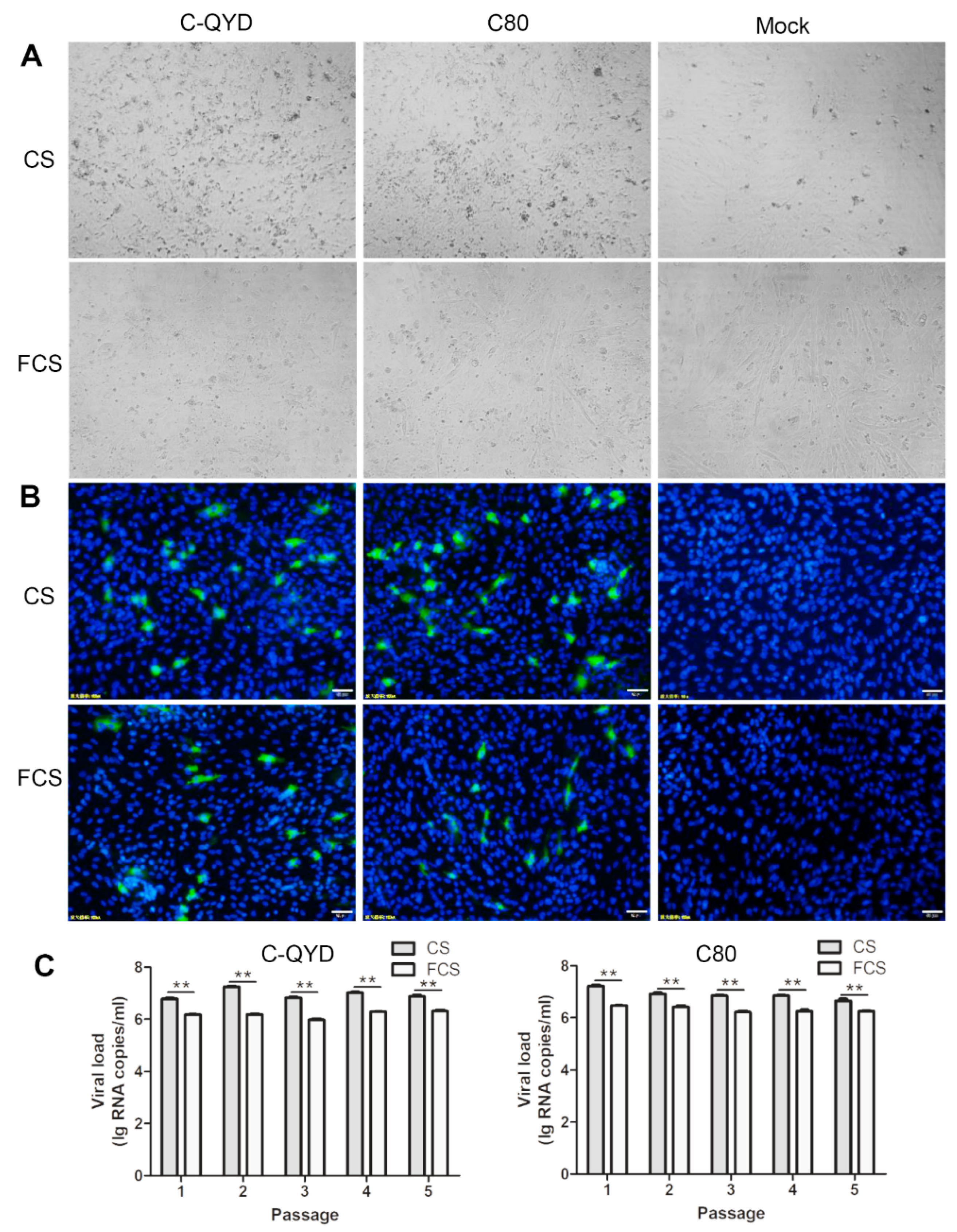
2.5. Immunofluorescence Test
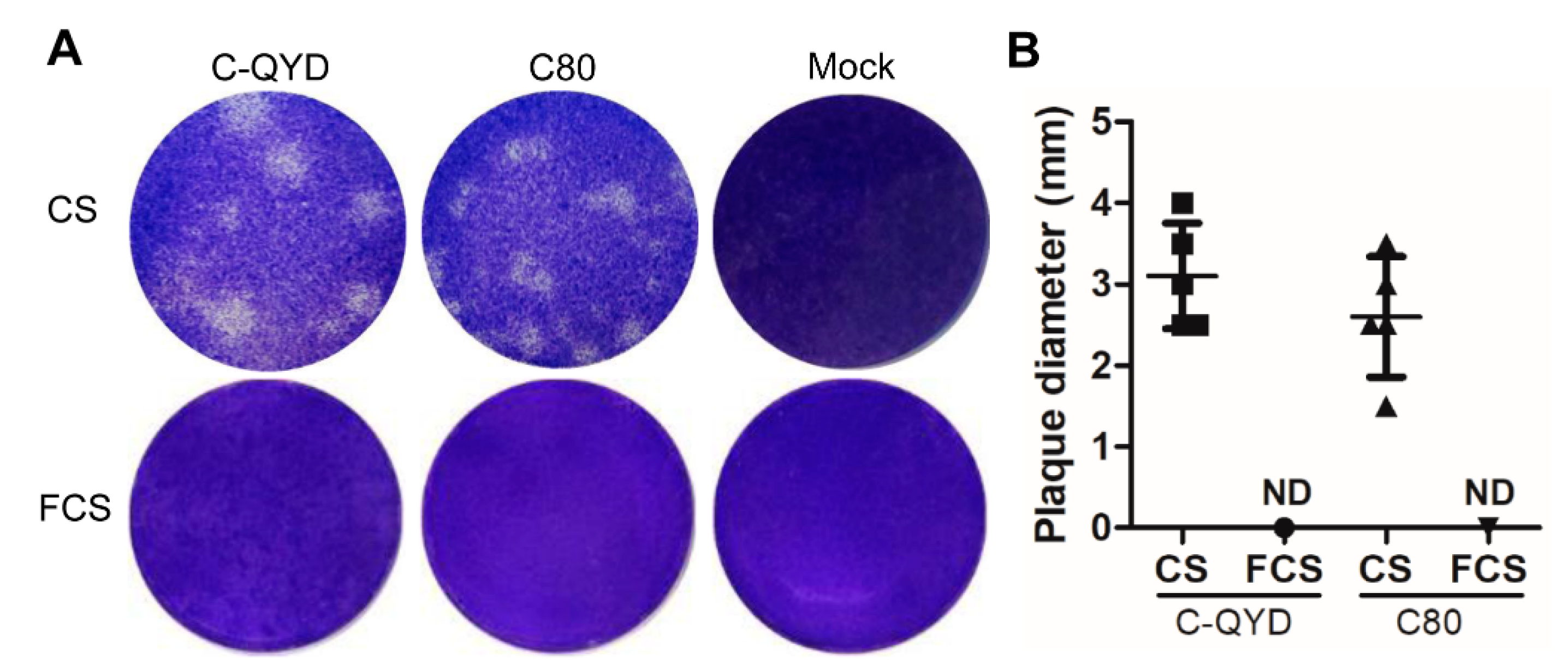
2.6. Plaque Formation Assay
2.7. Neutralization Test
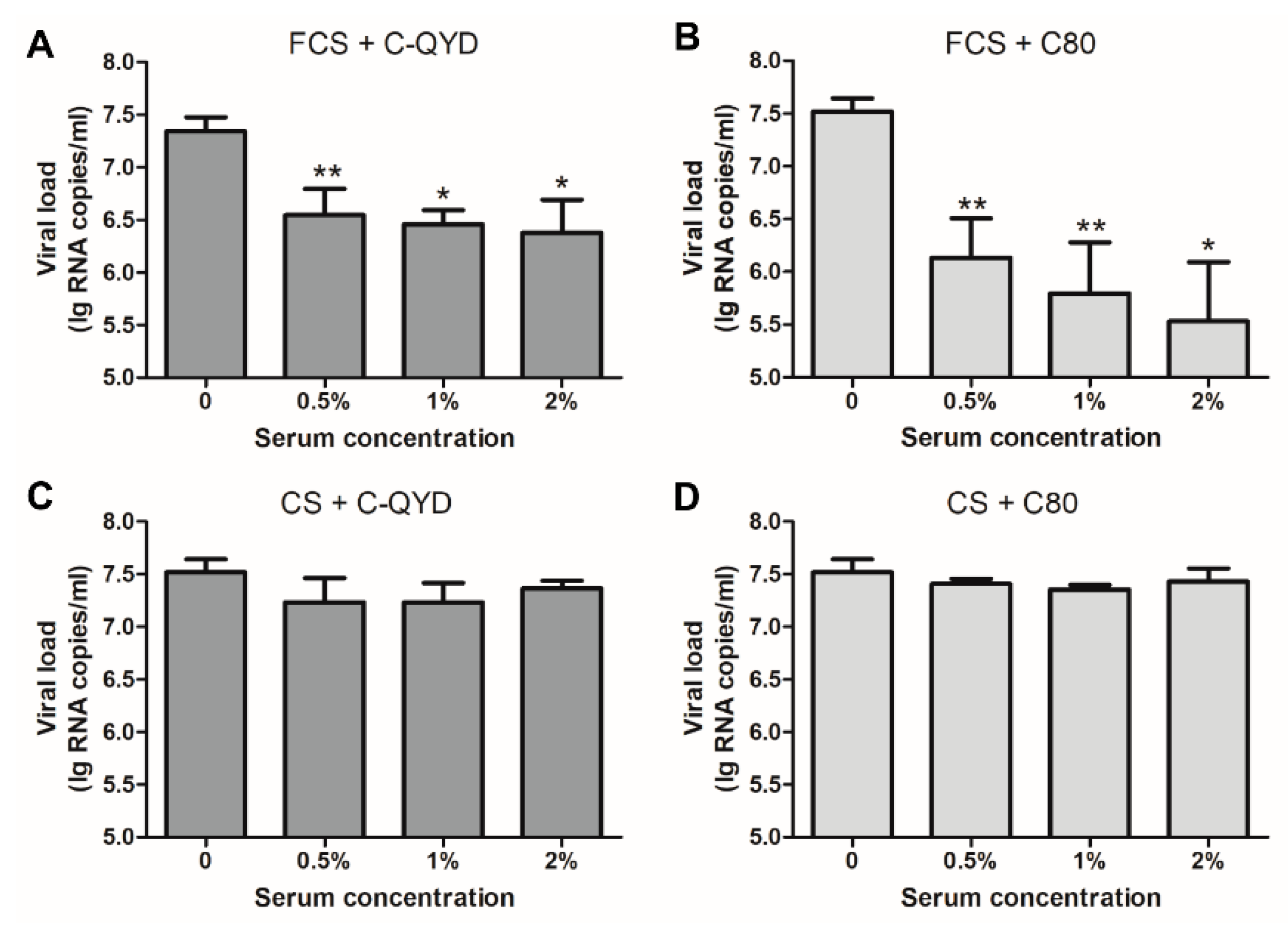
2.8. Assays for Effect of Pre-Incubation of Cells with FCS on Virus Infection
2.9. Virus Attachment Inhibition Assay
2.10. Virus Internalization Inhibition Assay
2.11. Assays for Effect of FCS on Viral RNA Replication
2.12. Assays for Effect of FCS on DHAV-1 IRES-Driven Translation
2.13. Assays for Effect of FCS on Extracellular Virus Yields
2.14. Statistical Analysis
3. Results
3.1. Replication Efficiency of DHAV-1 in DEF Cells Maintained in Medium Containing Chicken Serum (CS)
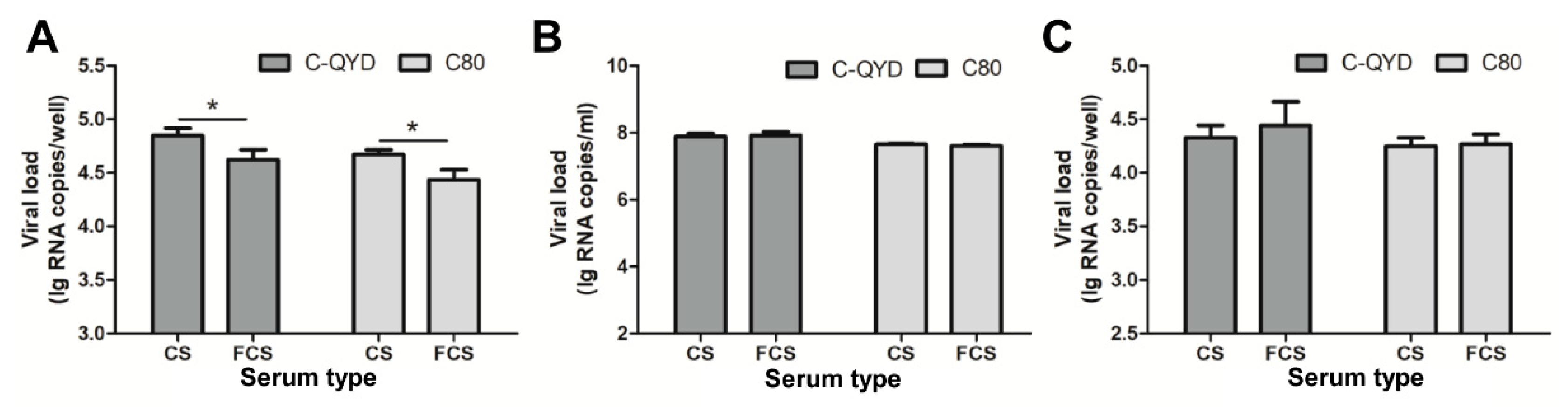
3.2. Effect of FCS on DHAV-1 Infection in DEF Cells in Comparison with CS
3.3. Effect of Serum Type on Formation of DHAV-1 Plaques in DEF Cells
3.4. Inhibitory Effect of FCS on DHAV-1
3.5. Effect of FCS on Virus Adsorption and Penetration
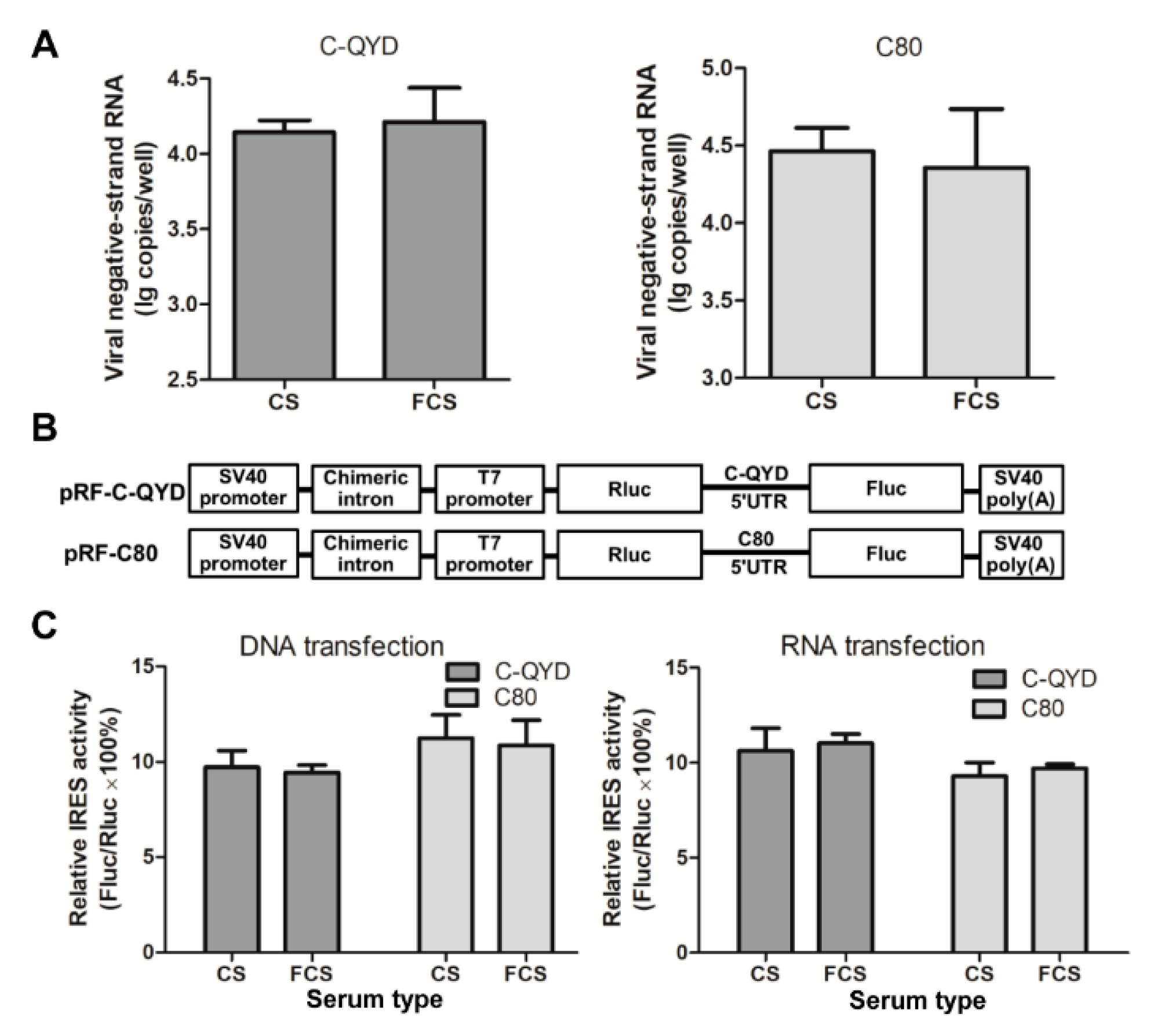
3.6. Effect of FCS on Gene Expression of DHAV-1
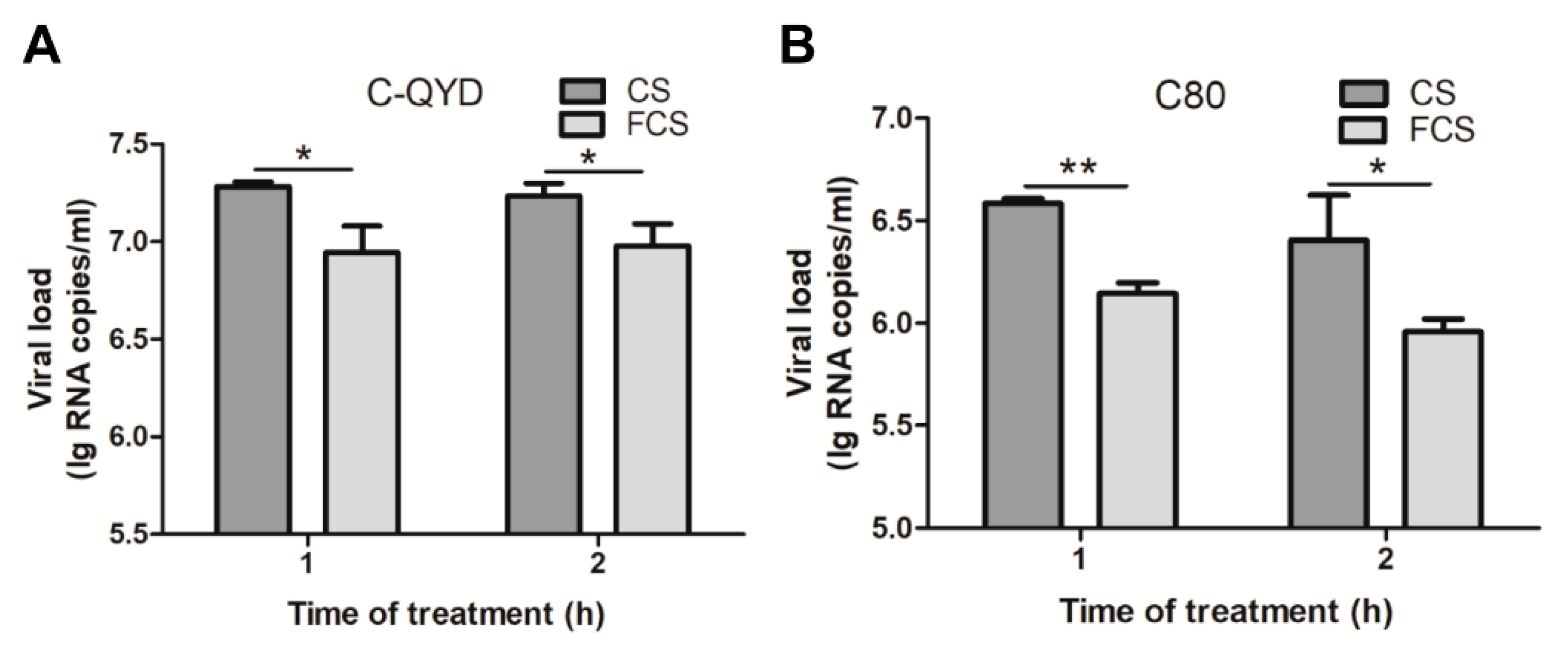
3.7. FCS-Mediated Inhibitory Effect on Extracellular Virus Yields
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Woolcock, P.R. Duck hepatitis. In Diseases of Poultry, 11th ed.; Saif, Y.M., Barnes, H.J., Glisson, J.R., Fadly, A.M., McDougald, L.R., Swayne, D.E., Eds.; Iowa State University Press: Ames, IA, USA, 2003; pp. 343–354. ISBN 081380423X. [Google Scholar]
- Tseng, C.H.; Tsai, H.J. Molecular characterization of a new serotype of duck hepatitis virus. Virus Res. 2007, 126, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.C.; Kwon, Y.K.; Joh, S.J.; Kim, S.J.; Tolf, C.; Kim, J.H.; Sung, H.W.; Lindberg, A.M.; Kwon, J.H. Recent Korean isolates of duck hepatitis virus reveal the presence of a new geno- and serotype when compared to duck hepatitis virus type 1 type strains. Arch. Virol. 2007, 152, 2059–2072. [Google Scholar] [CrossRef]
- Avihepatovirus. Available online: http://www.picornaviridae.com/avihepatovirus/avihepatovirus.htm (accessed on 13 August 2019).
- Asplin, F.D. Duck hepatitis: vaccination against two serological types. Vet. Rec. 1965, 77, 1529–1530. [Google Scholar] [CrossRef]
- Gough, R.E.; Borland, E.D.; Keymer, I.F.; Stuart, J.C. An outbreak of duck hepatitis type II in commercial ducks. Avian Pathol. 1985, 14, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Haider, S.A.; Calnek, B.W. In vitro isolation, propagation and characterization of duck hepatitis virus type III. Avian Dis. 1979, 23, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Todd, D.; Smyth, V.J.; Ball, N.W.; Donnelly, B.M.; Wylie, M.; Knowles, N.J.; Adair, B.M. Identification of chicken enterovirus-like viruses, duck hepatitis virus type 2 and duck hepatitis virus type 3 as astroviruses. Avian Pathol. 2009, 38, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Bosch, A.; Guix, S.; Krishna, N.K.; Méndez, E.; Monroe, S.S.; Pantin-Jackwood, M.; Schultz-Cherry, S. Astroviridae. In Virus Taxonomy. Classification and Nomenclature of Viruses: Ninth Report of the International Committee on the Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier Academic Press: London, UK, 2011; pp. 953–959. ISBN 978-0-12-384684-6. [Google Scholar]
- Woolcock, P.R.; Tsai, H.J. Duck hepatitis. In Diseases of Poultry, 13th ed.; Swayne, D.E., Glisson, J.R., McDougald, L.R., Lisa, K., Nolan, L.K., Suarez, D.L., Nair, V., Eds.; John Wiley & Sons, Inc: Hoboken, NJ, USA, 2013; pp. 422–431. ISBN 978-0-470-95899-5. [Google Scholar]
- Zhang, D.; Liu, N. Duck hepatitis virus. In Molecular Detection of Animal Viral Pathogens, 1st ed.; Liu, D., Ed.; CRC Press: Boca Raton, FL, USA, 2016; pp. 53–60. ISBN 9781498700368. [Google Scholar]
- Ding, C.; Zhang, D. Molecular analysis of duck hepatitis virus type 1. Virology 2007, 361, 9–17. [Google Scholar] [CrossRef]
- Kim, M.C.; Kwon, Y.K.; Joh, S.J.; Lindberg, A.M.; Kwon, J.H.; Kim, J.H.; Kim, S.J. Molecular analysis of duck hepatitis virus type 1 reveals a novel lineage close to the genus Parechovirus in the family Picornaviridae. J. Gen. Virol. 2006, 87, 3307–3316. [Google Scholar] [CrossRef]
- Tseng, C.H.; Knowles, N.J.; Tsai, H.J. Molecular analysis of duck hepatitis virus type 1 indicates that it should be assigned to a new genus. Virus Res. 2007, 123, 190–203. [Google Scholar] [CrossRef]
- Pan, M.; Yang, X.; Zhou, L.; Ge, X.; Guo, X.; Liu, J.; Zhang, D.; Yang, H. Duck Hepatitis A virus possesses a distinct type IV internal ribosome entry site element of picornavirus. J. Virol. 2012, 86, 1129–1144. [Google Scholar] [CrossRef]
- Pollard, M.; Starr, T.J. Propagation of Duck Hepatitis Virus in Tissue Culture. Proc. Soc. Exp. Biol. Med. 1959, 101, 521–524. [Google Scholar] [CrossRef] [PubMed]
- Kaeberle, M.L.; Drake, J.W.; Hanson, L.E. Cultivation of duck hepatitis virus in tissue culture. Proc. Soc. Exp. Biol. Med. 1961, 106, 755–757. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, J.E.; Hanson, L.E.; Wingard, M. Cytopathic effects of duck hepatitis virus in duck embryo kidney cell cultures. Proc. Soc. Exp. Biol. Med. 1963, 114, 814–816. [Google Scholar] [CrossRef]
- Fitzgerald, J.E.; Hanson, L.E. Certain properties of a cell-culture-modified duck hepatitis virus. Avian Dis. 1966, 10, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J. Duck hepatitis virus in duck embryo liver cell cultures. Avian Dis. 1966, 10, 508–512. [Google Scholar] [CrossRef] [PubMed]
- Mason, R.A.; Tauraso, N.M.; Ginn, R.K. Growth of Duck hepatitis virus in chicken embryos and in cell cultures derived from infected embryos. Avian Dis. 1972, 16, 973–979. [Google Scholar] [CrossRef] [PubMed]
- Davis, D.; Woolcock, P.R. Passage of duck hepatitis virus in cell cultures derived from avian embryos of different species. Res. Vet. Sci. 1986, 41, 133–134. [Google Scholar] [CrossRef]
- Woolcock, P.R. An assay for duck hepatitis virus type I in duck embryo liver cells and a comparison with other assays. Avian Pathol. 1986, 15, 75–82. [Google Scholar] [CrossRef]
- Kaleta, E.F. Duck viral hepatitis type 1 vaccination: Monitoring of the immune response with a microneutralisation test in pekin duck embryo kidney cell cultures. Avian Pathol. 1988, 17, 325–332. [Google Scholar] [CrossRef]
- Fu, Y.; Chen, Z.; Li, C.; Liu, G. Establishment of a duck cell line susceptible to duck hepatitis virus type 1. J. Virol. Methods 2012, 184, 41–45. [Google Scholar] [CrossRef]
- Mészáros, I.; Tóth, R.; Bálint, A.; Dán, A.; Jordan, I.; Zádori, Z. Propagation of viruses infecting waterfowl on continuous cell lines of Muscovy duck (Cairina moschata) origin. Avian Pathol. 2014, 43, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Said, A.; Wang, Y.; Fu, Q.; Xiao, Y.; Lv, S.; Shen, Z. Establishment and characterization of duck embryo epithelial (DEE) cell line and its use as a new approach toward DHAV-1 propagation and vaccine development. Virus Res. 2016, 213, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Woolcock, P.R.; Chalmers, W.S.; Davis, D. A plaque assay for duck hepatitis virus. Avian Pathol. 1982, 11, 607–610. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J. Duck hepatitis virus in duck embryo fibroblast cultures. Avian Dis. 1965, 9, 285–290. [Google Scholar] [CrossRef]
- Golubnichi, V.P.; Tishchenko, G.P.; Korolkov, V.I. Preparation of tissue culture antigens of duck hepatitis virus. Vet Nauk Proiz Tr. 1976, 14, 88–90. [Google Scholar]
- Chalmers, W.S.; Woolcock, P.R. The effect of animal sera on duck hepatitis virus. Avian Pathol. 1984, 13, 727–732. [Google Scholar] [CrossRef]
- Hernandez, R.; Brown, D.T. Growth and maintenance of chick embryo fibroblasts (CEF). Curr. Protoc. Microbiol. 2010, 17, 1–8. [Google Scholar] [CrossRef]
- Wang, L.; Pan, M.; Fu, Y.; Zhang, D. Classification of duck hepatitis virus into three genotypes based on molecular evolutionary analysis. Virus Genes 2008, 37, 52–59. [Google Scholar] [CrossRef]
- Yuan, Y. Development of the Original Seeds for an Attenuated Vaccine of Duck Hepatitis A Virus 3. Master’s Thesis, China Agricultural University, Beijing, China, 2013. [Google Scholar]
- Fu, Y.; Pan, M.; Wang, X.; Xu, Y.; Yang, H.; Zhang, D. Molecular detection and typing of duck hepatitis A virus directly from clinical specimens. Vet. Microbiol. 2008, 131, 247–257. [Google Scholar] [CrossRef]
- Chen, Y. Identities and Establishment of Real Time FQ-PCR for DHAV-1a and DHAV-1c. Master’s Thesis, Northeast Agricultural University, Harbin, China, 2013. [Google Scholar]
- Wang, M.; Li, Z.; Liu, H.; Wang, X.; Zhang, D. Effect of fetal calf serum on propagation of duck hepatitis A virus genotype 3 in duck embryo fibroblast cells. BMC Vet. Res. 2019, 15, 153. [Google Scholar] [CrossRef]
- Liang, T.; Liu, X.; Cui, S.; Qu, S.; Wang, D.; Liu, N.; Wang, F.; Ning, K.; Zhang, B.; Zhang, D. Generation of a reliable full-length cDNA of infectious Tembusu virus using a PCR-based protocol. Virus Res. 2016, 213, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Pan, M. Construction of an Infectious cDNA Clone of Duck Hepatitis A Virus Type 3 and Functional Analysis of Its 5′UTR. Ph.D. Thesis, China Agricultural University, Beijing, China, 2011. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Chai, L.; Liang, S.; Lv, J.; Yang, L.; Qu, S.; Jin, M.; Li, Q.; Wang, X.; Zhang, D. Fetal Calf Serum Exerts an Inhibitory Effect on Replication of Duck Hepatitis A Virus Genotype 1 in Duck Embryo Fibroblast Cells. Viruses 2020, 12, 80. https://doi.org/10.3390/v12010080
Wang M, Chai L, Liang S, Lv J, Yang L, Qu S, Jin M, Li Q, Wang X, Zhang D. Fetal Calf Serum Exerts an Inhibitory Effect on Replication of Duck Hepatitis A Virus Genotype 1 in Duck Embryo Fibroblast Cells. Viruses. 2020; 12(1):80. https://doi.org/10.3390/v12010080
Chicago/Turabian StyleWang, Minghang, Lili Chai, Suyun Liang, Junfeng Lv, Lixin Yang, Shenghua Qu, Meiling Jin, Qingxiangzi Li, Xiaoyan Wang, and Dabing Zhang. 2020. "Fetal Calf Serum Exerts an Inhibitory Effect on Replication of Duck Hepatitis A Virus Genotype 1 in Duck Embryo Fibroblast Cells" Viruses 12, no. 1: 80. https://doi.org/10.3390/v12010080
APA StyleWang, M., Chai, L., Liang, S., Lv, J., Yang, L., Qu, S., Jin, M., Li, Q., Wang, X., & Zhang, D. (2020). Fetal Calf Serum Exerts an Inhibitory Effect on Replication of Duck Hepatitis A Virus Genotype 1 in Duck Embryo Fibroblast Cells. Viruses, 12(1), 80. https://doi.org/10.3390/v12010080




