Global Gene Expression Analysis of the Brainstem in EV71- and CVA16-Infected Gerbils
Abstract
1. Introduction
2. Materials and Methods
2.1. Viral Isolation and Incubation
2.2. Gerbil Infection Experiments
2.3. Next-Generation Sequencing of mRNA and Data Processing
2.4. Quantitative RT-PCR (qRT-PCR) Validation
3. Results
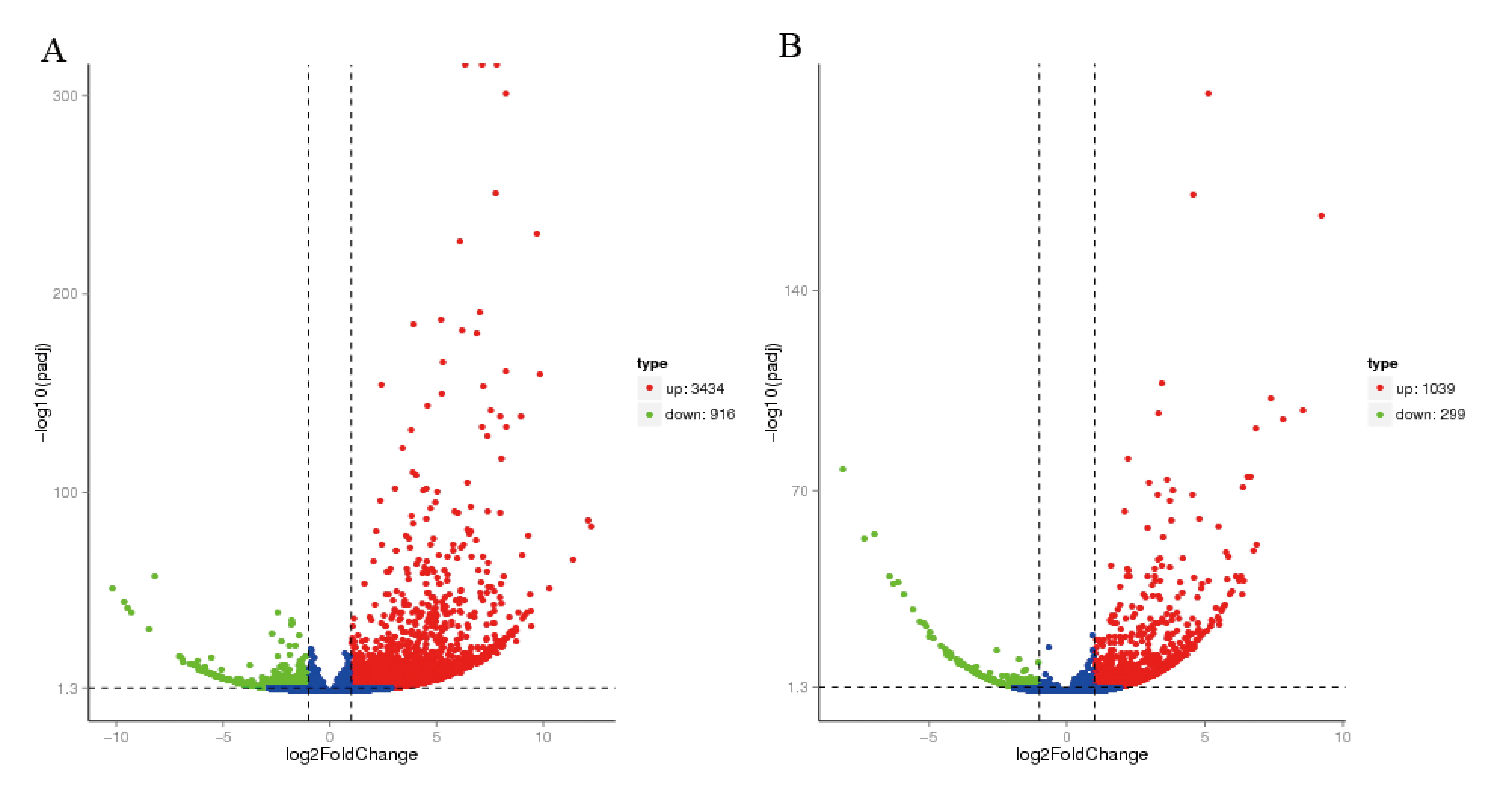
3.1. Different Gene Expressions in the Brainstem of Gerbils Infected by EV71 or CVA16
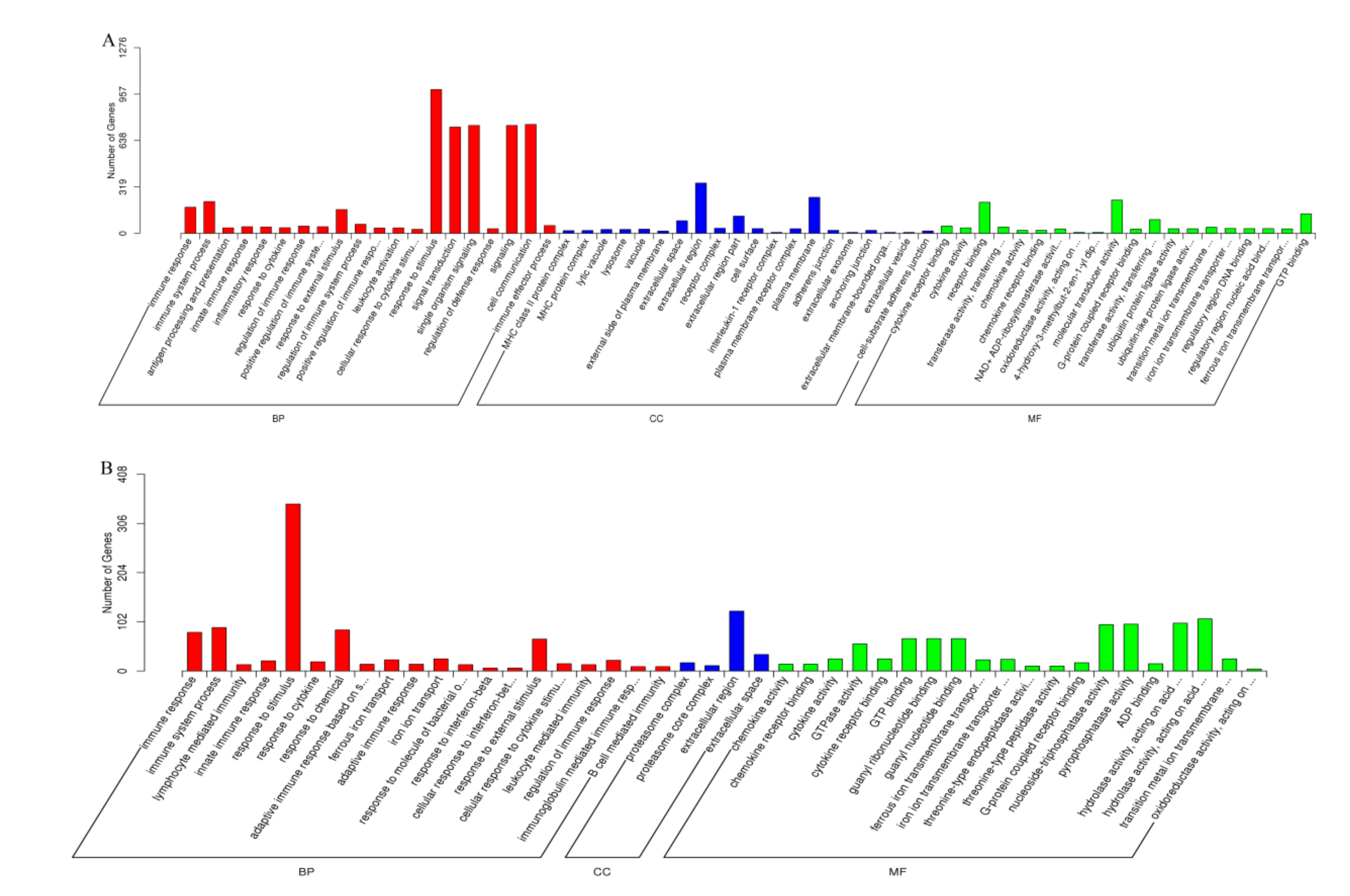
3.2. Target Gene Prediction and Functional Analysis
3.3. KEGG Pathway Scatter Plot
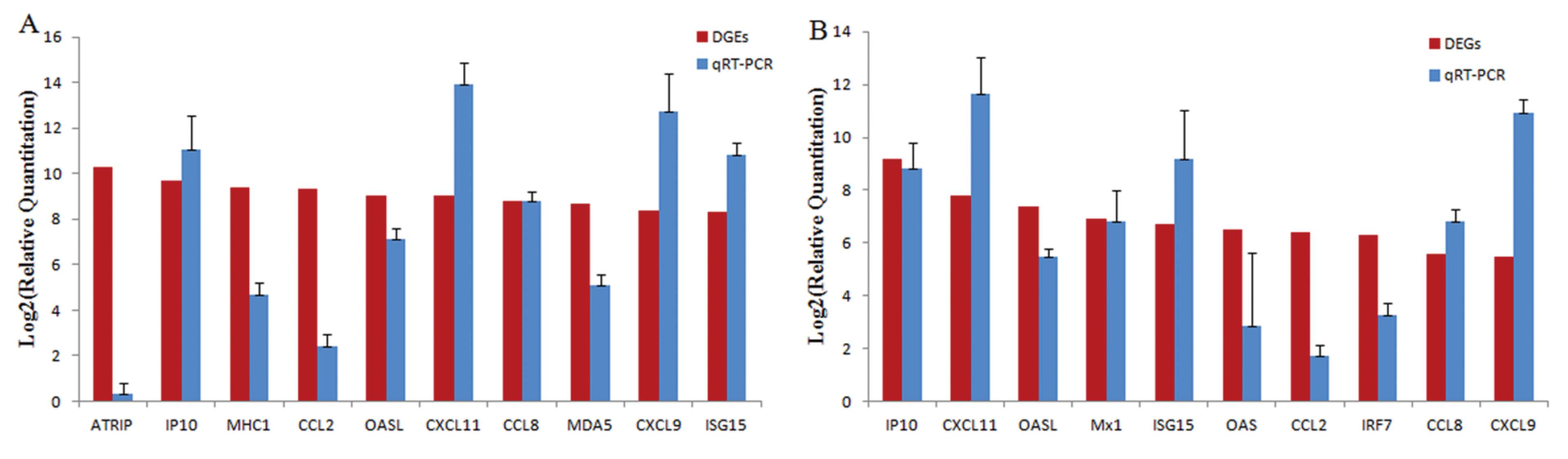
3.4. Gene Expression Validation by RT-PCR
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Koh, W.M.; Bogich, T.; Siegel, K.; Jin, J.; Chong, E.Y.; Tan, C.Y.; Chen, M.I.; Horby, P.; Cook, A.R. The Epidemiology of Hand, Foot and Mouth Disease in Asia: A Systematic Review and Analysis. Pediatr. Infect. Dis. J. 2016, 35, e285–e300. [Google Scholar] [CrossRef] [PubMed]
- Yao, P.P.; Qian, L.; Xia, Y.; Xu, F.; Yang, Z.N.; Xie, R.H.; Li, X.; Liang, W.F.; Huang, X.X.; Zhu, Z.Y.; et al. Enterovirus 71-induced neurological disorders in young gerbils, Meriones unguiculatus: Development and application of a neurological disease model. PLoS ONE 2012, 7, e51996. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.S.; Li, Y.J.; Xia, Y.; Xu, F.; Wang, W.W.; Yang, Z.N.; Lu, H.J.; Chen, Z.P.; Miao, Z.P.; Liang, W.F.; et al. Coxsackievirus A16 induced neurological disorders in young gerbils which could serve as a new animal model for vaccine evaluation. Sci. Rep. 2016, 6, 34299. [Google Scholar] [CrossRef] [PubMed]
- Mirand, A.; le Sage, F.V.; Pereira, B.; Cohen, R.; Levy, C.; Archimbaud, C.; Peigue-Lafeuille, H.; Bailly, J.L.; Henquell, C. Ambulatory Pediatric Surveillance of Hand, Foot and Mouth Disease as Signal of an Outbreak of Coxsackievirus A6 Infections, France, 2014–2015. Emerg. Infect. Dis. 2016, 22, 1884–1893. [Google Scholar] [CrossRef] [PubMed]
- Horsten, H.H.; Kemp, M.; Fischer, T.K.; Lindahl, K.H.; Bygum, A. Atypical Hand, Foot, and Mouth Disease Caused by Coxsackievirus A6 in Denmark: A Diagnostic Mimicker. Acta Derm. Venereol. 2018, 98, 350–354. [Google Scholar] [CrossRef]
- Yang, B.; Liu, F.; Liao, Q.; Wu, P.; Chang, Z.; Huang, J.; Long, L.; Luo, L.; Li, Y.; Leung, G.M.; et al. Epidemiology of hand, foot and mouth disease in China, 2008 to 2015 prior to the introduction of EV-A71 vaccine. Euro. Surveill. 2017, 22, 16–00824. [Google Scholar] [CrossRef]
- Too, I.H.; Yeo, H.; Sessions, O.M.; Yan, B.; Libau, E.A.; Howe, J.L.; Lim, Z.Q.; Suku-Maran, S.; Ong, W.Y.; Chua, K.B.; et al. Enterovirus 71 infection of motor neuron-like NSC-34 cells undergoes a non-lytic exit pathway. Sci. Rep. 2016, 6, 36983. [Google Scholar] [CrossRef]
- Mao, Q.; Wang, Y.; Yao, X.; Bian, L.; Wu, X.; Xu, M.; Liang, Z. Coxsackievirus A16: Epidemiology, diagnosis, and vaccine. Hum. Vaccin. Immunother. 2014, 10, 360–367. [Google Scholar] [CrossRef]
- Mao, Q.; Wang, Y.; Gao, R.; Shao, J.; Yao, X.; Lang, S.; Wang, C.; Mao, P.; Liang, Z.; Wang, J. A neonatal mouse model of coxsackievirus A16 for vaccine evaluation. J. Virol. 2012, 86, 11967–11976. [Google Scholar] [CrossRef]
- Liu, L.; Zhao, H.; Zhang, Y.; Wang, J.; Che, Y.; Dong, C.; Zhang, X.; Na, R.; Shi, H.; Jiang, L.; et al. Neonatal rhesus monkey is a potential animal model for studying pathogenesis of EV71 infection. Virology 2011, 412, 91–100. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.R.; Wang, P.S.; Wang, J.R.; Liu, H.S. Enterovirus 71-induced autophagy increases viral replication and pathogenesis in a suckling mouse model. J. Biomed. Sci. 2014, 21, 80. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.K.; Lin, T.; Zhu, S.L.; Xianyu, L.Z.; Lu, S.Y. Global quantitative proteomic analysis of human glioma cells profiled host protein expression in response to enterovirus type 71 infection. Proteomics 2015, 15, 3784–3796. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Song, J.; Liu, L.; Li, J.; Tang, B.; Zhang, Y.; Wang, J.; Wang, L.; Fan, S.; Feng, M.; et al. Comparison analysis of microRNAs in response to EV71 and CA16 infection in human bronchial epithelial cells by high-throughput sequencing to reveal differential infective mechanisms. Virus Res. 2017, 228, 90–101. [Google Scholar] [CrossRef][Green Version]
- Song, J.; Hu, Y.; Hu, Y.; Wang, J.; Zhang, X.; Wang, L.; Guo, L.; Wang, Y.; Ning, R.; Liao, Y.; et al. Global gene expression analysis of peripheral blood mononuclear cells in rhesus monkey infants with CA16 infection-induced HFMD. Virus Res. 2016, 214, 1–10. [Google Scholar] [CrossRef]
- Jin, J.; Li, R.; Jiang, C.; Zhang, R.; Ge, X.; Liang, F.; Sheng, X.; Dai, W.; Chen, M.; Wu, J.; et al. Transcriptome analysis reveals dynamic changes in coxsackievirus A16 infected HEK 293T cells. BMC Genomics 2017, 18, 933. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, E.; Pu, J.; Liu, L.; Che, Y.; Wang, J.; Liao, Y.; Wang, L.; Ding, D.; Zhao, T.; et al. The gene expression profile of peripheral blood mononuclear cells from EV71-infected rhesus infants and the significance in viral pathogenesis. PLoS ONE 2014, 9, e83766. [Google Scholar] [CrossRef]
- Xun, M.; Ma, C.F.; Du, Q.L.; Ji, Y.H.; Xu, J.R. Differential expression of miRNAs in enterovirus 71-infected cells. Virol. J. 2015, 12, 56. [Google Scholar] [CrossRef]
- Liu, J.; Li, S.; Cai, C.; Xu, Y.; Jiang, Y.; Chen, Z. Cerebrospinal fluid chemokine patterns in children with enterovirus 71-related encephalitis. Sci. Rep. 2018, 8, 1658. [Google Scholar] [CrossRef]
- Fallahi, P.; Elia, G. Interferon-gamma-induced protein 10 in Dengue Virus infection. Clin. Ter. 2016, 167, e186–e191. [Google Scholar] [CrossRef]
- Zeng, M.; Zheng, X.; Wei, R.; Zhang, N.; Zhu, K.; Xu, B.; Yang, C.H.; Yang, C.F.; Deng, C.; Pu, D.; et al. The cytokine and chemokine profiles in patients with hand, foot and mouth disease of different severities in Shanghai, China, 2010. PLoS Negl. Trop. Dis. 2013, 7, e2599. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, D.; Mukhopadhyay, A.; Ojha, D.; Sadhukhan, P.; Dutta, S. Immuno-metabolic changes in herpes virus infection. Cytokine 2018, 112, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Shen, F.H.; Tsai, C.C.; Wang, L.C.; Chang, K.C.; Tung, Y.Y.; Su, I.J.; Chen, S.H. Enterovirus 71 infection increases expression of interferon-gamma-inducible protein 10 which protects mice by reducing viral burden in multiple tissues. J. Gen. Virol. 2013, 94, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Marques, C.P.; Hu, S.; Sheng, W.; Lokensgard, J.R. Microglial cells initiate vigorous yet non-protective immune responses during HSV-1 brain infection. Virus Res. 2006, 121, 1–10. [Google Scholar] [CrossRef]
- Zheng, M.; Conrady, C.D.; Ward, J.M.; Bryant-Hudson, K.M.; Carr, D.J. Comparison of the host immune response to herpes simplex virus 1 (HSV-1) and HSV-2 at two different mucosal sites. J. Virol. 2012, 86, 7454–7458. [Google Scholar] [CrossRef]
- Sun, Y.S.; Yao, P.P.; Yang, Z.N.; Xu, F.; Lv, H.J.; Wu, L.Z.; Zhu, H.P. The killing mechanism of primary gerbil muscle cells induced by Coxsackievirus A16. Prev. Med. 2018, 30, 325–328. [Google Scholar] [CrossRef]


| Gene | Sequences | |
|---|---|---|
| Forward Primer (5′-3′) | Reverse Primer (5′-3′) | |
| ATRIP | GATGCAGGACCTCAACGCAAT | CCCACCAGCAGAAGCAGACAG |
| IP10 | CCTCAGCGTAGCTTCCAATAC | AAGTCAAGCCCAGGAACAAG |
| MHC1 | CAGGGAGATGTCAGCAGGGTA | GGAAGTGGGAAAACAGCGAGT |
| CCL2 | AGGTTCAAGGATGCCGAGTT | GTGTTGGCTAGGCCAGATTC |
| OASL | TGGAGTATCTGGCTCGGGTT | CGCTTTGAGTCGGCTATCTT |
| CXCL11 | CAAGCACGCCTTATACGACA | TTCCGGTTGCCAGTCACTTT |
| CCL8 | TGAGAAGTGGGTCCAGTCATA | TCGTGGGTCAAGTTAGCATT |
| MDA5 | GCAGGTCGGTGAGTGTGGGT | GGGTGGGGGCAGATGTTTGT |
| CXCL9 | CCAGATTCGGCAAATGTGAA | CAGTGAAGGCATTCCGCTAA |
| ISG15 | GGTCTTCTTGTACTTGCTCCTTCT | ACAGCGTCACCCTTATTAGCC |
| MxA-1 | AAGGCGAAGACCTCTATTGC | GATGATTAAAGGGATGTGGC |
| OAS | TGGAGTATCTGGCTCGGGTTAA | ATCGCTTTGAGTCGGCTATCTT |
| IRF7 | TGGCTTTATGGTTCAGTTTGTGA | CCAAGGCTCTGACTGGGAAG |
| β-actin | AACACCCCAGCCATGTACGTA | TCTCCGGAGTCCATCACAATG |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.-S.; Yang, Z.-N.; Xu, F.; Chen, C.; Lu, H.-J.; Jiang, J.-M.; Zhang, Y.-J.; Zhu, H.-P.; Yao, P.-P. Global Gene Expression Analysis of the Brainstem in EV71- and CVA16-Infected Gerbils. Viruses 2020, 12, 46. https://doi.org/10.3390/v12010046
Sun Y-S, Yang Z-N, Xu F, Chen C, Lu H-J, Jiang J-M, Zhang Y-J, Zhu H-P, Yao P-P. Global Gene Expression Analysis of the Brainstem in EV71- and CVA16-Infected Gerbils. Viruses. 2020; 12(1):46. https://doi.org/10.3390/v12010046
Chicago/Turabian StyleSun, Yi-Sheng, Zhang-Nv Yang, Fang Xu, Chen Chen, Hang-Jing Lu, Jian-Min Jiang, Yan-Jun Zhang, Han-Ping Zhu, and Ping-Ping Yao. 2020. "Global Gene Expression Analysis of the Brainstem in EV71- and CVA16-Infected Gerbils" Viruses 12, no. 1: 46. https://doi.org/10.3390/v12010046
APA StyleSun, Y.-S., Yang, Z.-N., Xu, F., Chen, C., Lu, H.-J., Jiang, J.-M., Zhang, Y.-J., Zhu, H.-P., & Yao, P.-P. (2020). Global Gene Expression Analysis of the Brainstem in EV71- and CVA16-Infected Gerbils. Viruses, 12(1), 46. https://doi.org/10.3390/v12010046




