Vesiculopolins, a New Class of Anti-Vesiculoviral Compounds, Inhibit Transcription Initiation of Vesiculoviruses
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Viruses
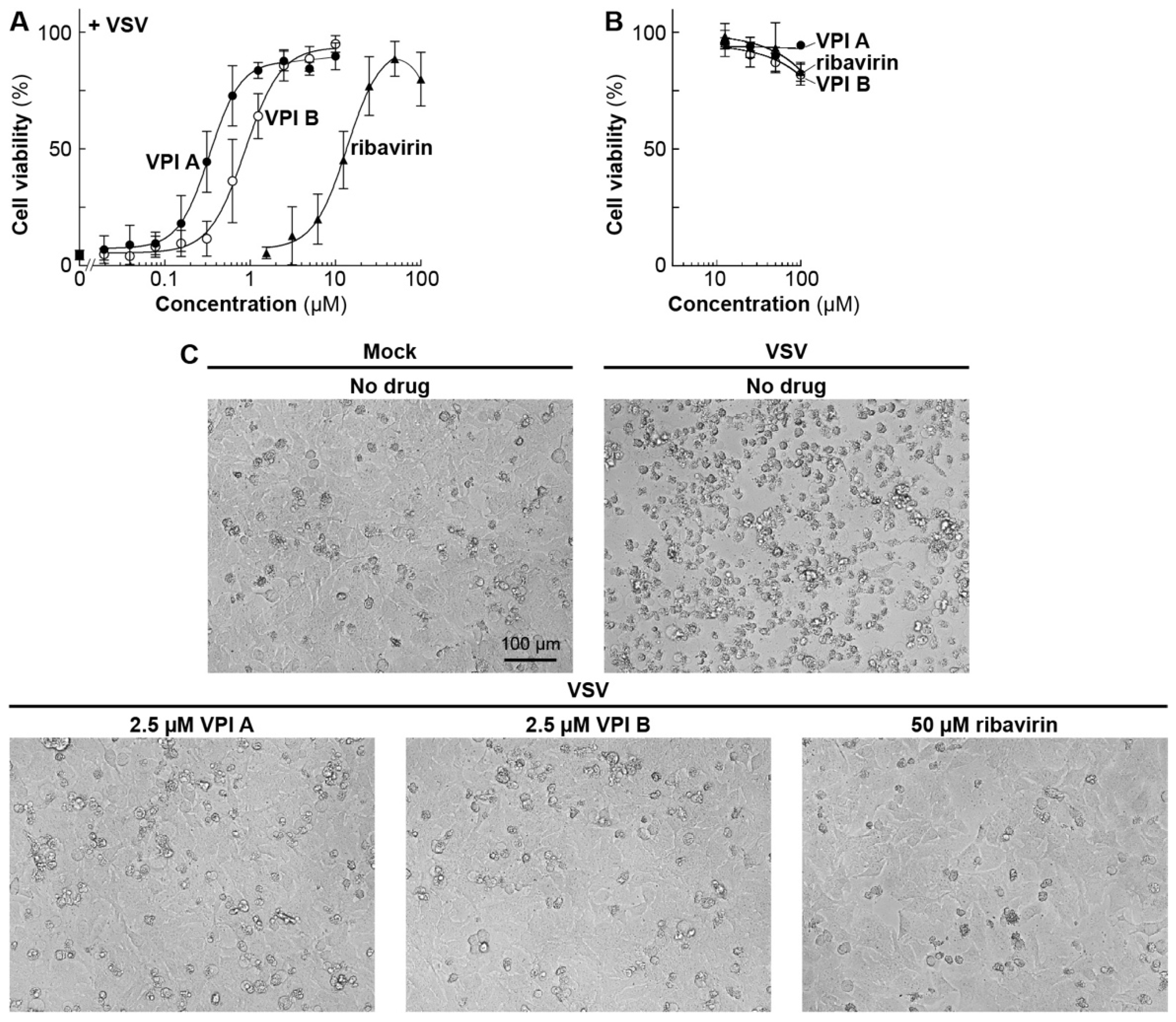
2.3. VSV Cell Killing Assay in a 384-Well Format
2.4. VSV Cell Killing Assay in a 96-Well Format
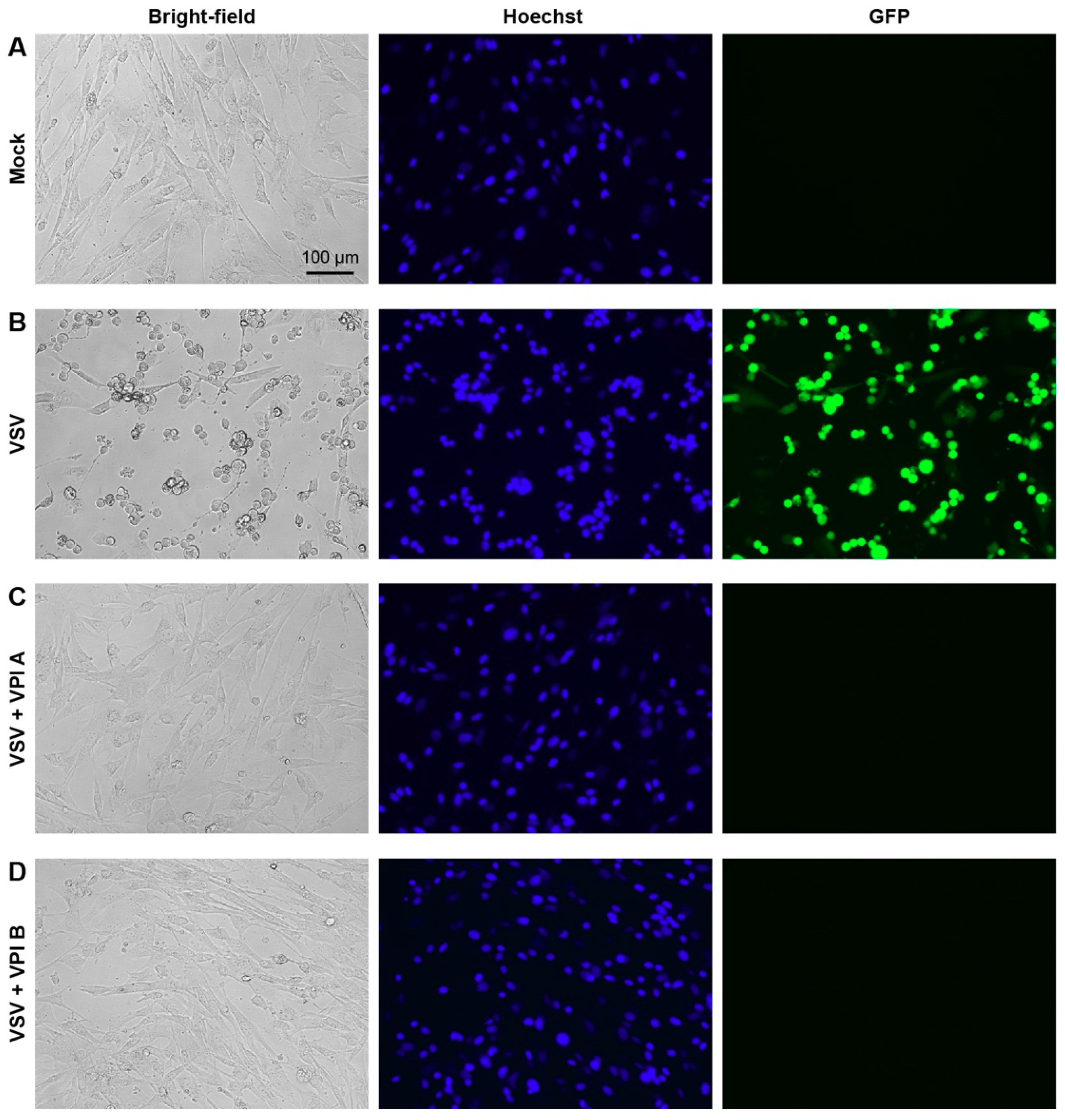
2.5. Other VSV Cell Infection Assays
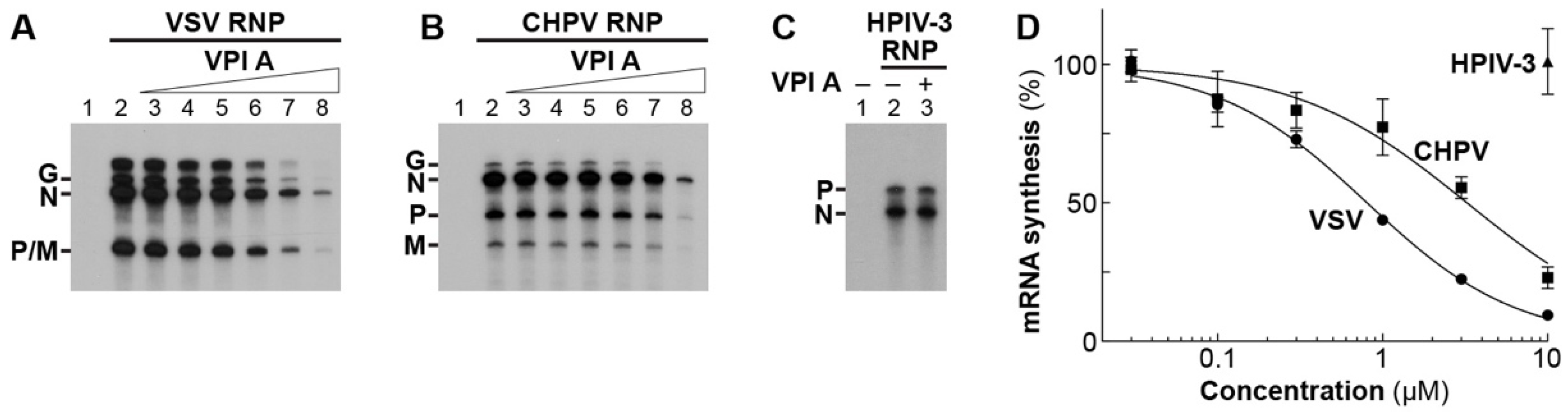
2.6. Preparation of Viral RNPs and Proteins
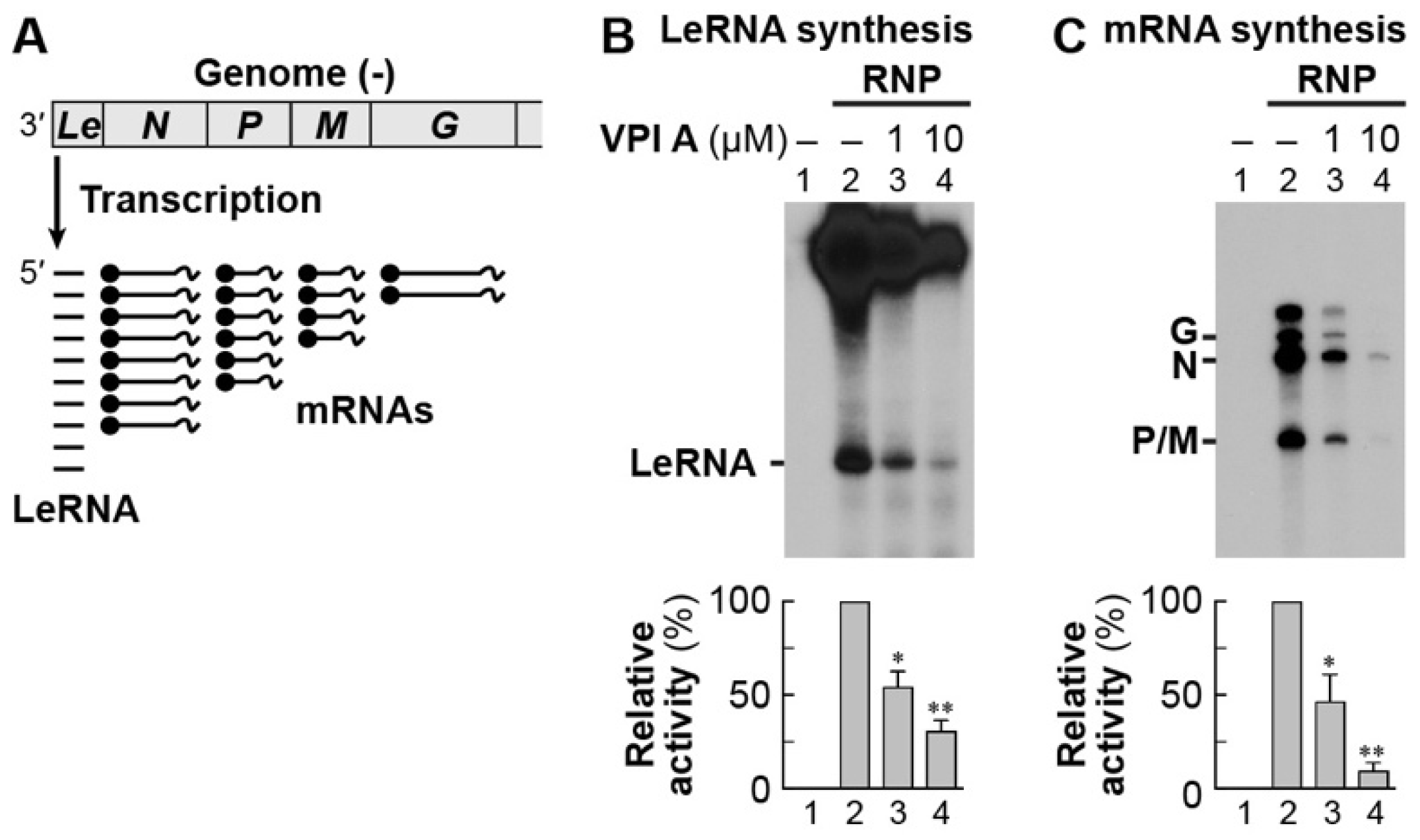
2.7. In Vitro Transcription and Capping Assays
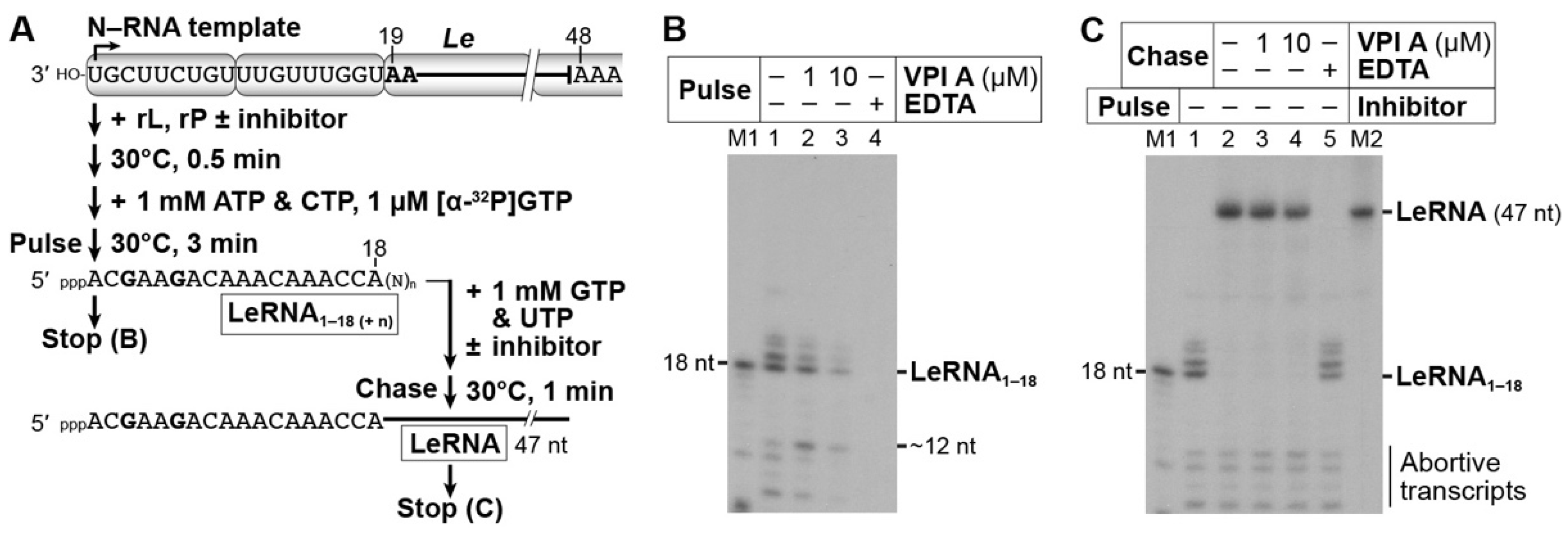
2.8. In Vitro Pulse-Chase Lerna Synthesis Assay
3. Results
3.1. Discovery of Vesiculopolins as VSV RdRp Inhibitors
3.2. Antiviral Activities of Vesiculopolins A and B against Rhabdoviruses
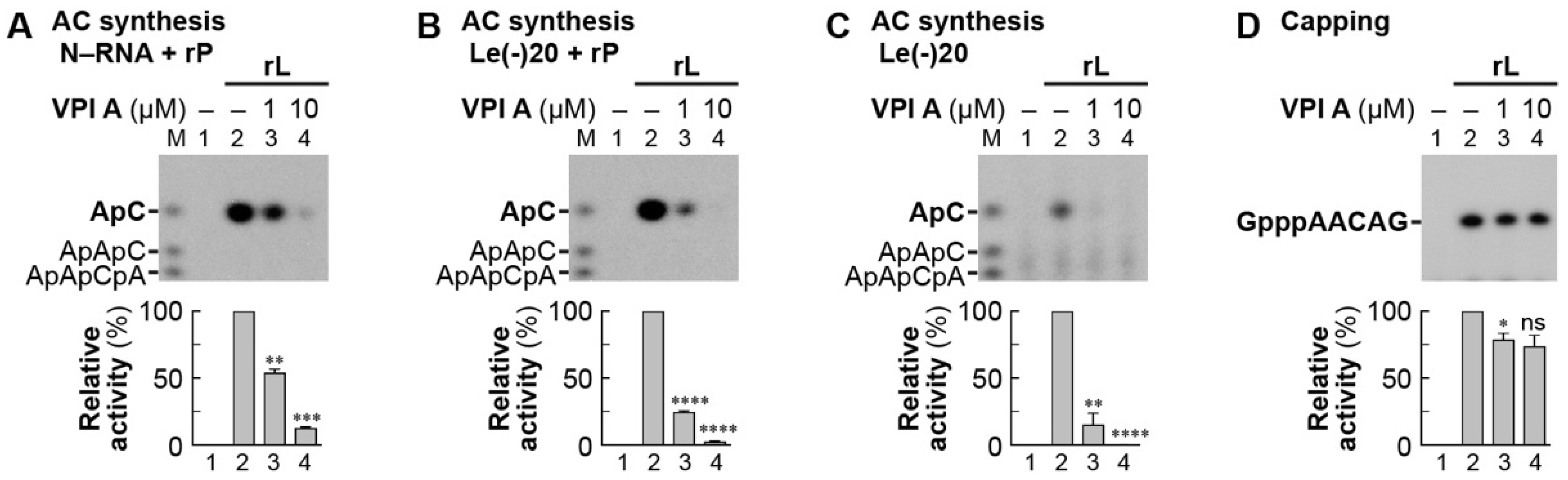
3.3. The Mode of Action of Vesiculopolin A in Inhibition of VSV RdRp
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stojdl, D.F.; Lichty, B.; Knowles, S.; Marius, R.; Atkins, H.; Sonenberg, N.; Bell, J.C. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat. Med. 2000, 6, 821–825. [Google Scholar] [CrossRef] [PubMed]
- Stojdl, D.F.; Lichty, B.D.; tenOever, B.R.; Paterson, J.M.; Power, A.T.; Knowles, S.; Marius, R.; Reynard, J.; Poliquin, L.; Atkins, H.; et al. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 2003, 4, 263–275. [Google Scholar] [CrossRef]
- Lichty, B.D.; Power, A.T.; Stojdl, D.F.; Bell, J.C. Vesicular stomatitis virus: re-inventing the bullet. Trends Mol. Med. 2004, 10, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Hastie, E.; Grdzelishvili, V.Z. Vesicular stomatitis virus as a flexible platform for oncolytic virotherapy against cancer. J. Gen. Virol. 2012, 93, 2529–2545. [Google Scholar] [CrossRef] [PubMed]
- Bourgeois-Daigneault, M.C.; Roy, D.G.; Falls, T.; Twumasi-Boateng, K.; St-Germain, L.E.; Marguerie, M.; Garcia, V.; Selman, M.; Jennings, V.A.; Pettigrew, J.; et al. Oncolytic vesicular stomatitis virus expressing interferon-gamma has enhanced therapeutic activity. Mol. Ther. Oncolytics 2016, 3, 16001. [Google Scholar] [CrossRef] [PubMed]
- Felt, S.A.; Grdzelishvili, V.Z. Recent advances in vesicular stomatitis virus-based oncolytic virotherapy: A 5-year update. J. Gen. Virol. 2017, 98, 2895–2911. [Google Scholar] [CrossRef] [PubMed]
- Mire, C.E.; Matassov, D.; Geisbert, J.B.; Latham, T.E.; Agans, K.N.; Xu, R.; Ota-Setlik, A.; Egan, M.A.; Fenton, K.A.; Clarke, D.K.; et al. Single-dose attenuated Vesiculovax vaccines protect primates against Ebola Makona virus. Nature 2015, 520, 688–691. [Google Scholar] [CrossRef]
- Henao-Restrepo, A.M.; Camacho, A.; Longini, I.M.; Watson, C.H.; Edmunds, W.J.; Egger, M.; Carroll, M.W.; Dean, N.E.; Diatta, I.; Doumbia, M.; et al. Efficacy and effectiveness of an rVSV-vectored vaccine in preventing Ebola virus disease: final results from the Guinea ring vaccination, open-label, cluster-randomised trial (Ebola Ca Suffit!). Lancet 2017, 389, 505–518. [Google Scholar] [CrossRef]
- Regules, J.A.; Beigel, J.H.; Paolino, K.M.; Voell, J.; Castellano, A.R.; Hu, Z.; Munoz, P.; Moon, J.E.; Ruck, R.C.; Bennett, J.W.; et al. A recombinant vesicular stomatitis virus Ebola vaccine. N. Engl. J. Med. 2017, 376, 330–341. [Google Scholar] [CrossRef]
- Huttner, A.; Dayer, J.A.; Yerly, S.; Combescure, C.; Auderset, F.; Desmeules, J.; Eickmann, M.; Finckh, A.; Goncalves, A.R.; Hooper, J.W.; et al. The effect of dose on the safety and immunogenicity of the VSV Ebola candidate vaccine: a randomised double-blind, placebo-controlled phase 1/2 trial. Lancet Infect. Dis. 2015, 15, 1156–1166. [Google Scholar] [CrossRef]
- Agnandji, S.T.; Huttner, A.; Zinser, M.E.; Njuguna, P.; Dahlke, C.; Fernandes, J.F.; Yerly, S.; Dayer, J.A.; Kraehling, V.; Kasonta, R.; et al. Phase 1 trials of rVSV Ebola vaccine in Africa and Europe. N. Engl. J. Med. 2016, 374, 1647–1660. [Google Scholar] [CrossRef] [PubMed]
- Quiroz, E.; Moreno, N.; Peralta, P.H.; Tesh, R.B. A human case of encephalitis associated with vesicular stomatitis virus (Indiana serotype) infection. Am. J. Trop. Med. Hyg. 1988, 39, 312–314. [Google Scholar] [CrossRef] [PubMed]
- van den Pol, A.N.; Dalton, K.P.; Rose, J.K. Relative neurotropism of a recombinant rhabdovirus expressing a green fluorescent envelope glycoprotein. J. Virol. 2002, 76, 1309–1327. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.E.; Nasar, F.; Coleman, J.W.; Price, R.E.; Javadian, A.; Draper, K.; Lee, M.; Reilly, P.A.; Clarke, D.K.; Hendry, R.M.; et al. Neurovirulence properties of recombinant vesicular stomatitis virus vectors in non-human primates. Virology 2007, 360, 36–49. [Google Scholar] [CrossRef] [PubMed]
- McWilliams, I.L.; Kielczewski, J.L.; Ireland, D.D.C.; Sykes, J.S.; Lewkowicz, A.P.; Konduru, K.; Xu, B.C.; Chan, C.C.; Caspi, R.R.; Manangeeswaran, M.; et al. Pseudovirus rVSVDeltaG-ZEBOV-GP infects neurons in retina and CNS, causing apoptosis and neurodegeneration in neonatal mice. Cell Rep. 2019, 26, 1718–1726 e4. [Google Scholar] [CrossRef] [PubMed]
- Ogino, T.; Banerjee, A.K. An unconventional pathway of mRNA cap formation by vesiculoviruses. Virus Res. 2011, 162, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Ogino, T.; Green, T.J. RNA synthesis and capping by non-segmented negative strand RNA viral polymerases: lessons from a prototypic virus. Front. Microbiol. 2019, 10, 1490. [Google Scholar] [CrossRef]
- Abraham, G.; Rhodes, D.P.; Banerjee, A.K. The 5′ terminal structure of the methylated mRNA synthesized in vitro by vesicular stomatitis virus. Cell 1975, 5, 51–58. [Google Scholar] [CrossRef]
- Abraham, G.; Banerjee, A.K. Sequential transcription of the genes of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 1976, 73, 1504–1508. [Google Scholar] [CrossRef]
- Ball, L.A.; White, C.N. Order of transcription of genes of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 1976, 73, 442–446. [Google Scholar] [CrossRef]
- Testa, D.; Chanda, P.K.; Banerjee, A.K. Unique mode of transcription in vitro by Vesicular stomatitis virus. Cell 1980, 21, 267–275. [Google Scholar] [CrossRef]
- Ogino, T.; Banerjee, A.K. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol. Cell 2007, 25, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Ogino, T.; Yadav, S.P.; Banerjee, A.K. Histidine-mediated RNA transfer to GDP for unique mRNA capping by vesicular stomatitis virus RNA polymerase. Proc. Natl. Acad. Sci. USA 2010, 107, 3463–3468. [Google Scholar] [CrossRef] [PubMed]
- Ogino, T.; Banerjee, A.K. The HR motif in the RNA-dependent RNA polymerase L protein of Chandipura virus is required for unconventional mRNA-capping activity. J. Gen. Virol. 2010, 91, 1311–1314. [Google Scholar] [CrossRef] [PubMed]
- Ogino, M.; Ito, N.; Sugiyama, M.; Ogino, T. The rabies virus L protein catalyzes mRNA capping with GDP polyribonucleotidyltransferase activity. Viruses 2016, 8, 144. [Google Scholar] [CrossRef] [PubMed]
- Ogino, T. Capping of vesicular stomatitis virus pre-mRNA is required for accurate selection of transcription stop-start sites and virus propagation. Nucleic. Acids. Res. 2014, 42, 12112–12125. [Google Scholar] [CrossRef] [PubMed]
- Neubauer, J.; Ogino, M.; Green, T.J.; Ogino, T. Signature motifs of GDP polyribonucleotidyltransferase, a non-segmented negative strand RNA viral mRNA capping enzyme, domain in the L protein are required for covalent enzyme-pRNA intermediate formation. Nucleic. Acids. Res. 2016, 44, 330–341. [Google Scholar] [CrossRef]
- Ogino, M.; Gupta, N.; Green, T.J.; Ogino, T. A dual-functional priming-capping loop of rhabdoviral RNA polymerases directs terminal de novo initiation and capping intermediate formation. Nucleic Acids Res. 2019, 47, 299–309. [Google Scholar] [CrossRef]
- Ogino, T.; Green, T.J. Transcriptional control and mRNA capping by the GDP polyribonucleotidyltransferase domain of the rabies virus large protein. Viruses 2019, 11, 504. [Google Scholar] [CrossRef]
- Grdzelishvili, V.Z.; Smallwood, S.; Tower, D.; Hall, R.L.; Hunt, D.M.; Moyer, S.A. A single amino acid change in the L-polymerase protein of vesicular stomatitis virus completely abolishes viral mRNA cap methylation. J. Virol. 2005, 79, 7327–7337. [Google Scholar] [CrossRef]
- Li, J.; Fontaine-Rodriguez, E.C.; Whelan, S.P. Amino acid residues within conserved domain VI of the vesicular stomatitis virus large polymerase protein essential for mRNA cap methyltransferase activity. J. Virol. 2005, 79, 13373–13384. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, T.; Min, W.; Lillehoj, H.S. Lymphocyte proliferation response during Eimeria tenella infection assessed by a new, reliable, nonradioactive colorimetric assay. Avian Dis. 2002, 46, 10–16. [Google Scholar] [CrossRef]
- Chadha, M.S.; Arankalle, V.A.; Jadi, R.S.; Joshi, M.V.; Thakare, J.P.; Mahadev, P.V.; Mishra, A.C. An outbreak of Chandipura virus encephalitis in the eastern districts of Gujarat state, India. Am. J. Trop. Med. Hyg. 2005, 73, 566–570. [Google Scholar] [CrossRef] [PubMed]
- Tandale, B.V.; Tikute, S.S.; Arankalle, V.A.; Sathe, P.S.; Joshi, M.V.; Ranadive, S.N.; Kanojia, P.C.; Eshwarachary, D.; Kumarswamy, M.; Mishra, A.C. Chandipura virus: a major cause of acute encephalitis in children in North Telangana, Andhra Pradesh, India. J. Med. Virol. 2008, 80, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Schnell, M.J.; Buonocore, L.; Whitt, M.A.; Rose, J.K. The minimal conserved transcription stop-start signal promotes stable expression of a foreign gene in vesicular stomatitis virus. J. Virol. 1996, 70, 2318–2323. [Google Scholar]
- Lawson, N.D.; Stillman, E.A.; Whitt, M.A.; Rose, J.K. Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. USA 1995, 92, 4477–4481. [Google Scholar] [CrossRef]
- Obuchi, M.; Fernandez, M.; Barber, G.N. Development of recombinant vesicular stomatitis viruses that exploit defects in host defense to augment specific oncolytic activity. J. Virol. 2003, 77, 8843–8856. [Google Scholar] [CrossRef]
- Naik, S.; Nace, R.; Federspiel, M.J.; Barber, G.N.; Peng, K.W.; Russell, S.J. Curative one-shot systemic virotherapy in murine myeloma. Leukemia 2012, 26, 1870–1878. [Google Scholar] [CrossRef]
- Tober, R.; Banki, Z.; Egerer, L.; Muik, A.; Behmuller, S.; Kreppel, F.; Greczmiel, U.; Oxenius, A.; von Laer, D.; Kimpel, J. VSV-GP: A potent viral vaccine vector that boosts the immune response upon repeated applications. J. Virol. 2014, 88, 4897–4907. [Google Scholar] [CrossRef]
- Garbutt, M.; Liebscher, R.; Wahl-Jensen, V.; Jones, S.; Moller, P.; Wagner, R.; Volchkov, V.; Klenk, H.D.; Feldmann, H.; Stroher, U. Properties of replication-competent vesicular stomatitis virus vectors expressing glycoproteins of filoviruses and arenaviruses. J. Virol. 2004, 78, 5458–5465. [Google Scholar] [CrossRef]
- Jones, S.M.; Feldmann, H.; Stroher, U.; Geisbert, J.B.; Fernando, L.; Grolla, A.; Klenk, H.D.; Sullivan, N.J.; Volchkov, V.E.; Fritz, E.A.; et al. Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nat. Med. 2005, 11, 786–790. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.; Wright, K.J.; Calderon, P.C.; Guo, M.; Nasar, F.; Johnson, J.E.; Coleman, J.W.; Lee, M.; Kotash, C.; Yurgelonis, I.; et al. Attenuation of recombinant vesicular stomatitis virus-human immunodeficiency virus type 1 vaccine vectors by gene translocations and g gene truncation reduces neurovirulence and enhances immunogenicity in mice. J. Virol. 2008, 82, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, S.; Ito, N.; Ohka, S.; Kaneda, S.; Nakamura, H.; Agari, T.; Masatani, T.; Nakagawa, K.; Okada, K.; Okadera, K.; et al. Involvement of the rabies virus phosphoprotein gene in neuroinvasiveness. J. Virol. 2013, 87, 12327–12338. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.H.; Chung, T.D.; Oldenburg, K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening sssays. J. Biomol. Screen. 1999, 4, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Walters, W.P.; Namchuk, M. Designing screens: How to make your hits a hit. Nat. Rev. Drug Discov. 2003, 2, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Crowley, L.C.; Marfell, B.J.; Waterhouse, N.J. Analyzing cell death by nuclear staining with Hoechst 33342. Cold Spring Harb. Protoc. 2016, 2016, 778–781. [Google Scholar] [CrossRef] [PubMed]
- Ogino, T. In vitro capping and transcription of rhabdoviruses. Methods 2013, 59, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Obijeski, J.F.; Marchenko, A.T.; Bishop, D.H.; Cann, B.W.; Murphy, F.A. Comparative electrophoretic analysis of the virus proteins of four rhabdoviruses. J. Gen. Virol. 1974, 22, 21–33. [Google Scholar] [CrossRef]
- Ogino, T.; Fukuda, H.; Imajoh-Ohmi, S.; Kohara, M.; Nomoto, A. Membrane binding properties and terminal residues of the mature hepatitis C virus capsid protein in insect cells. J. Virol. 2004, 78, 11766–11777. [Google Scholar] [CrossRef]
- Ogino, M.; Ogino, T. 5′-Phospho-RNA acceptor specificity of GDP polyribonucleotidyltransferase of vesicular stomatitis virus in mRNA capping. J. Virol. 2017, 91, e02322-02316. [Google Scholar] [CrossRef]
- Fensterl, V.; Wetzel, J.L.; Ramachandran, S.; Ogino, T.; Stohlman, S.A.; Bergmann, C.C.; Diamond, M.S.; Virgin, H.W.; Sen, G.C. Interferon-induced Ifit2/ISG54 protects mice from lethal VSV neuropathogenesis. PLoS Pathog. 2012, 8, e1002712. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.R.; Sunderland, A.; Grdzelishvili, V.Z. Cell type mediated resistance of vesicular stomatitis virus and Sendai virus to ribavirin. PLoS ONE 2010, 5, e11265. [Google Scholar] [CrossRef] [PubMed]
- Emerson, S.U. Reconstitution studies detect a single polymerase entry site on the vesicular stomatitis virus genome. Cell 1982, 31, 635–642. [Google Scholar] [CrossRef]
- Tomei, L.; Altamura, S.; Bartholomew, L.; Biroccio, A.; Ceccacci, A.; Pacini, L.; Narjes, F.; Gennari, N.; Bisbocci, M.; Incitti, I.; et al. Mechanism of action and antiviral activity of benzimidazole-based allosteric inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 2003, 77, 13225–13231. [Google Scholar] [CrossRef] [PubMed]
- McKercher, G.; Beaulieu, P.L.; Lamarre, D.; LaPlante, S.; Lefebvre, S.; Pellerin, C.; Thauvette, L.; Kukolj, G. Specific inhibitors of HCV polymerase identified using an NS5B with lower affinity for template/primer substrate. Nucleic Acids Res. 2004, 32, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Rigat, K.L.; Lu, H.; Wang, Y.K.; Argyrou, A.; Fanslau, C.; Beno, B.; Wang, Y.; Marcinkeviciene, J.; Ding, M.; Gentles, R.G.; et al. Mechanism of inhibition for BMS-791325, a novel non-nucleoside inhibitor of hepatitis C virus NS5B polymerase. J. Biol. Chem. 2014, 289, 33456–33468. [Google Scholar] [CrossRef]
- Conry, R.M.; Westbrook, B.; McKee, S.; Norwood, T.G. Talimogene laherparepvec: First in class oncolytic virotherapy. Hum. Vaccin. Immunother. 2018, 14, 839–846. [Google Scholar] [CrossRef]
- Pol, J.G.; Atherton, M.J.; Bridle, B.W.; Stephenson, K.B.; Le Boeuf, F.; Hummel, J.L.; Martin, C.G.; Pomoransky, J.; Breitbach, C.J.; Diallo, J.S.; et al. Development and applications of oncolytic Maraba virus vaccines. Oncolytic Virother 2018, 7, 117–128. [Google Scholar] [CrossRef]
- Grard, G.; Fair, J.N.; Lee, D.; Slikas, E.; Steffen, I.; Muyembe, J.J.; Sittler, T.; Veeraraghavan, N.; Ruby, J.G.; Wang, C.; et al. A novel rhabdovirus associated with acute hemorrhagic fever in central Africa. PLoS Pathog. 2012, 8, e1002924. [Google Scholar] [CrossRef]



| % Inhibition | |||||
|---|---|---|---|---|---|
| ChemBridge ID # | VPI | Metabolic Shut-down 1 (CCK Assay) | Nuclear Condensation 1 (Cell Imaging) | mRNA Synthesis 2 (In vitro Transcription) | |
| 1 | 53425003 | A | 105 | 98 | 99 |
| 2 | 20208871 | B | 102 | 92 | 89 |
| 3 | 66896553 | C | 58 | 63 | 72 |
| 4 | 35540582 | D | 55 | 57 | 72 |
| 5 | 80285323 | E | 50 | 94 | 72 |
| 6 | 10443614 | F | 48 | 69 | 34 |
| 7 | 37324373 | G | 46 | 54 | 68 |
| 8 | 52194062 | 44 | 51 | 10 | |
| 9 | 69805228 | 39 | 0 | n.d. | |
| 10 | 43534388 | 35 | 10 | n.d. | |
| 11 | 90353655 | H | 34 | 53 | 73 |
| 12 | 81699442 | 31 | 0 | n.d. | |
| 13 | 66349773 | 30 | 9 | n.d. | |
| Compound | BHK-21 Cells | HeLa Cells | ||||||
|---|---|---|---|---|---|---|---|---|
| Cytotoxicity | Anti-VSV Activity | Anti-CHPV Activity | Cytotoxicity | Anti-VSV Activity | ||||
| CC50 1 | IC50 1 | SI 2 | IC50 1 | SI 2 | CC50 1 | IC50 1 | SI 2 | |
| VPI A | >100 | 2.0 ± 0.1 | >50 | 7.9 ± 0.4 | >13 | >100 | 0.42 ± 0.02 | >240 |
| VPI B | >100 | 6.2 ± 0.2 | >16 | 11 ± 1 | >9 | >100 | 1.1 ± 0.1 | >91 |
| Ribavirin | 86 ± 0.5 | >100 | <1 | >100 | <1 | >100 | 15 ± 1 | >7 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogino, M.; Fedorov, Y.; Adams, D.J.; Okada, K.; Ito, N.; Sugiyama, M.; Ogino, T. Vesiculopolins, a New Class of Anti-Vesiculoviral Compounds, Inhibit Transcription Initiation of Vesiculoviruses. Viruses 2019, 11, 856. https://doi.org/10.3390/v11090856
Ogino M, Fedorov Y, Adams DJ, Okada K, Ito N, Sugiyama M, Ogino T. Vesiculopolins, a New Class of Anti-Vesiculoviral Compounds, Inhibit Transcription Initiation of Vesiculoviruses. Viruses. 2019; 11(9):856. https://doi.org/10.3390/v11090856
Chicago/Turabian StyleOgino, Minako, Yuriy Fedorov, Drew J. Adams, Kazuma Okada, Naoto Ito, Makoto Sugiyama, and Tomoaki Ogino. 2019. "Vesiculopolins, a New Class of Anti-Vesiculoviral Compounds, Inhibit Transcription Initiation of Vesiculoviruses" Viruses 11, no. 9: 856. https://doi.org/10.3390/v11090856
APA StyleOgino, M., Fedorov, Y., Adams, D. J., Okada, K., Ito, N., Sugiyama, M., & Ogino, T. (2019). Vesiculopolins, a New Class of Anti-Vesiculoviral Compounds, Inhibit Transcription Initiation of Vesiculoviruses. Viruses, 11(9), 856. https://doi.org/10.3390/v11090856





