Presence of a Novel Subtype of Bovine Hepacivirus in China and Expanded Classification of Bovine Hepacivirus Strains Worldwide into 7 Subtypes
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Virus Detection
2.3. Viral Genome Sequencing and Analysis
2.4. Phylogenetic Analysis
3. Results
3.1. BovHepV Detection in Commercial Bovine Serum Samples
3.2. Viral Genome Sequencing and Analysis
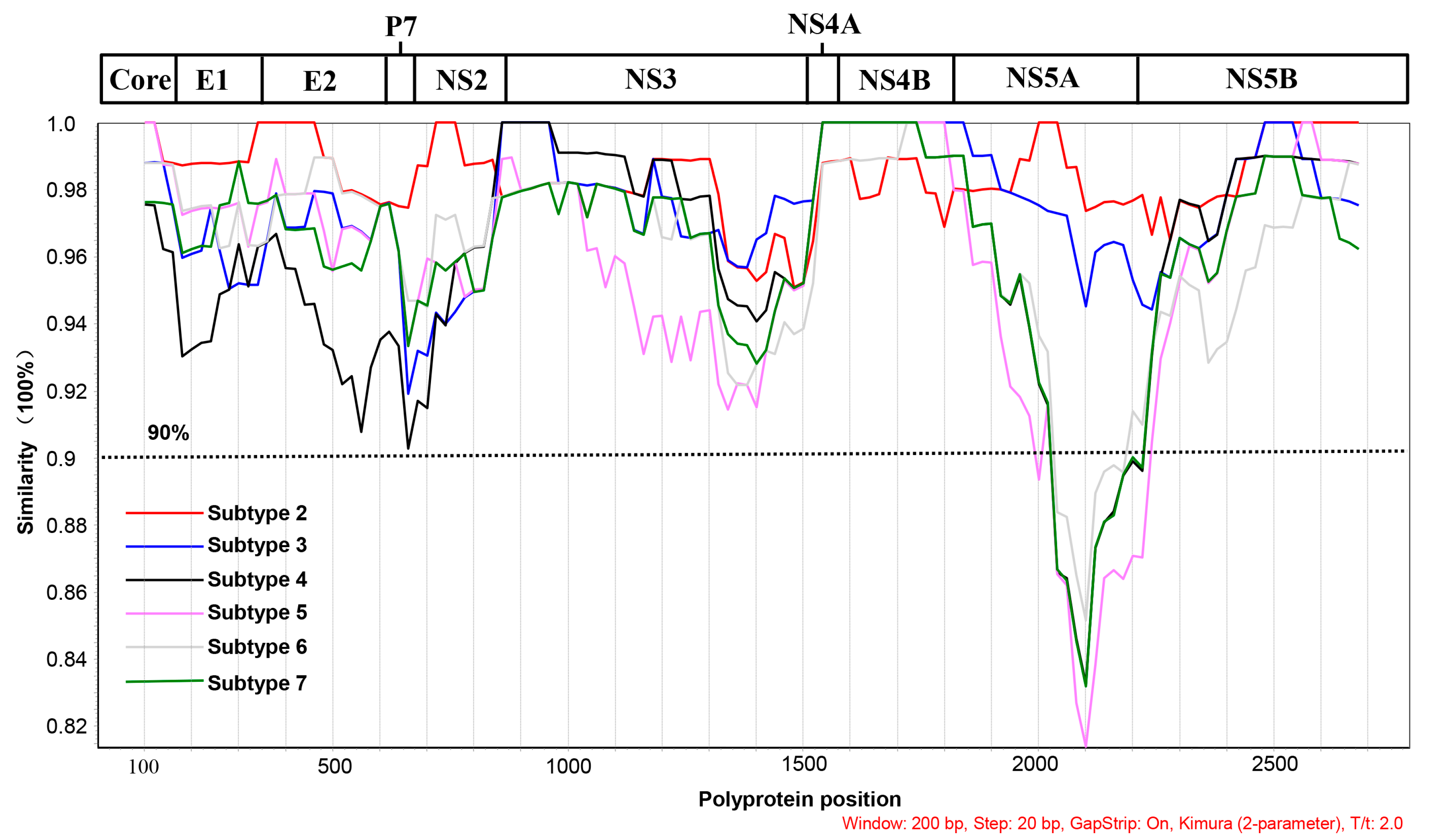
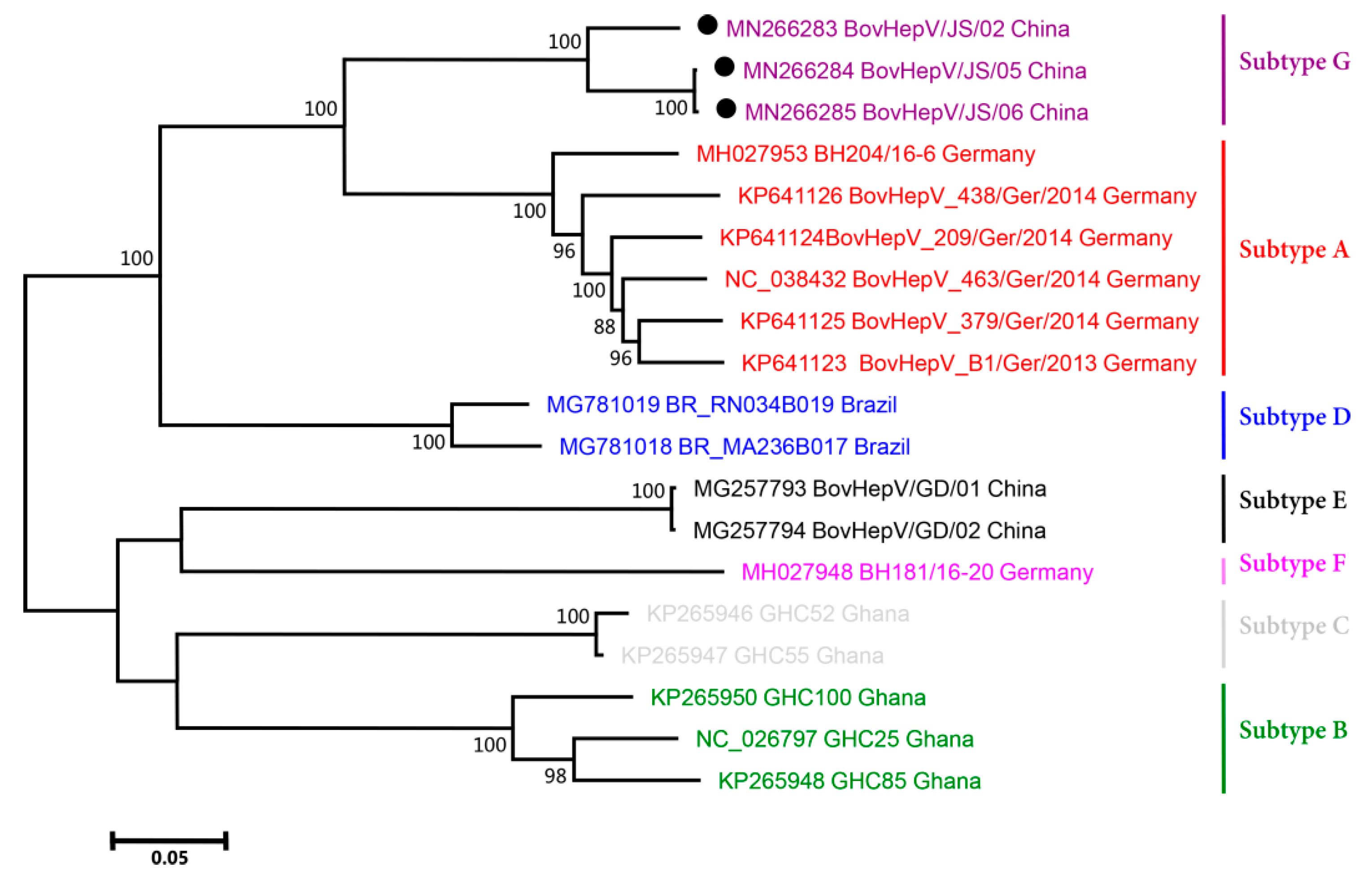
3.3. Phylogenetic Analysis
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Thomas, D.L. Global control of hepatitis C: Where challenge meets opportunity. Nat. Med. 2013, 19, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, A.; Simmonds, P.; Gerold, G.; Qaisar, N.; Jain, K.; Henriquez, J.A.; Firth, C.; Hirschberg, D.L.; Rice, C.M.; Shields, S.; et al. Characterization of a canine homolog of hepatitis C virus. Proc. Natl. Acad. Sci. USA 2011, 108, 11608–11613. [Google Scholar] [CrossRef] [PubMed]
- Burbelo, P.D.; Dubovi, E.J.; Simmonds, P.; Medina, J.L.; Henriquez, J.A.; Mishra, N.; Wagner, J.; Tokarz, R.; Cullen, J.M.; Iadarola, M.J.; et al. Serology-enabled discovery of genetically diverse hepaciviruses in a new host. J. Virol. 2012, 86, 6171–6178. [Google Scholar] [CrossRef] [PubMed]
- Quan, P.L.; Firth, C.; Conte, J.M.; Williams, S.H.; Zambrana-Torrelio, C.M.; Anthony, S.J.; Ellison, J.A.; Gilbert, A.T.; Kuzmin, I.V.; Niezgoda, M.; et al. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc. Natl. Acad. Sci. USA 2013, 110, 8194–8199. [Google Scholar] [CrossRef] [PubMed]
- Drexler, J.F.; Corman, V.M.; Muller, M.A.; Lukashev, A.N.; Gmyl, A.; Coutard, B.; Adam, A.; Ritz, D.; Leijten, L.M.; van Riel, D.; et al. Evidence for novel hepaciviruses in rodents. PLoS Pathog. 2013, 9, e1003438. [Google Scholar] [CrossRef] [PubMed]
- Baechlein, C.; Fischer, N.; Grundhoff, A.; Alawi, M.; Indenbirken, D.; Postel, A.; Baron, A.L.; Offinger, J.; Becker, K.; Beineke, A.; et al. Identification of a novel hepacivirus in domestic cattle from Germany. J. Virol. 2015, 89, 7007–7015. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Grundhoff, A.; Baechlein, C.; Fischer, N.; Gmyl, A.; Wollny, R.; Dei, D.; Ritz, D.; Binger, T.; Adankwah, E.; et al. Highly divergent hepaciviruses from African cattle. J. Virol. 2015, 89, 5876–5882. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lauck, M.; Sibley, S.D.; Lara, J.; Purdy, M.A.; Khudyakov, Y.; Hyeroba, D.; Tumukunde, A.; Weny, G.; Switzer, W.M.; Chapman, C.A.; et al. A novel hepacivirus with an unusually long and intrinsically disordered NS5A protein in a wild Old World primate. J. Virol. 2013, 87, 8971–8981. [Google Scholar] [CrossRef]
- Guo, H.; Cai, C.; Wang, B.; Zhuo, F.; Jiang, R.; Wang, N.; Li, B.; Zhang, W.; Zhu, Y.; Fan, Y.; et al. Novel hepacivirus in Asian house shrew, China. Sci. China Life Sci. 2019, 62, 701–704. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.D.; Vasilakis, N.; Tian, J.H.; Li, C.X.; Chen, L.J.; Eastwood, G.; Diao, X.N.; Chen, M.H.; Chen, X.; et al. Divergent viruses discovered in arthropods and vertebrates revise the evolutionary history of the Flaviviridae and related viruses. J. Virol. 2016, 90, 659–669. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Lin, X.D.; Chen, X.; Tian, J.H.; Chen, L.J.; Li, K.; Wang, W.; Eden, J.S.; Shen, J.J.; Liu, L.; et al. The evolutionary history of vertebrate RNA viruses. Nature 2018, 556, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.B.; Becher, P.; Bukh, J.; Gould, E.A.; Meyers, G.; Monath, T.; Muerhoff, A.S.; Pletnev, A.; Rico-Hesse, R.; Stapleton, J.T.; et al. Proposed update to the taxonomy of the genera Hepacivirus and Pegivirus within the Flaviviridae family. J. Gen. Virol. 2016, 97, 2894–2907. [Google Scholar] [CrossRef] [PubMed]
- Canal, C.W.; Weber, M.N.; Cibulski, S.P.; Silva, M.S.; Puhl, D.E.; Stalder, H.; Peterhans, E. A novel genetic group of bovine hepacivirus in archival serum samples from Brazilian cattle. Biomed. Res. Int. 2017, 2017, 4732520. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, M.S.; Junqueira, D.M.; Baumbach, L.F.; Cibulski, S.P.; Mosena, A.C.S.; Weber, M.N.; Silveira, S.; de Moraes, G.M.; Maia, R.D.; Coimbra, V.C.S.; et al. Comprehensive evolutionary and phylogenetic analysis of Hepacivirus N (HNV). J. Gen. Virol. 2018, 99, 890–896. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Jia, K.; Ping, X.; Huang, J.; Luo, A.; Wu, P.; Li, S. Novel bovine hepacivirus in dairy cattle, China. Emerg. Microbes Infect. 2018, 7, 54. [Google Scholar] [CrossRef]
- Deng, Y.; Guan, S.H.; Wang, S.; Hao, G.; Rasmussen, T.B. The detection and phylogenetic analysis of bovine hepacivirus in China. Biomed. Res. Int. 2018, 2018, 6216853. [Google Scholar] [CrossRef]
- Sadeghi, M.; Kapusinszky, B.; Yugo, D.M.; Phan, T.G.; Deng, X.; Kanevsky, I.; Opriessnig, T.; Woolums, A.R.; Hurley, D.J.; Meng, X.J.; et al. Virome of US bovine calf serum. Biol. J. Int. Assoc. Biol. Stand. 2017, 46, 64–67. [Google Scholar] [CrossRef]
- Yesilbag, K.; Baechlein, C.; Kadiroglu, B.; Baldan Toker, E.; Alpay, G.; Becher, P. Presence of bovine hepacivirus in Turkish cattle. Vet. Microbiol. 2018, 225, 1–5. [Google Scholar] [CrossRef]
- Baechlein, C.; Baron, A.L.; Meyer, D.; Gorriz-Martin, L.; Pfankuche, V.M.; Baumgartner, W.; Polywka, S.; Peine, S.; Fischer, N.; Rehage, J.; et al. Further characterization of bovine hepacivirus: Antibody response, course of infection, and host tropism. Transbound. Emerg. Dis. 2019, 66, 195–206. [Google Scholar] [CrossRef]
- Smith, D.B.; Bukh, J.; Kuiken, C.; Muerhoff, A.S.; Rice, C.M.; Stapleton, J.T.; Simmonds, P. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: Updated criteria and genotype assignment web resource. Hepatology 2014, 59, 318–327. [Google Scholar] [CrossRef]
- Schlottau, K.; Wernike, K.; Forth, L.; Holsteg, M.; Hoper, D.; Beer, M.; Hoffmann, B. Presence of two different bovine hepacivirus clusters in Germany. Transbound. Emerg. Dis. 2018, 65, 1705–1711. [Google Scholar] [CrossRef] [PubMed]
- The NCBI Website. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 8 August 2019).
- Toohey-Kurth, K.; Sibley, S.D.; Goldberg, T.L. Metagenomic assessment of adventitious viruses in commercial bovine sera. Biol. J. Int. Assoc. Biol. Stand. 2017, 47, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Ou, J.; Sun, Y.; Wu, L.; Xu, H.; Zhang, G.; Li, S. Natural recombination of equine hepacivirus subtype 1 within the NS5A and NS5B genes. Virology 2019, 533, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, S.; Nichols, A.K.; Saravanabalaji, D.; Welsch, C.; Yi, M. HCV NS5A dimer interface residues regulate HCV replication by controlling its self-interaction, hyperphosphorylation, subcellular localization and interaction with cyclophilin A. PLoS Pathog. 2018, 14, e1007177. [Google Scholar] [CrossRef] [PubMed]
| Serum Sample ID | Batch | Origin Country/Continent |
|---|---|---|
| A | Uruguay/America | |
| B | France/Europe | |
| C | New Zealand/Oceania | |
| D | Brazil/America | |
| E | Brazil/America | |
| F | Uruguay/America | |
| G | Israel/Asia | |
| H | New Zealand/Oceania | |
| I | Australia/Oceania | |
| J | batch 1 | China/Asia |
| batch 2 | ||
| batch 3 | ||
| K | batch 1 | China/Asia |
| batch 2 | ||
| batch 3 |
| Primer Sequence (5′→3′) | Targeted Genomic Region § | Product Length (bp) |
|---|---|---|
| TAGTAGGAGGCGCCTATCCC; TTNACCTTRTAYTCRCACCC | 64–1583 | 1520 |
| CTCCGTGGGTGCGAATACAA; TACAGCGYTGVACCATRGCR | 1558–3394 | 1837 |
| ACTTGGAYGTCTGCACTTGT; ACCAHGCCATACCRCTGTC | 3200–4423 | 1204 |
| GGGTGCTCCTGGRGTTTAC; CGAGCGATCCGACARTCRGT | 4320–6321 | 2002 |
| CCGCTCCRTGACTACGAGAC; CGAGACGGTATCCACAGCYC | 6283–8739 | 2457 |
| A | B | C | D | E | F | G | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KP641123 | KP641125 | KP641127 | KP641124 | KP641126 | MH027953 | KP265950 | KP265943 | KP265948 | KP265946 | KP265947 | MG781018 | MG781019 | MG257793 | MG257794 | MH027948 | MN266283 | MN266284 | MN266285 | |
| KP641123 BovHepV/B1/2013 | 93.8 | 93.6 | 93.3 | 91.0 | 90.8 | 80.1 | 79.9 | 79.8 | 80.3 | 80.4 | 82.4 | 82.5 | 79.5 | 79.5 | 80.1 | 84.4 | 84.5 | 84.5 | |
| KP641125 BovHepV/379/2014 | 98.2 | 93.6 | 93.1 | 91.4 | 91.0 | 80.2 | 80.0 | 79.8 | 80.3 | 80.5 | 82.5 | 82.8 | 79.5 | 79.5 | 80.1 | 84.3 | 84.1 | 84.0 | |
| KP641127 BovHepV/463/2014 | 98.0 | 97.9 | 93.5 | 91.7 | 90.9 | 80.0 | 80.2 | 79.9 | 80.3 | 80.6 | 82.5 | 82.9 | 79.6 | 79.6 | 80.3 | 84.4 | 84.5 | 84.5 | |
| KP641124 BovHepV/209/2014 | 97.9 | 98.1 | 98.0 | 91.6 | 90.9 | 79.9 | 80.0 | 79.8 | 80.5 | 80.7 | 82.5 | 82.5 | 79.5 | 79.5 | 80.1 | 84.5 | 84.5 | 84.5 | |
| KP641126 BovHepV/438/2014 | 97.5 | 97.4 | 97.4 | 97.4 | 90.5 | 79.6 | 80.1 | 79.7 | 80.2 | 80.3 | 82.2 | 82.3 | 78.9 | 78.9 | 79.6 | 84.3 | 83.9 | 83.9 | |
| MH027953 BH204/16-6 | 97.3 | 97.2 | 97.6 | 97.3 | 97.1 | 79.5 | 79.8 | 79.7 | 79.8 | 80.1 | 82.3 | 82.5 | 79.5 | 79.5 | 79.6 | 84.0 | 83.9 | 83.8 | |
| KP265950 GHC100 | 92.9 | 93.3 | 93.0 | 93.1 | 92.7 | 93.0 | 90.9 | 90.4 | 82.7 | 82.8 | 80.7 | 80.5 | 82.0 | 82.0 | 81.6 | 80.2 | 80.0 | 79.9 | |
| KP265943 GHC25 | 93.0 | 93.1 | 93.1 | 93.0 | 92.8 | 93.0 | 98.6 | 92.0 | 82.7 | 82.9 | 80.4 | 80.7 | 81.7 | 81.7 | 81.4 | 80.3 | 80.0 | 79.9 | |
| KP265948 GHC85 | 93.1 | 93.4 | 93.2 | 93.3 | 92.8 | 93.1 | 98.7 | 99.0 | 82.4 | 82.7 | 80.2 | 80.5 | 81.7 | 81.7 | 81.1 | 80.0 | 79.8 | 79.8 | |
| KP265946 GHC52 | 92.3 | 92.5 | 92.3 | 92.4 | 92.3 | 92.0 | 95.5 | 95.6 | 95.5 | 98.4 | 80.9 | 80.9 | 81.5 | 81.4 | 81.3 | 79.8 | 79.9 | 79.9 | |
| KP265947 GHC55 | 93.0 | 93.2 | 93.1 | 93.1 | 92.9 | 92.7 | 96.2 | 96.3 | 96.3 | 98.8 | 81.1 | 81.1 | 81.6 | 81.6 | 81.5 | 80.0 | 80.1 | 80.1 | |
| MG781018 BR_MA236B017 | 93.8 | 94.4 | 93.9 | 94.1 | 93.9 | 94.3 | 93.5 | 93.1 | 93.3 | 92.8 | 93.4 | 93.9 | 80.2 | 80.2 | 80.0 | 82.2 | 82.1 | 82.1 | |
| MG781019 BR_RN034B019 | 93.6 | 94.2 | 93.7 | 93.9 | 93.8 | 94.0 | 93.5 | 93.2 | 93.4 | 92.5 | 93.2 | 98.1 | 80.7 | 80.7 | 80.0 | 82.4 | 82.4 | 82.4 | |
| MG257793 BovHepV/GD/01 | 92.1 | 92.2 | 92.1 | 92.1 | 91.7 | 92.0 | 95.4 | 95.3 | 95.2 | 94.4 | 94.8 | 92.7 | 92.6 | 99.8 | 81.0 | 79.8 | 79.3 | 79.3 | |
| MG257794 BovHepV/GD/02 | 92.1 | 92.3 | 92.2 | 92.1 | 91.7 | 92.1 | 95.4 | 95.3 | 95.3 | 94.5 | 94.9 | 92.8 | 92.6 | 100.0 | 81.0 | 79.8 | 79.2 | 79.2 | |
| MH027948 BH181/16-20 | 92.0 | 92.3 | 92.2 | 92.1 | 92.0 | 92.2 | 95.2 | 94.8 | 94.8 | 94.4 | 95.0 | 92.6 | 92.9 | 94.3 | 94.4 | 79.4 | 79.7 | 79.7 | |
| MN266283 BovHepV/JS/02 | 95.3 | 95.6 | 95.7 | 95.6 | 95.6 | 95.8 | 93.4 | 93.2 | 93.4 | 92.7 | 93.2 | 94.7 | 94.7 | 92.5 | 92.5 | 92.5 | 92.8 | 92.7 | |
| MN266284 BovHepV/JS/05 | 95.4 | 95.6 | 95.6 | 95.6 | 95.4 | 95.6 | 92.9 | 92.8 | 93.1 | 92.4 | 92.9 | 94.5 | 94.4 | 92.3 | 92.3 | 92.2 | 98.0 | 99.9 | |
| MN266285 BovHepV/JS/06 | 95.3 | 95.5 | 95.6 | 95.5 | 95.3 | 95.5 | 92.8 | 92.8 | 93.0 | 92.4 | 92.9 | 94.4 | 94.3 | 92.3 | 92.3 | 92.1 | 97.9 | 99.8 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, G.; Ou, J.; Zhao, J.; Li, S. Presence of a Novel Subtype of Bovine Hepacivirus in China and Expanded Classification of Bovine Hepacivirus Strains Worldwide into 7 Subtypes. Viruses 2019, 11, 843. https://doi.org/10.3390/v11090843
Lu G, Ou J, Zhao J, Li S. Presence of a Novel Subtype of Bovine Hepacivirus in China and Expanded Classification of Bovine Hepacivirus Strains Worldwide into 7 Subtypes. Viruses. 2019; 11(9):843. https://doi.org/10.3390/v11090843
Chicago/Turabian StyleLu, Gang, Jiajun Ou, Jiawei Zhao, and Shoujun Li. 2019. "Presence of a Novel Subtype of Bovine Hepacivirus in China and Expanded Classification of Bovine Hepacivirus Strains Worldwide into 7 Subtypes" Viruses 11, no. 9: 843. https://doi.org/10.3390/v11090843
APA StyleLu, G., Ou, J., Zhao, J., & Li, S. (2019). Presence of a Novel Subtype of Bovine Hepacivirus in China and Expanded Classification of Bovine Hepacivirus Strains Worldwide into 7 Subtypes. Viruses, 11(9), 843. https://doi.org/10.3390/v11090843





