Comparison of Transcriptome Differences in Soybean Response to Soybean Mosaic Virus under Normal Light and in the Shade
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material, Virus Inoculation, and Light Treatment
2.2. Sample Collection and Illumina Sequencing
2.3. Read Alignment and Expression Analysis
2.4. Functional Enrichment Analysis of DEGs
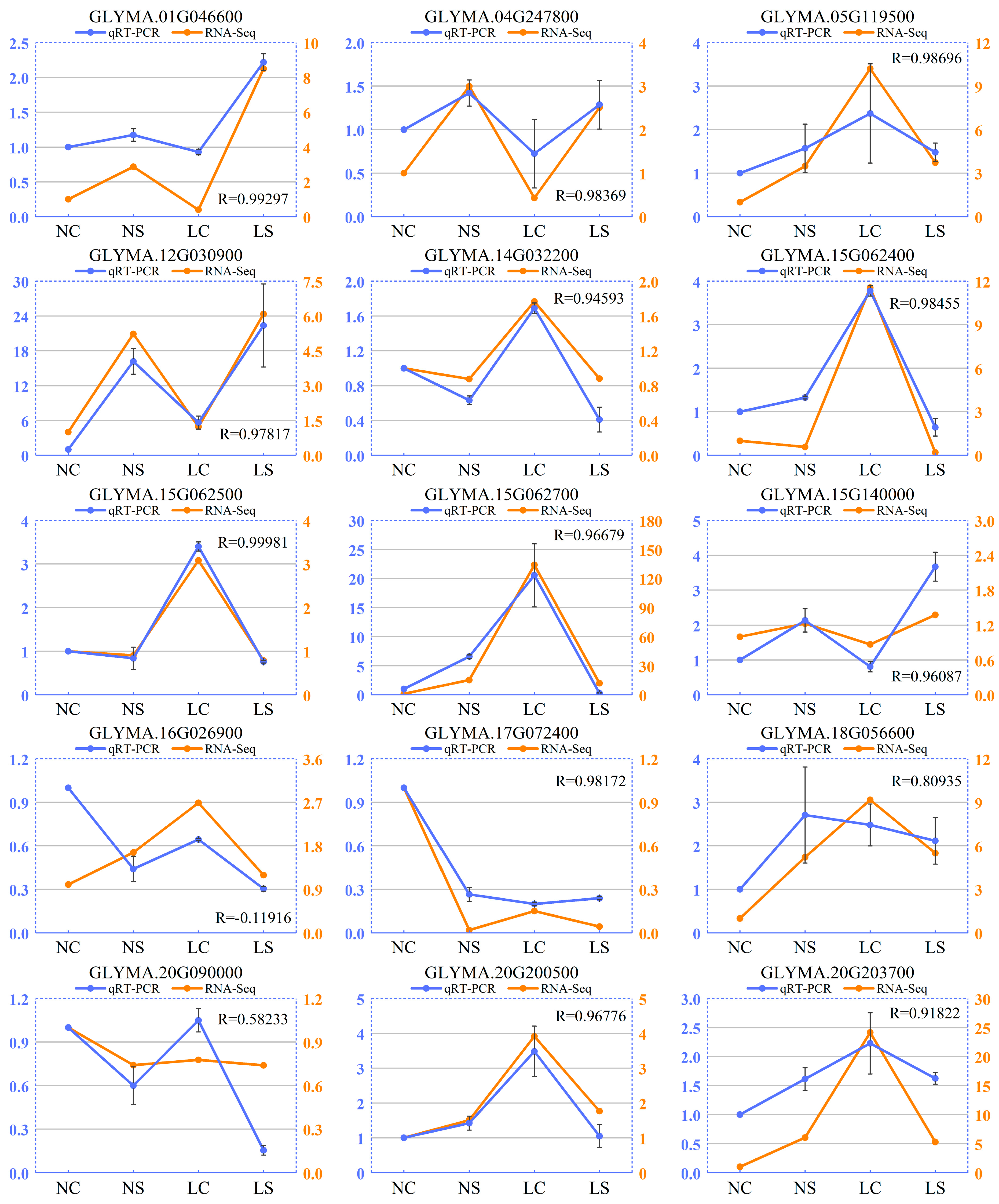
2.5. Validation of Gene Expression by qRT-PCR
3. Results and Discussions
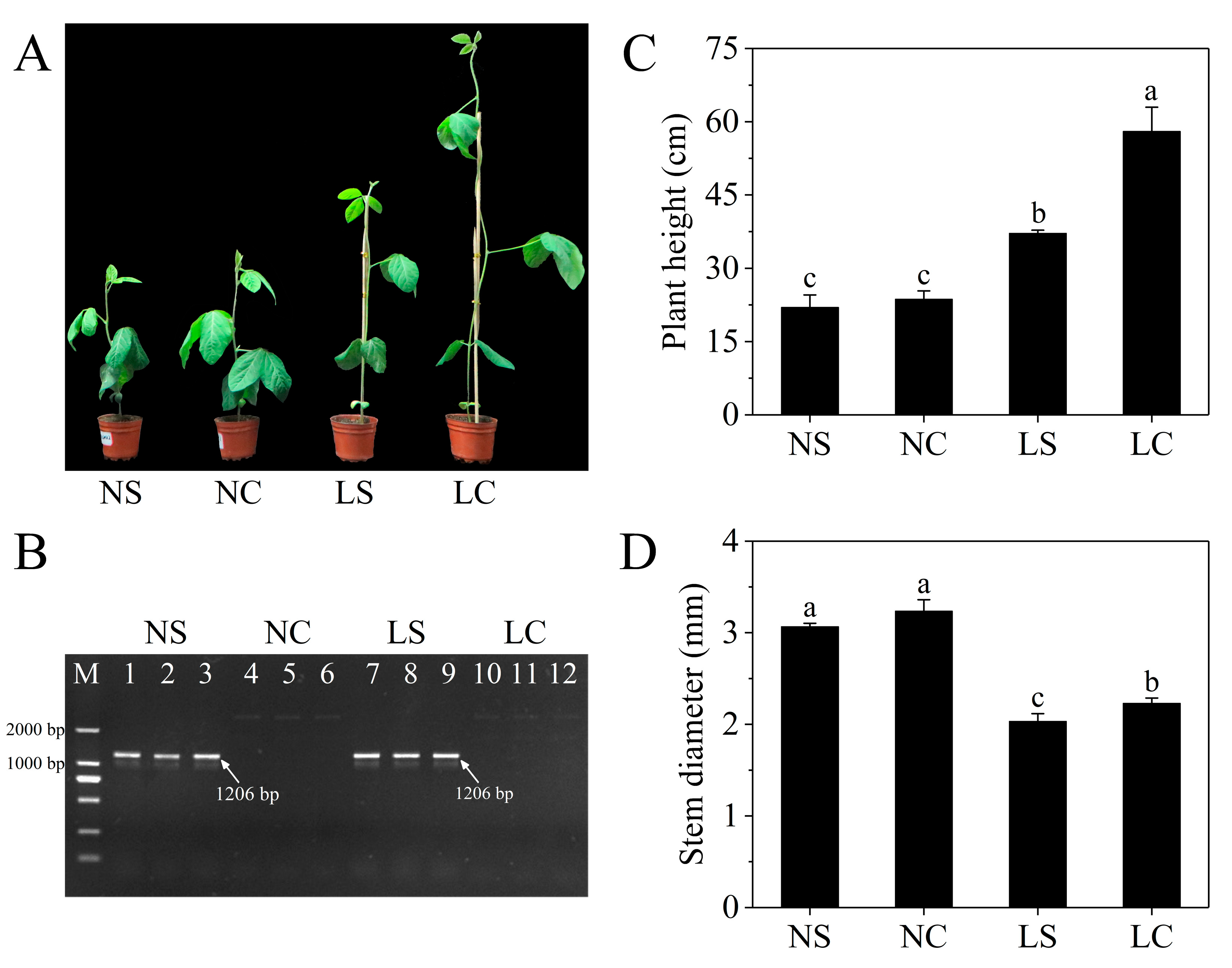
3.1. Plant Phenotypes and Virus Detection after Inoculation
3.2. Evaluation of RNA-Seq Data
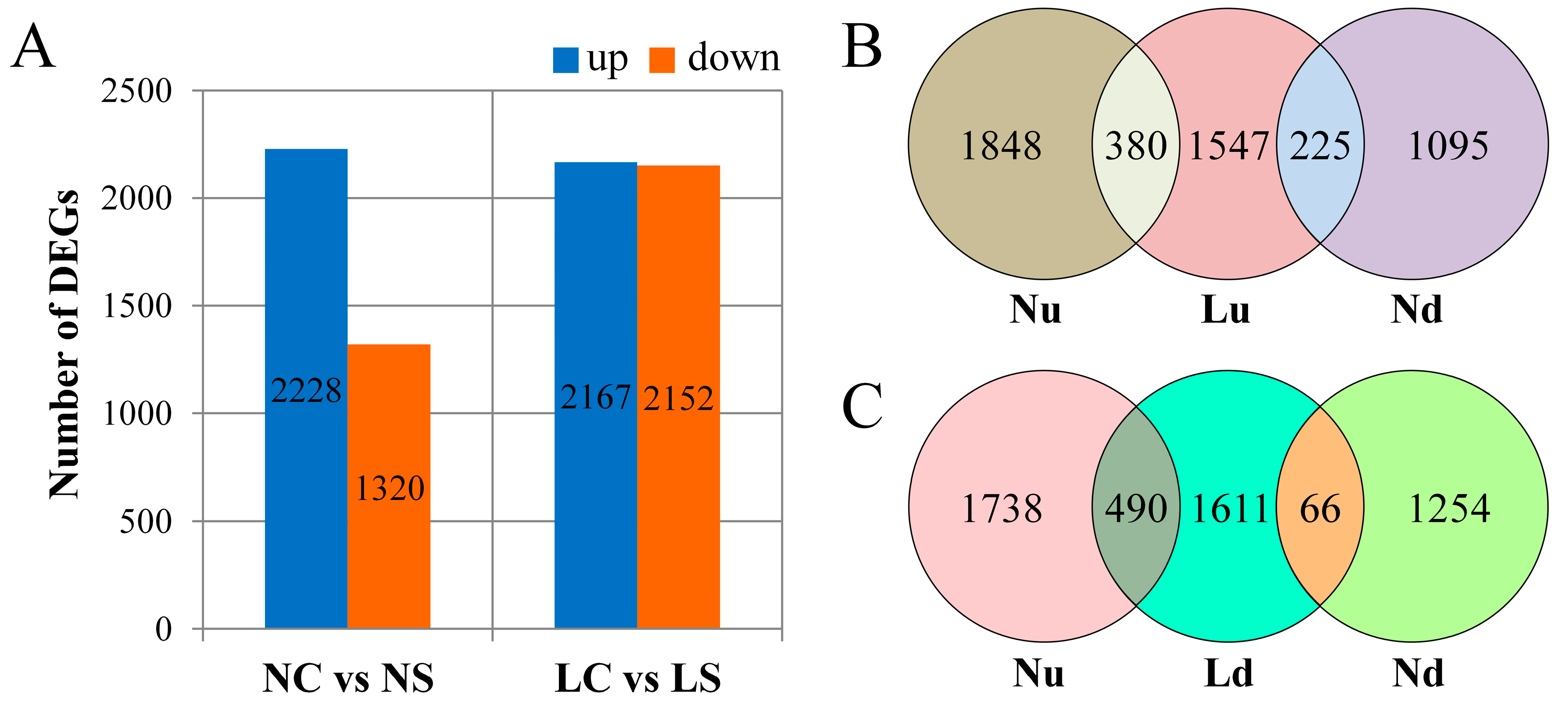
3.3. Statistics on the Number of DEGs
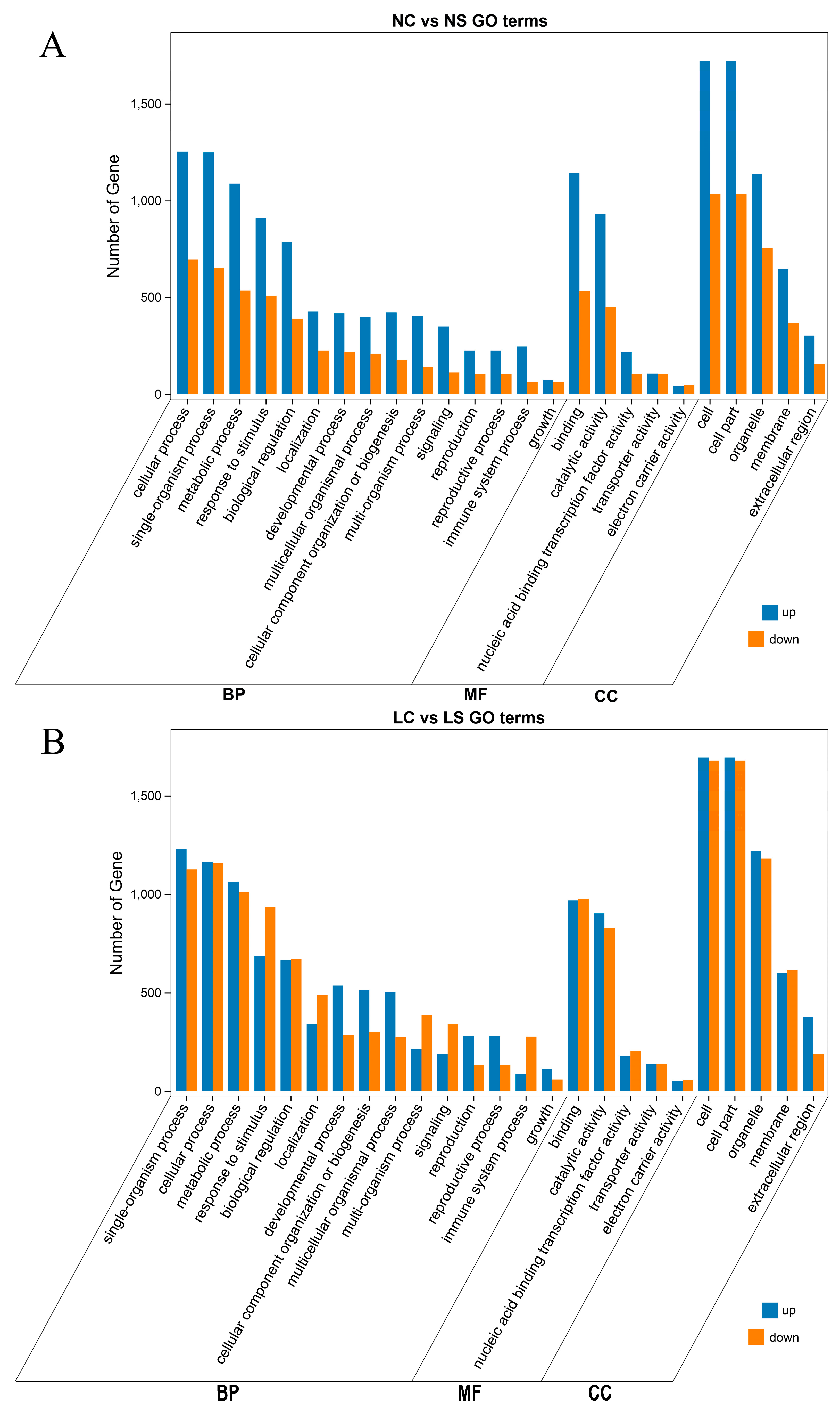
3.4. GO Function Enrichment Analysis of DEGs
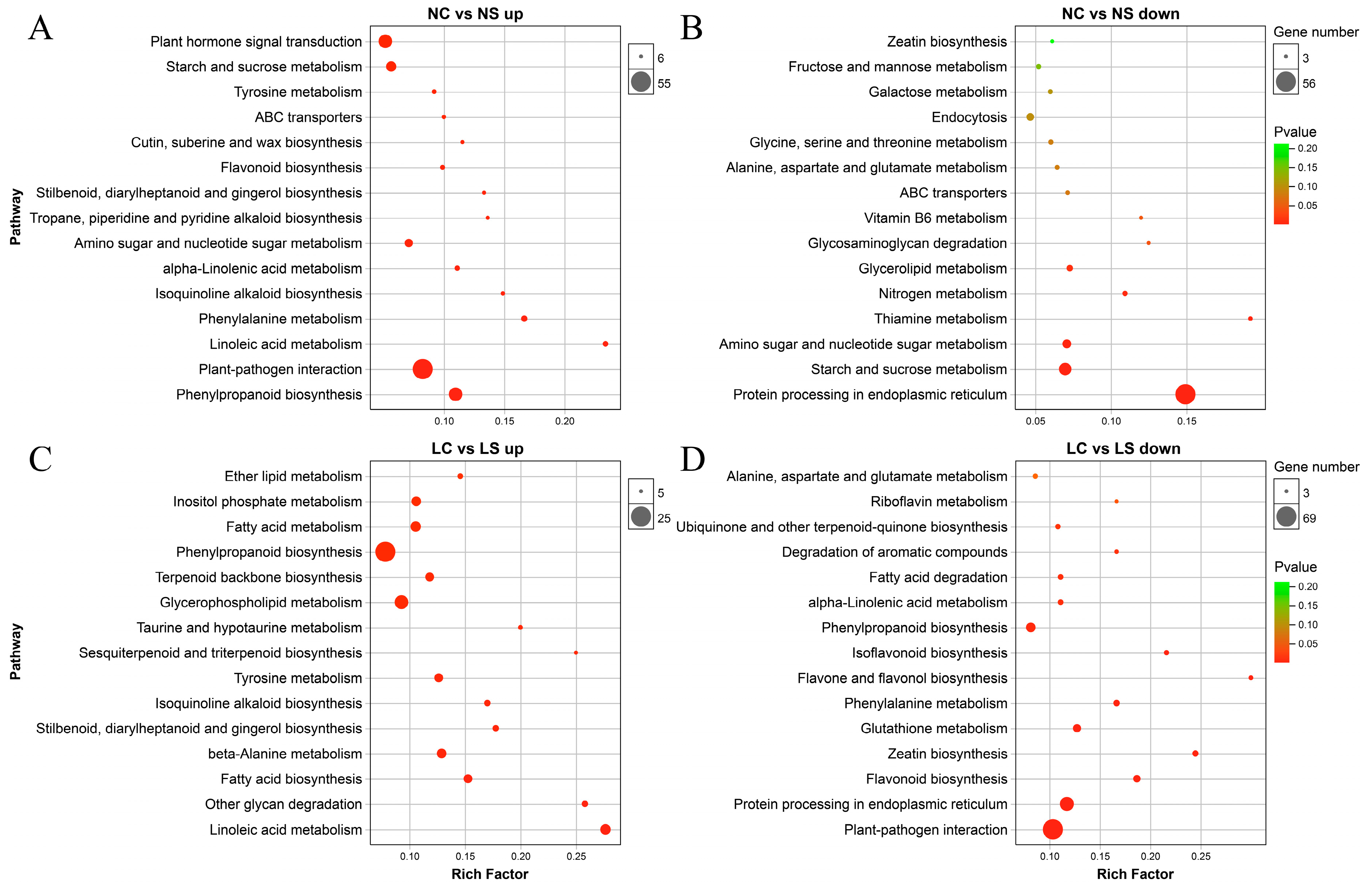
3.5. KEGG Pathway Enrichment Analysis of DEGs
3.6. DEGs Involved in Plant-Pathogen Interaction
3.7. DEGs Involved in Plant Hormone Signal Transduction
3.8. Validation of RNA-Seq Data by qRT-PCR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CaM | calmodulin |
| ChiVMV | chilli veinal mottle virus |
| CK | control |
| CMV | cucumber mosaic virus |
| DEGs | differentially expressed genes |
| DPI | days post-inoculation |
| EDS1 | enhanced disease susceptibility 1 |
| ERF | ethylene-responsive transcription factor |
| ETH | ethylene |
| GO | Gene Ontology |
| JA | jasmonic acid |
| JAZ | jasmonate ZIM-domain |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| log2FC | log2Fold Change |
| LC | Control in the shade |
| LS | SMV infection in the shade |
| NC | Control under normal light |
| NS | SMV infection under normal light |
| PAR | photosynthetically active radiation |
| phyB | phytochrome B |
| PR1 | pathogenesis-related 1 |
| qRT-PCR | quantitative real-time PCR |
| R:FR | red:far red-light ratio |
| RPKM | reads per kb per million reads |
| SA | salicylic acid |
| SAS | shade avoidance syndrome |
| SMV | soybean mosaic virus |
| TF | transcription factor |
| TuMV | turnip mosaic virus |
References
- Chen, P.; Song, C.; Liu, X.; Zhou, L.; Yang, H.; Zhang, X.; Zhou, Y.; Du, Q.; Pang, T.; Fu, Z.; et al. Yield advantage and nitrogen fate in an additive maize-soybean relay intercropping system. Sci. Total Environ. 2019, 657, 987–999. [Google Scholar] [CrossRef]
- Du, J.; Han, T.; Gai, J.; Yong, T.; Sun, X.; Wang, X.; Yang, F.; Liu, J.; Shu, K.; Liu, W.; et al. Maize-soybean strip intercropping: Achieved a balance between high productivity and sustainability. J. Integr. Agr. 2018, 17, 747–754. [Google Scholar] [CrossRef]
- Yang, F.; Feng, L.; Liu, Q.; Wu, X.; Fan, Y.; Raza, M.A.; Cheng, Y.; Chen, J.; Wang, X.; Yong, T.; et al. Effect of interactions between light intensity and red-to- far-red ratio on the photosynthesis of soybean leaves under shade condition. Environ. Exp. Bot. 2018, 150, 79–87. [Google Scholar] [CrossRef]
- Wu, Y.; Gong, W.; Yang, W. Shade inhibits leaf size by controlling cell proliferation and enlargement in soybean. Sci. Rep. 2017, 7, 9259. [Google Scholar] [CrossRef]
- Hill, J.H.; Whitham, S.A. Control of virus diseases in soybeans. Adv. Virus Res. 2014, 90, 355–390. [Google Scholar]
- Hajimorad, M.R.; Domier, L.L.; Tolin, S.A.; Whitham, S.A.; Saghai Maroof, M.A. Soybean mosaic virus: a successful potyvirus with a wide distribution but restricted natural host range. Mol. Plant Pathol. 2018, 19, 1563–1579. [Google Scholar] [CrossRef]
- Chung, B.Y.W.; Miller, W.A.; Atkins, J.F.; Firth, A.E. An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. USA. 2008, 105, 5897–5902. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, X.; Hong, Y.; Liu, Y. Chloroplast in plant-virus interaction. Front. Microbiol. 2016, 7, 1565. [Google Scholar] [CrossRef]
- Lu, J.; Du, Z.X.; Kong, J.; Chen, L.N.; Qiu, Y.H.; Li, G.F.; Meng, X.H.; Zhu, S.F. Transcriptome analysis of Nicotiana tabacum infected by Cucumber mosaic virus during systemic symptom development. PLoS ONE 2012, 7, e43447. [Google Scholar] [CrossRef]
- Li, K.; Wu, G.; Li, M.; Ma, M.; Du, J.; Sun, M.; Sun, X.; Qing, L. Transcriptome analysis of Nicotiana benthamiana infected by Tobacco curly shoot virus. Virol. J. 2018, 15, 138. [Google Scholar] [CrossRef]
- Li, X.; An, M.; Xia, Z.; Bai, X.; Wu, Y. Transcriptome analysis of watermelon (Citrullus lanatus) fruits in response to Cucumber green mottle mosaic virus (CGMMV) infection. Sci. Rep. 2017, 7, 16747. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.; Singh, J.; Hill, J.H.; Whitham, S.A.; Cannon, S.B. Dynamic transcriptome profiling of Bean Common Mosaic Virus (BCMV) infection in common bean (Phaseolus vulgaris L.). BMC Genom. 2016, 17, 613. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Fan, M.; He, Y. Transcriptome analysis of watermelon leaves reveals candidate genes responsive to Cucumber green mottle mosaic virus infection. Int. J. Mol. Sci. 2019, 20, 610. [Google Scholar] [CrossRef] [PubMed]
- Babu, M.; Gagarinova, A.G.; Brandle, J.E.; Wang, A. Association of the transcriptional response of soybean plants with soybean mosaic virus systemic infection. J. Gen. Virol. 2008, 89, 1069–1080. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Li, H.; Sun, H.; Li, A.; Liu, S.; Yu, R.; Cui, X.; Zhang, D.; Wuriyanghan, H. Salicylic acid and broad spectrum of NBS-LRR family genes are involved in SMV-soybean interactions. Plant Physiol. Bioch. 2018, 123, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Cruz, G.A.; Cassone, B.J. A tale of survival: Molecular defense mechanisms of soybean to overcome Soybean mosaic virus infection. Physiol. Mol. Plant Pathol. 2018, 102, 79–87. [Google Scholar] [CrossRef]
- Yang, F.; Huang, S.; Gao, R.; Liu, W.; Yong, T.; Wang, X.; Wu, X.; Yang, W. Growth of soybean seedlings in relay strip intercropping systems in relation to light quantity and red:far-red ratio. Field Crop. Res. 2014, 155, 245–253. [Google Scholar] [CrossRef]
- Smith, H. Physiological and ecological function within the phytochrome family. Annu. Rev. Plant Physiol. Plant Mol. Biol 1995, 46, 289–315. [Google Scholar] [CrossRef]
- Quail, P.H. Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Bio. 2002, 3, 85–93. [Google Scholar] [CrossRef]
- Ballare, C.L.; Pierik, R. The shade-avoidance syndrome: multiple signals and ecological consequences. Plant Cell Environ. 2017, 40, 2530–2543. [Google Scholar] [CrossRef]
- Ballaré, C.L. Light regulation of plant defense. Annu. Rev. Plant Biol. 2014, 65, 335–363. [Google Scholar] [CrossRef] [PubMed]
- Chico, J.M.; Fernandez-Barbero, G.; Chini, A.; Fernandez-Calvo, P.; Diez-Diaz, M.; Solano, R. Repression of jasmonate-dependent defenses by shade involves differential regulation of protein stability of MYC transcription factors and their JAZ repressors in Arabidopsis. Plant Cell 2014, 26, 1967–1980. [Google Scholar] [CrossRef] [PubMed]
- Cerrudo, I.; Keller, M.M.; Cargnel, M.D.; Demkura, P.V.; de Wit, M.; Patitucci, M.S.; Pierik, R.; Pieterse, C.M.J.; Ballare, C.L. Low red/far-red ratios reduce Arabidopsis resistance to Botrytis cinerea and jasmonate responses via a COI1-JAZ10-dependent, salicylic acid-independent mechanism. Plant Physiol. 2012, 158, 2042–2052. [Google Scholar] [CrossRef] [PubMed]
- Manfre, A.; Glenn, M.; Nuñez, A.; Moreau, R.A.; Dardick, C. Light quantity and photosystem function mediate host susceptibility to Turnip mosaic virus via a salicylic acid-independent mechanism. Mol. Plant-Microbe Interact. 2011, 24, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Xue, Y.; Zhou, J.; Zhang, B.; Chang, H.; Takano, M. Phytochromes regulate SA and JA signaling pathways in rice and are required for developmentally controlled resistance to Magnaporthe grisea. Mol. Plant 2011, 4, 688–696. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.J.; Fei, C.Y.; Xu, Z.P.; Wu, G.; Lin, H.H.; Xi, D.H. Positive role of phytochromes in Nicotiana tabacum against Cucumber mosaic virus via a salicylic acid-dependent pathway. Plant Pathol. 2018, 67, 488–498. [Google Scholar] [CrossRef]
- Fei, C.; Chen, L.; Yang, T.; Zou, W.; Lin, H.; Xi, D. The role of phytochromes in Nicotiana tabacum against Chilli veinal mottle virus. Plant Physiol. Bioch. 2019, 139, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Shang, J.; Jia, Q.; Li, K.; Yang, H.; Liu, H.; Tang, Z.; Chang, X.; Zhang, M.; Wang, W.; et al. Genetic evolutionary analysis of soybean mosaic virus populations from three geographic locations in China based on the P1 and CP genes. Arch. Virol. 2019, 164, 1037–1048. [Google Scholar] [CrossRef] [PubMed]
- Ballaré, C.L.; Austin, A.T. Recalculating growth and defense strategies under competition: key roles of photoreceptors and jasmonates. J. Exp. Bot. 2019. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Gao, F.; Cao, X.; Chen, M.; Ye, G.; Wei, C.; Li, Y. The rice dwarf virus P2 protein interacts with ent-kaurene oxidases in vivo, leading to reduced biosynthesis of gibberellins and rice dwarf symptoms. Plant Physiol. 2005, 139, 1935–1945. [Google Scholar] [CrossRef]
- Sun, F.; Fang, P.; Li, J.; Du, L.; Lan, Y.; Zhou, T.; Fan, Y.; Shen, W.; Zhou, Y. RNA-seq-based digital gene expression analysis reveals modification of host defense responses by rice stripe virus during disease symptom development in Arabidopsis. Virol. J. 2016, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Lozovaya, V.V.; Lygin, A.V.; Zernova, O.V.; Li, S.; Hartman, G.L.; Widholm, J.M. Isoflavonoid accumulation in soybean hairy roots upon treatment with Fusarium solani. Plant Physiol. Bioch. 2004, 42, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Morkunas, I.; Marczak, A.; Stachowiak, J.; Stobiecki, M. Sucrose-induced lupine defense against Fusarium oxysporum. Plant Physiol. Bioch. 2005, 43, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhao, L.; Yan, B.; Zhu, Y.; Ma, H.; Chen, W.; Ruan, S. Comparative transcriptome analysis reveals the transcriptional alterations in growth- and development-related genes in sweet potato plants infected and non-infected by SPFMV, SPV2, and SPVG. Int. J. Mol. Sci. 2019, 20, 1012. [Google Scholar] [CrossRef] [PubMed]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Liu, Y.; Schiff, M.; Dinesh-Kumar, S.P. Involvement of MEK1 MAPKK, NTF6 MAPK, WRKY/MYB transcription factors, COI1 and CTR1 in N-mediated resistance to tobacco mosaic virus. Plant J. 2004, 38, 800–809. [Google Scholar] [CrossRef]
- Pandey, S.P.; Somssich, I.E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef]
- Li, T.; Zhang, X.; Huang, Y.; Xu, Z.; Wang, F.; Xiong, A. An R2R3-MYB transcription factor, SlMYB28, involved in the regulation of TYLCV infection in tomato. Sci. Hortic. 2018, 237, 192–200. [Google Scholar] [CrossRef]
- Yamakawa, H.; Mitsuhara, I.; Ito, N.; Seo, S.; Kamada, H.; Ohashi, Y. Transcriptionally and post-transcriptionally regulated response of 13 calmodulin genes to tobacco mosaic virus-induced cell death and wounding in tobacco plant. Eur. J. Biochem. 2001, 268, 3916–3929. [Google Scholar] [CrossRef]
- Cui, H.; Gobbato, E.; Kracher, B.; Qiu, J.; Bautor, J.; Parker, J.E. A core function of EDS1 with PAD4 is to protect the salicylic acid defense sector in Arabidopsis immunity. New Phytol. 2017, 213, 1802–1817. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Qiu, J.; Zhou, Y.; Bhandari, D.D.; Zhao, C.; Bautor, J.; Parker, J.E. Antagonism of transcription factor MYC2 by EDS1/PAD4 complexes bolsters salicylic acid defense in Arabidopsis effector-triggered immunity. Mol. Plant 2018, 11, 1053–1066. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.J.; Leon-Reyes, A.; Van der Ent, S.; Van Wees, S.C.M. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 2009, 5, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Vlot, A.C.; Dempsey, D.A.; Klessig, D.F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef] [PubMed]
- Itagaki, K.; Shibuya, T.; Tojo, M.; Endo, R.; Kitaya, Y. Early development of powdery mildew on cucumber leaves acclimatized to illumination with different red-to-far-red ratios. HortScience 2016, 51, 530–536. [Google Scholar] [CrossRef]
- Wit, M.D.; Spoel, S.H.; Sanchez-Perez, G.F.; Gommers, C.M.M.; Pieterse, C.M.J.; Voesenek, L.A.C.J.; Pierik, R. Perception of low red:far-red ratio compromises both salicylic acid- and jasmonic acid-dependent pathogen defences in Arabidopsis. Plant J. 2013, 75, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Berens, M.L.; Berry, H.M.; Mine, A.; Argueso, C.T.; Tsuda, K. Evolution of hormone signaling networks in plant defense. Annu. Rev. Phytopathol. 2017, 55, 401–425. [Google Scholar] [CrossRef]
| Gene ID | Description | Log2FC (NS/NC) | Log2FC (LS/LC) |
|---|---|---|---|
| GLYMA_05G234600 | MYB transcription factor MYB84 | 5.33 | −3.24 |
| GLYMA_18G056600 | WRKY transcription factor 62 | 5.21 | −3.66 |
| GLYMA_09G038900 | MYB transcription factor MYB13 | 5.06 | 1.74 |
| GLYMA_06G187300 | protein EDS1L | 4.01 | −2.16 |
| GLYMA_20G034200 | uncharacterized LOC100526868 | 3.94 | −2.59 |
| GLYMA_16G218300 | probable cyclic nucleotide-gated ion channel 20 | 3.83 | −1.29 |
| GLYMA_02G270700 | chitin elicitor receptor kinase 1-like | 3.76 | −1.16 |
| GLYMA_19G214900 | MYB transcription factor MYB111 | 3.20 | 2.40 |
| GLYMA_09G210600 | disease resistance protein RPM1 | 3.10 | −1.92 |
| GLYMA_06G034700 | probable calcium-binding protein CML41 | 2.90 | −2.53 |
| GLYMA_20G209700 | MYB/HD-like transcription factor | 2.81 | −3.06 |
| GLYMA_10G180800 | MYB29 protein | 2.25 | −3.85 |
| GLYMA_18G208800 | probable WRKY transcription factor 33 | 2.23 | −3.83 |
| GLYMA_03G042700 | probable WRKY transcription factor 33 | 1.99 | −2.70 |
| GLYMA_14G222000 | calcium-dependent protein kinase 29 | 1.97 | 1.79 |
| GLYMA_05G119500 | BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 | 1.80 | −1.45 |
| GLYMA_10G230000 | protein SGT1 homolog B-like | 1.73 | −1.51 |
| GLYMA_02G244600 | MYB transcription factor MYB20 | 1.65 | 1.14 |
| GLYMA_06G187400 | protein EDS1-like | 1.55 | −1.01 |
| GLYMA_19G255300 | cyclic nucleotide-gated ion channel 1 | 1.35 | −1.20 |
| GLYMA_02G059600 | putative calcium-binding protein | 1.13 | −1.11 |
| GLYMA_14G156300 | calcium-binding EF-hand family protein | −2.18 | −1.76 |
| GLYMA_16G178800 | heat shock protein 90-A2 | −5.23 | −1.33 |
| GLYMA_09G131500 | heat shock protein 83 | −7.64 | −2.66 |
| Gene ID | Description | Log2FC (NS/NC) | Log2FC (LS/LC) | Pathway |
|---|---|---|---|---|
| GLYMA_10G186800 | ethylene-responsive transcription factor 1B | 2.71 | −1.21 | ETH |
| GLYMA_04G147000 | EIN3-binding F-box protein 1 | 2.70 | - | |
| GLYMA_20G203700 | ethylene-responsive transcription factor 1B | 2.59 | −2.19 | |
| GLYMA_02G006200 | ethylene-responsive transcription factor 1B | - | −6.71 | |
| GLYMA_10G036700 | ethylene-responsive transcription factor 1B | - | −2.88 | |
| GLYMA_10G007000 | ethylene-responsive transcription factor 1B | - | −2.68 | |
| GLYMA_19G248900 | ethylene-responsive transcription factor 1B | - | −1.55 | |
| GLYMA_18G018400 | putative ETHYLENE INSENSITIVE 3 | 1.67 | 2.24 | |
| GLYMA_13G166200 | EIN3-binding F-box protein 1 | 1.58 | - | |
| GLYMA_20G202200 | ethylene receptor 2 | 1.23 | - | |
| GLYMA_13G076800 | ETHYLENE INSENSITIVE 3-like 1 protein | 1.15 | - | |
| GLYMA_10G188500 | ethylene receptor | 1.07 | - | |
| GLYMA_16G020500 | transcription factor MYC2 | 2.49 | - | JA |
| GLYMA_17G209000 | transcription factor MYC2 | - | −1.52 | |
| GLYMA_13G112000 | Jasmonate ZIM domain-containing protein | 1.69 | - | |
| GLYMA_16G026900 | jasmonic acid-amido synthetase JAR1 | - | −1.17 | |
| GLYMA_18G030200 | coronatine-insensitive protein 1 | −1.31 | - | |
| GLYMA_15G062300 | pathogenesis-related protein 1-like protein | 2.7 | - | SA |
| GLYMA_15G062400 | pathogenesis-related protein 1 | - | −5.91 | |
| GLYMA_15G062700 | pathogenesis-related protein 1 | - | −3.48 | |
| GLYMA_15G062500 | pathogenesis-related protein 1 | - | −1.97 | |
| GLYMA_09G020800 | NPR1-1 protein | - | −1.05 | |
| GLYMA_14G031300 | regulatory protein NPR3 | 2.31 | - | |
| GLYMA_02G283300 | regulatory protein NPR3 | 1.04 | - | |
| GLYMA_03G128600 | regulatory protein NPR5 | - | 1.99 | |
| GLYMA_05G182500 | transcription factor TGA1-like | −1.06 | - | |
| GLYMA_18G020900 | transcription factor TGA4-like | - | −1.1 | |
| GLYMA_14G167000 | transcription factor TGA7 | - | 1.44 | |
| GLYMA_13G085100 | transcription factor bZIP83 | - | 1.08 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Shang, J.; Wang, W.; Du, J.; Li, K.; Wu, X.; Yu, L.; Liu, C.; Khaskheli, M.I.; Yang, W. Comparison of Transcriptome Differences in Soybean Response to Soybean Mosaic Virus under Normal Light and in the Shade. Viruses 2019, 11, 793. https://doi.org/10.3390/v11090793
Zhang L, Shang J, Wang W, Du J, Li K, Wu X, Yu L, Liu C, Khaskheli MI, Yang W. Comparison of Transcriptome Differences in Soybean Response to Soybean Mosaic Virus under Normal Light and in the Shade. Viruses. 2019; 11(9):793. https://doi.org/10.3390/v11090793
Chicago/Turabian StyleZhang, Lei, Jing Shang, Wenming Wang, Junbo Du, Kai Li, Xiaoling Wu, Liang Yu, Chunyan Liu, Muhammad Ibrahim Khaskheli, and Wenyu Yang. 2019. "Comparison of Transcriptome Differences in Soybean Response to Soybean Mosaic Virus under Normal Light and in the Shade" Viruses 11, no. 9: 793. https://doi.org/10.3390/v11090793
APA StyleZhang, L., Shang, J., Wang, W., Du, J., Li, K., Wu, X., Yu, L., Liu, C., Khaskheli, M. I., & Yang, W. (2019). Comparison of Transcriptome Differences in Soybean Response to Soybean Mosaic Virus under Normal Light and in the Shade. Viruses, 11(9), 793. https://doi.org/10.3390/v11090793





