Identification of Key Residues Required for RNA Silencing Suppressor Activity of p23 Protein from a Mild Strain of Citrus Tristeza Virus
Abstract
1. Introduction
2. Materials and Methods
2.1. Virus Source and Plant Materials
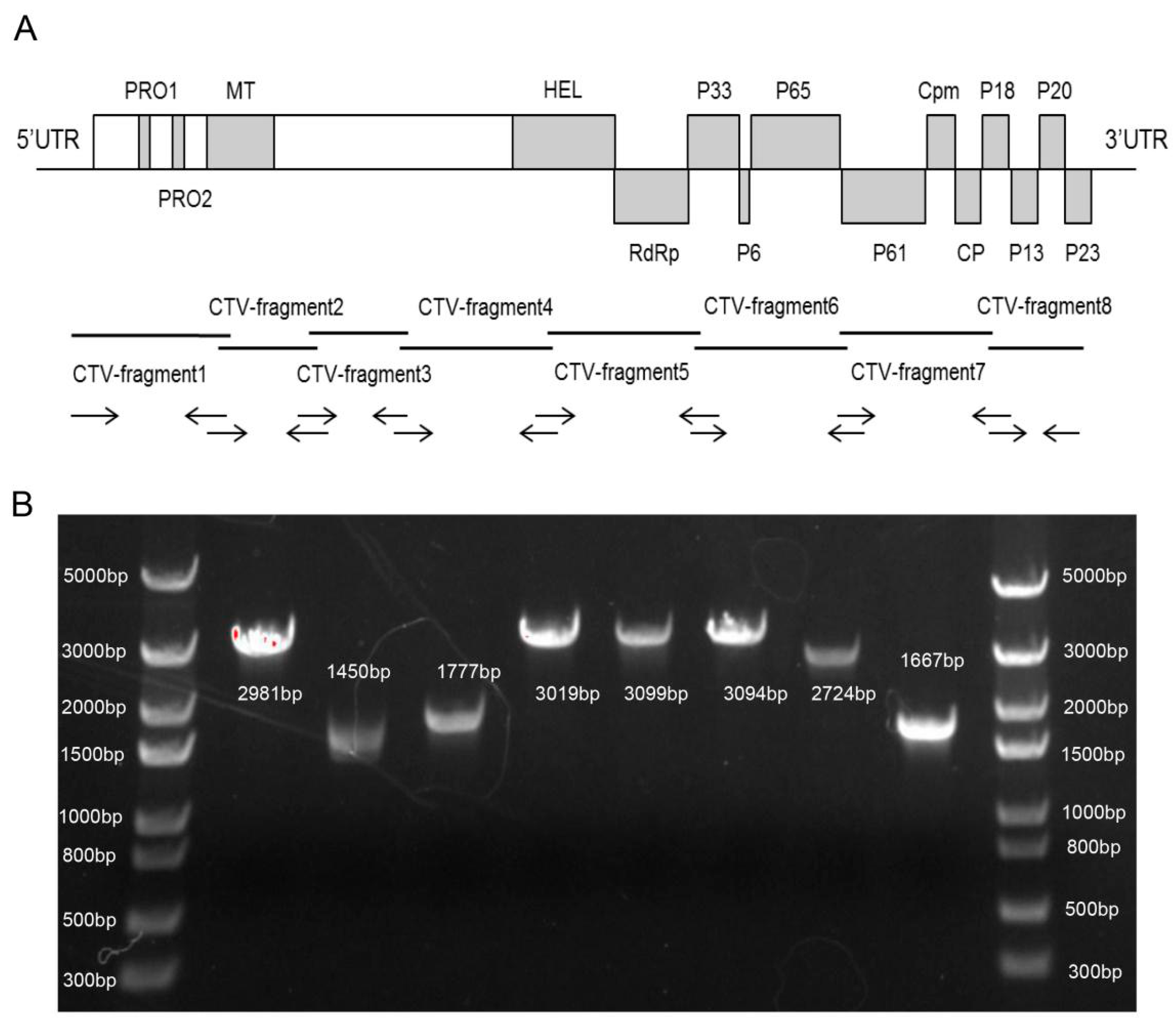
2.2. Cloning of N4 Sequence
2.3. CTV-N4VSR Expression Vector Construction and Transient Assays
2.4. Agroinfiltration, Subcellular Localization, and Live Cell Imaging
2.5. Amino Acid Sequence Alignment, Classification, and Phylogenetic Tree Analysis
2.6. Construction and Transient Expression of CTV-N4p23 Mutants with Point Mutation
2.7. Western Blotting
2.8. Northern Blotting
3. Results
3.1. Whole-Genome Sequencing and Cloning of CTV-N4
3.2. CTV-N4p23 is a Strong Silencing Suppressor
3.3. Subcellular Localization of N4p23
3.4. Identification of Conserved Amino Acids in CTV-p23
3.5. Conserved Amino Acids of p23 were Important for Its Suppressor Activity
3.6. Conserved Amino Acids in p23 Protein Played Differential Roles in Protein Stability and Suppressor Activity
4. Discussion
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Moreno, P.; Ambrós, S.; Albiach-Martí, M.R.; Guerri, J.; Peña, L. Citrus tristeza virus: A pathogen that changed the course of the citrus industry. Mol. Plant Pathol. 2008, 9, 251–268. [Google Scholar] [CrossRef] [PubMed]
- Harper, S.J. Citrus tristeza virus: Evolution of Complex and Varied Genotypic Groups. Front. Microbiol. 2013, 4, 93. [Google Scholar] [CrossRef] [PubMed]
- Hilf, M.E.; Karasev, A.V.; Albiach-Marti, M.R.; Dawson, W.O.; Garnsey, S.M. Two paths of sequence divergence in the citrus tristeza virus complex. Phytopathology 1999, 89, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Karasev, A.V.; Boyko, V.P.; Gowda, S.; Nikolaeva, O.V.; Hilf, M.E.; Koonin, E.V.; Niblett, C.L.; Cline, K.; Gumpf, D.J.; Lee, R.F. Complete sequence of the citrus tristeza virus RNA genome. Virology 1995, 208, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Albiach-Martí, M.R.; Mawassi, M.; Gowda, S.; Satyanarayana, T.; Hilf, M.E.; Shanker, S.; Almira, E.C.; Vives, M.C.; López, C.; Guerri, J.; et al. Sequences of Citrus tristeza virus separated in time and space are essentially identical. J. Virol. 2000, 74, 6856–6865. [Google Scholar] [CrossRef] [PubMed]
- Mawassi, M.; Gafny, R.; Gagliardi, D.; Bar-Joseph, M. Populations of citrus tristeza virus contain smaller-than-full-length particles which encapsidate sub-genomic RNA molecules. J. Gen. Virol. 1995, 76 Pt 3, 651–659. [Google Scholar] [CrossRef]
- Mawassi, M.; Mietkiewska, E.; Gofman, R.; Yang, G.; Bar-Joseph, M. Unusual sequence relationships between two isolates of citrus tristeza virus. J. Gen. Virol. 1996, 77 Pt 9, 2359–2364. [Google Scholar] [CrossRef]
- Roy, A.; Brlansky, R.H. Genome analysis of an orange stem pitting citrus tristeza virus isolate reveals a novel recombinant genotype. Virus Res. 2010, 151, 118–130. [Google Scholar] [CrossRef]
- Harper, S.J.; Dawson, T.E.; Pearson, M.N. Isolates of Citrus tristeza virus that overcome Poncirus trifoliata resistance comprise a novel strain. Arch. Virol. 2010, 155, 471–480. [Google Scholar] [CrossRef]
- Guo, Z.X.; Li, Y.; Ding, S.W. Small RNA-based antimicrobial immunity. Nat. Rev. Immunol. 2019, 19, 31–44. [Google Scholar] [CrossRef]
- Khalid, A.; Zhang, Q.; Yasir, M.; Li, F. Small RNA Based Genetic Engineering for Plant Viral Resistance: Application in Crop Protection. Front. Microbiol. 2017, 8, 43. [Google Scholar] [CrossRef] [PubMed]
- Hajeri, S.; Killiny, N.; El-Mohtar, C.; Dawson, W.O.; Gowda, S. Citrus tristeza virus-based RNAi in citrus plants induces gene silencing in Diaphorina citri, a phloem-sap sucking insect vector of citrus greening disease (Huanglongbing). J. Biotechnol. 2014, 176, 42–49. [Google Scholar] [CrossRef]
- Burgyan, J.; Havelda, Z. Viral suppressors of RNA silencing. Trends Plant Sci. 2011, 16, 265–272. [Google Scholar] [CrossRef]
- Li, F.; Ding, S.W. Virus counterdefense: Diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 2006, 60, 503–531. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Folimonov, A.; Shintaku, M.; Li, W.X.; Falk, B.W.; Dawson, W.O.; Ding, S.W. Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc. Natl. Acad. Sci. USA 2004, 101, 15742–15747. [Google Scholar] [CrossRef]
- Folimonov, A.S.; Folimonova, S.Y.; Bar-Joseph, M.; Dawson, W.O. A stable RNA virus-based vector for citrus trees. Virology 2007, 368, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ruiz, S.; Navarro, B.; Gisel, A.; Peña, L.; Navarro, L.; Moreno, P.; Di Serio, F.; Flores, R. Citrus tristeza virus infection induces the accumulation of viral small RNAs (21-24-nt) mapping preferentially at the 3′-terminal region of the genomic RNA and affects the host small RNA profile. Plant Mol. Biol. 2011, 75, 607–619. [Google Scholar] [CrossRef]
- Diaz-Pendon, J.A.; Ding, S.W. Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu. Rev. Phytopathol. 2008, 46, 303–326. [Google Scholar] [CrossRef] [PubMed]
- Cevik, B.; Yardimci, N.; Korkmaz, S. The First Identified Citrus tristeza virus Isolate of Turkey Contains a Mixture of Mild and Severe Strains. Plant Pathol. J. 2013, 29, 31–41. [Google Scholar] [CrossRef]
- Silhavy, D.; Molnár, A.; Lucioli, A.; Szittya, G.; Hornyik, C.; Tavazza, M.; Burgyán, J. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 2002, 21, 3070–3080. [Google Scholar] [CrossRef]
- Lucy, A.P.; Guo, H.S.; Li, W.X.; Ding, S.W. Suppression of post-transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J. 2000, 19, 1672–1680. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Ruiz-Ruiz, S.; Soler, N.; Sánchez-Navarro, J.; Fagoaga, C.; López, C.; Navarro, L.; Moreno, P.; Peña, L. Citrus tristeza virus p23: A unique protein mediating key virus-host interactions. Front. Microbiol. 2013, 4, 98. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.F.; Keremane, M.L. Mild strain cross protection of tristeza: A review of research to protect against decline on sour orange in Florida. Front. Microbiol. 2013, 4, 259. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ruiz, S.; Soler, N.; Sánchez-Navarro, J.; Fagoaga, C.; López, C.; Navarro, L.; Moreno, P.; Peña, L.; Flores, R. Citrus tristeza virus p23: Determinants for nucleolar localization and their influence on suppression of RNA silencing and pathogenesis. Mol. Plant Microbe Interact. 2013, 26, 306–318. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ruiz, S.; Spanò, R.; Navarro, L.; Moreno, P.; Peña, L.; Flores, R. Citrus tristeza virus co-opts glyceraldehyde 3-phosphate dehydrogenase for its infectious cycle by interacting with the viral-encoded protein p23. Plant Mol. Biol. 2018, 98, 363–373. [Google Scholar] [CrossRef] [PubMed]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; He, Y.; Luo, T.; Zhang, X.; Wan, H.; Ur Rehman, A.; Bao, X.; Zhang, Q.; Chen, J.; Xu, R.; et al. Identification of Key Residues Required for RNA Silencing Suppressor Activity of p23 Protein from a Mild Strain of Citrus Tristeza Virus. Viruses 2019, 11, 782. https://doi.org/10.3390/v11090782
Li Z, He Y, Luo T, Zhang X, Wan H, Ur Rehman A, Bao X, Zhang Q, Chen J, Xu R, et al. Identification of Key Residues Required for RNA Silencing Suppressor Activity of p23 Protein from a Mild Strain of Citrus Tristeza Virus. Viruses. 2019; 11(9):782. https://doi.org/10.3390/v11090782
Chicago/Turabian StyleLi, Zhuoran, Yizhong He, Tao Luo, Xi Zhang, Haoliang Wan, Atta Ur Rehman, Xinru Bao, Qian Zhang, Jia Chen, Rangwei Xu, and et al. 2019. "Identification of Key Residues Required for RNA Silencing Suppressor Activity of p23 Protein from a Mild Strain of Citrus Tristeza Virus" Viruses 11, no. 9: 782. https://doi.org/10.3390/v11090782
APA StyleLi, Z., He, Y., Luo, T., Zhang, X., Wan, H., Ur Rehman, A., Bao, X., Zhang, Q., Chen, J., Xu, R., Deng, Y., Zeng, Y., Xu, J., Hong, N., Li, F., & Cheng, Y. (2019). Identification of Key Residues Required for RNA Silencing Suppressor Activity of p23 Protein from a Mild Strain of Citrus Tristeza Virus. Viruses, 11(9), 782. https://doi.org/10.3390/v11090782







