Virophages of Giant Viruses: An Update at Eleven
Abstract
1. Introduction
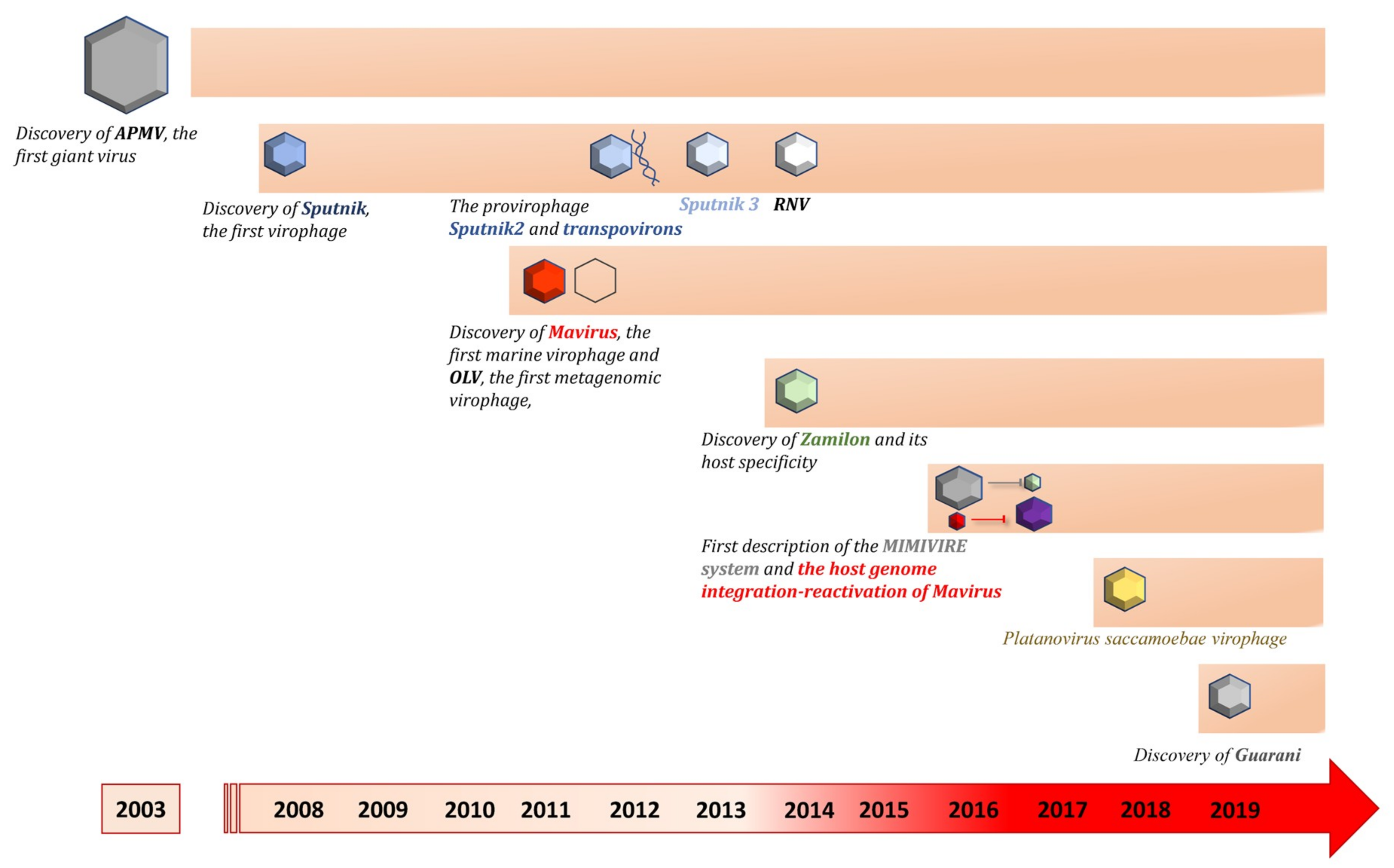
2. Giant Virus Discovery
3. Virophage Discovery over a Decade
3.1. Virophages Isolated by Culture
3.1.1. Sputnik, the First Virophage
3.1.2. Other Virophages of the Sputnik Clade
3.1.3. Mavirus Virophage
3.1.4. Zamilon Virophage
3.1.5. Virophages Isolated by Culture Partially Characterized
3.2. Virophages Discovered by Genomic
3.3. Virophages from Metagenomic Datasets
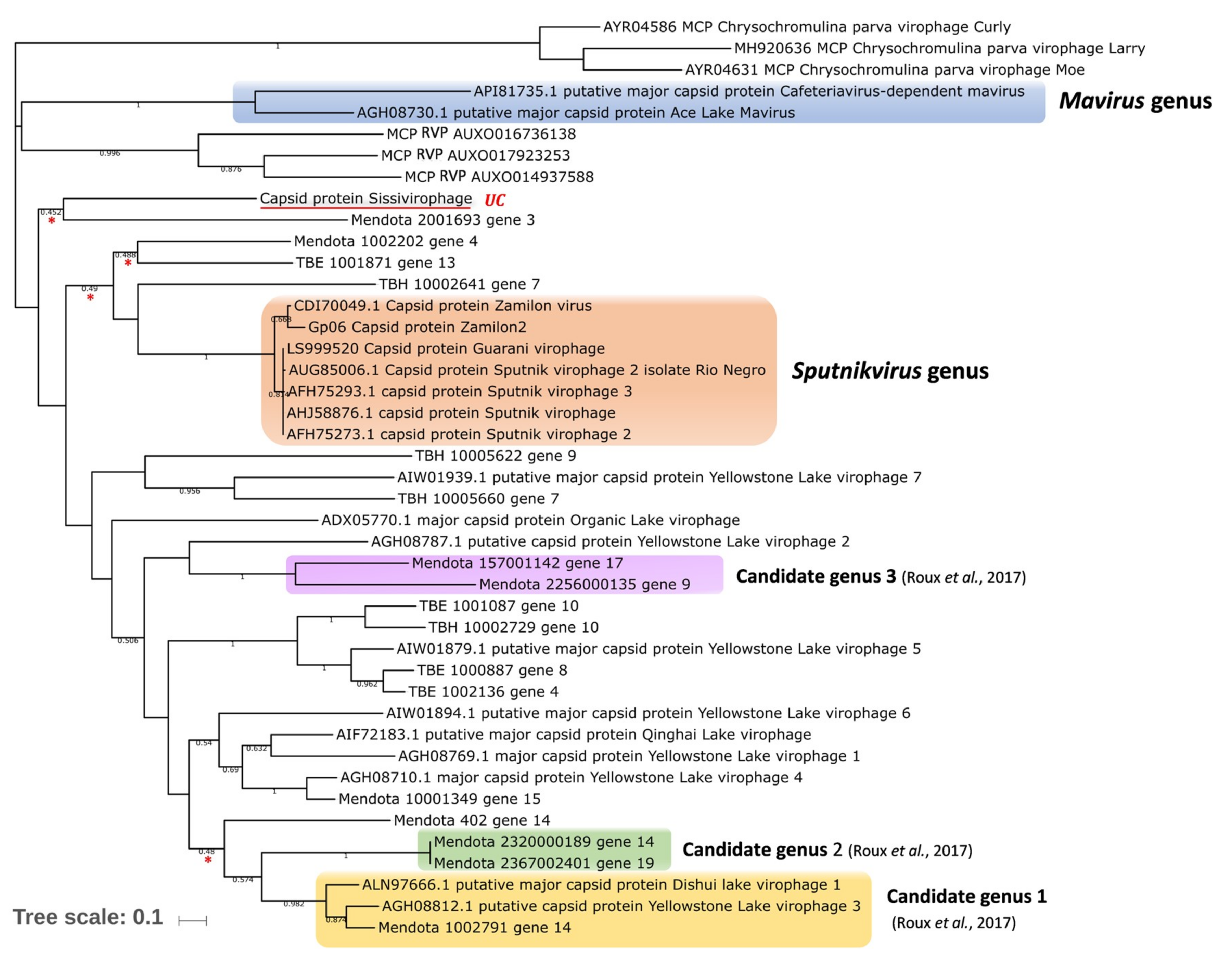
4. Virophages in the Context of Evolution
5. The Virophage Lifestyle
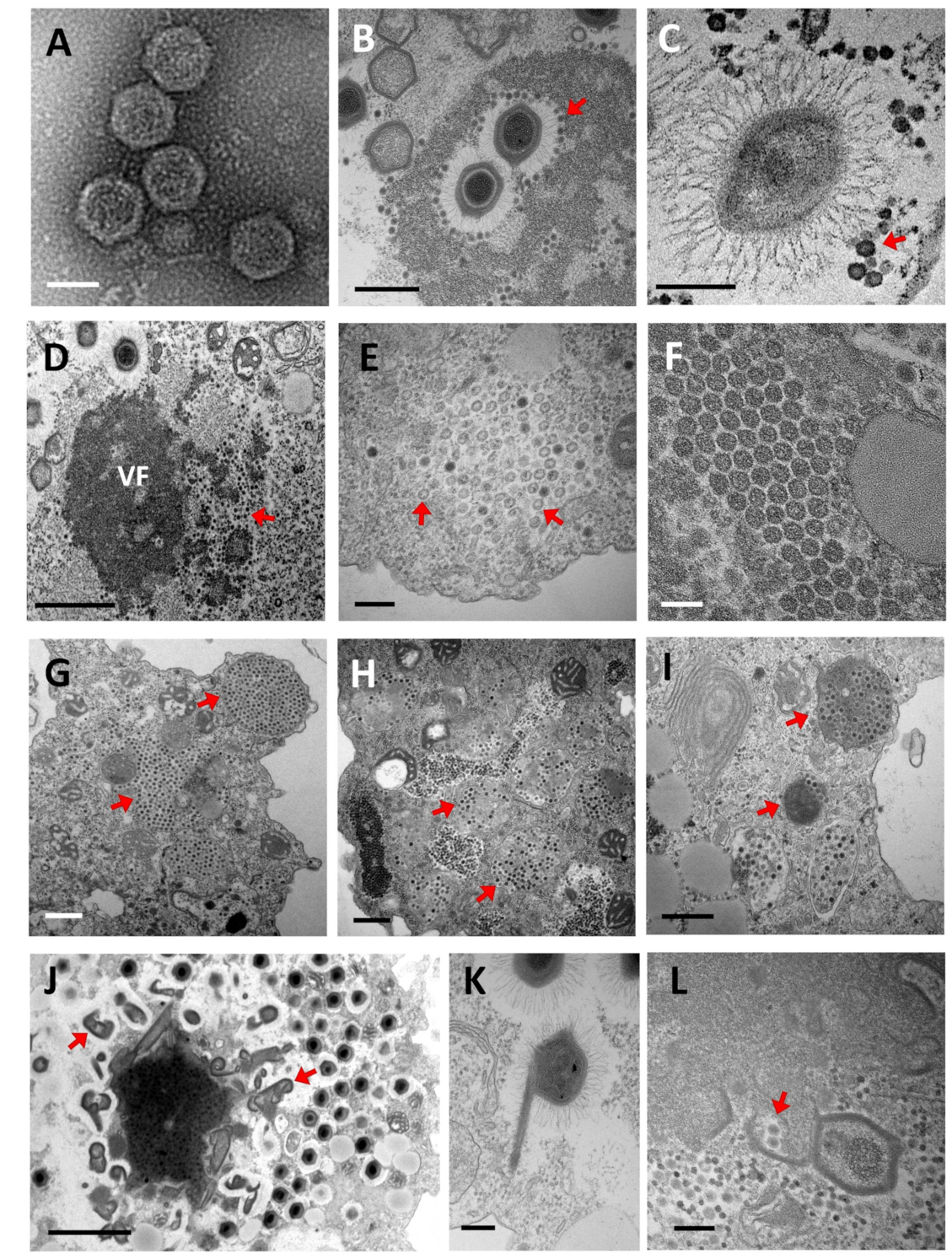
5.1. Replication Cycle
5.1.1. Entry
5.1.2. Genome Release, Expression, Replication, and Viral Morphogenesis
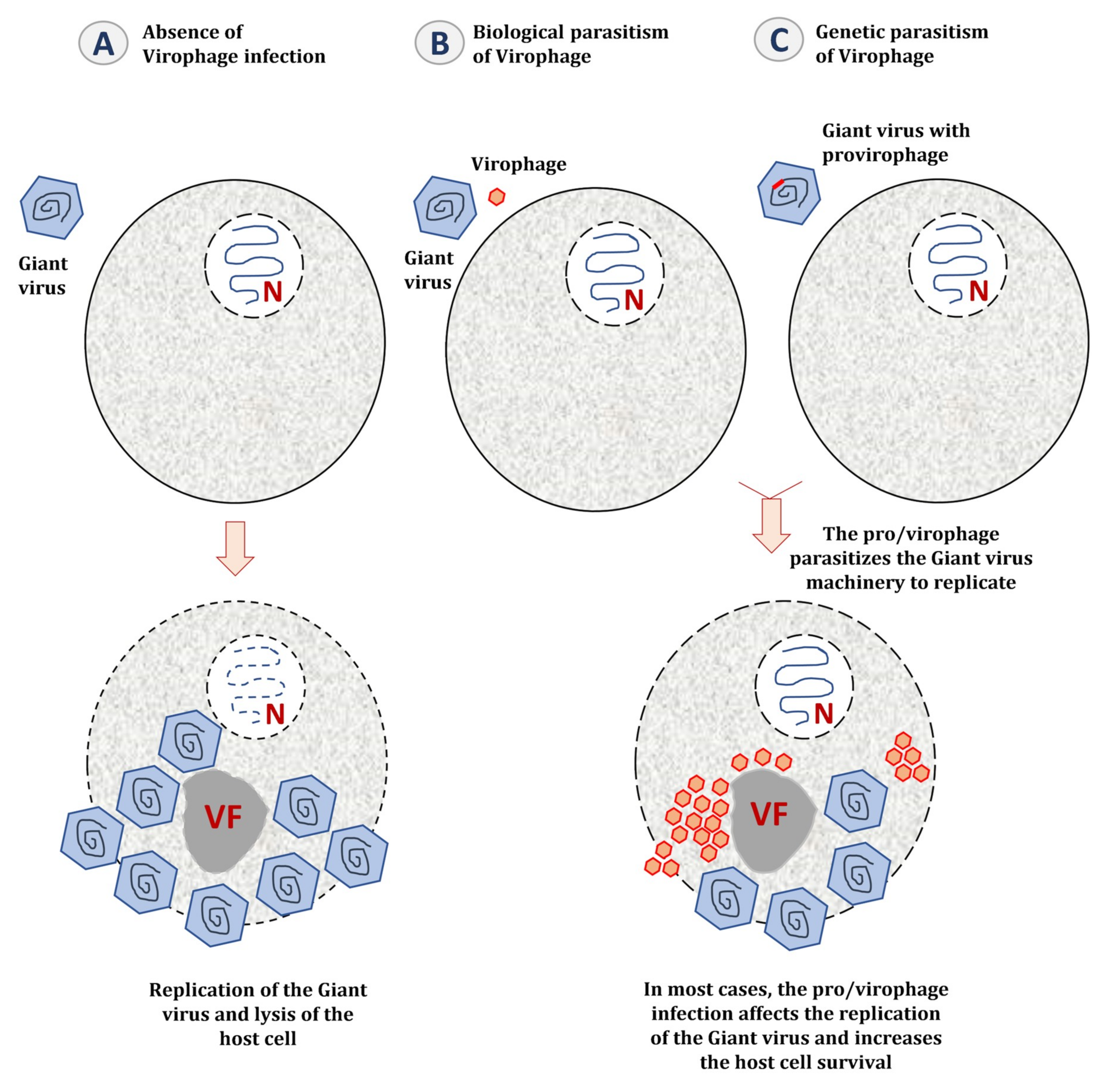
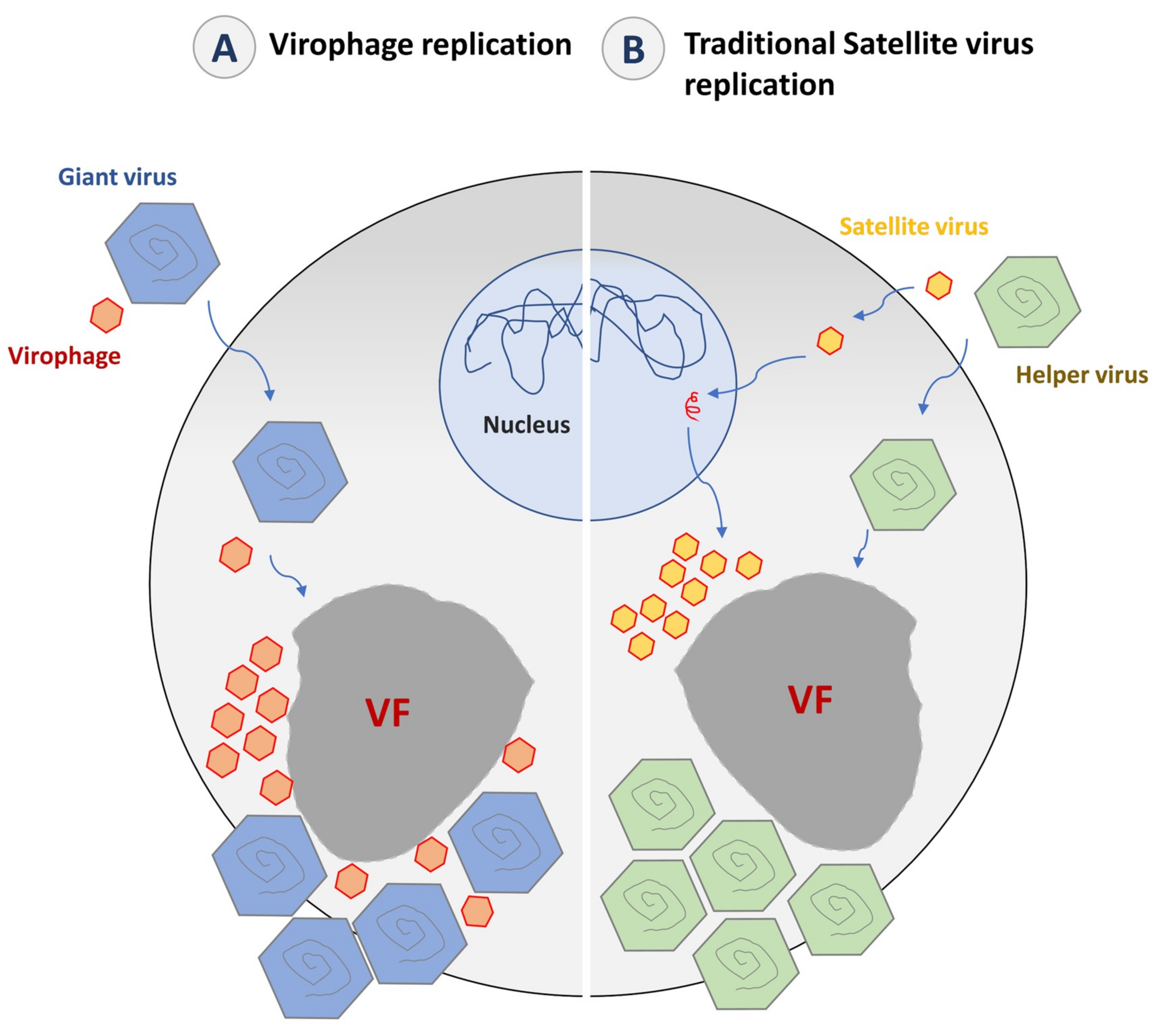
5.2. The Impact of Virophage Infection on the Giant Virus Cycle
6. Virophages as Mobile Genetic Elements of Giant Viruses and Their Host Cells
6.1. Genetic Parasitism of the Giant Virus Genome
6.2. Integration of Virophage Genome in the Host Cell Genomes
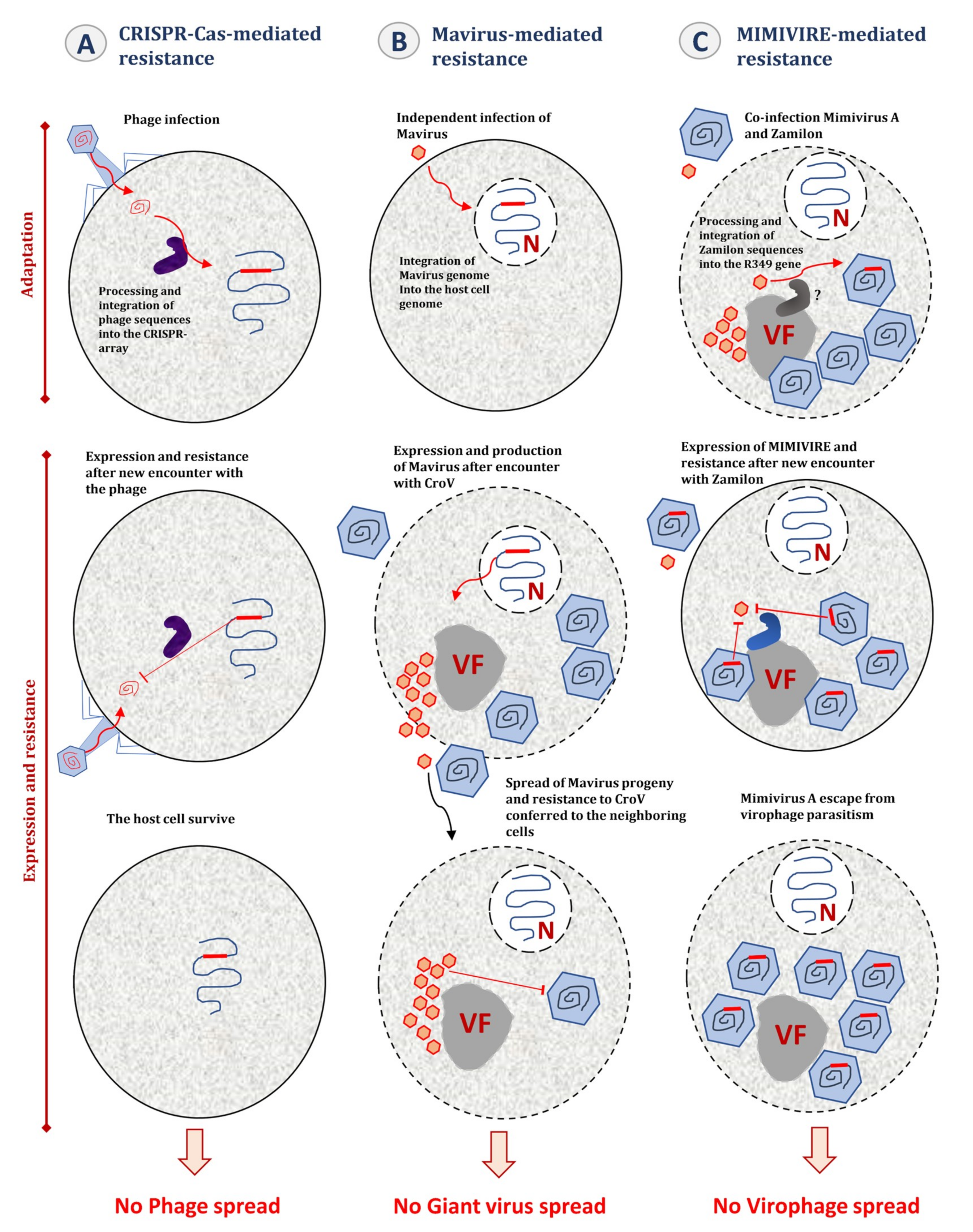
7. MIMIVIRE, a Giant Virus Defense System
8. Virophages, a Source of Controversy
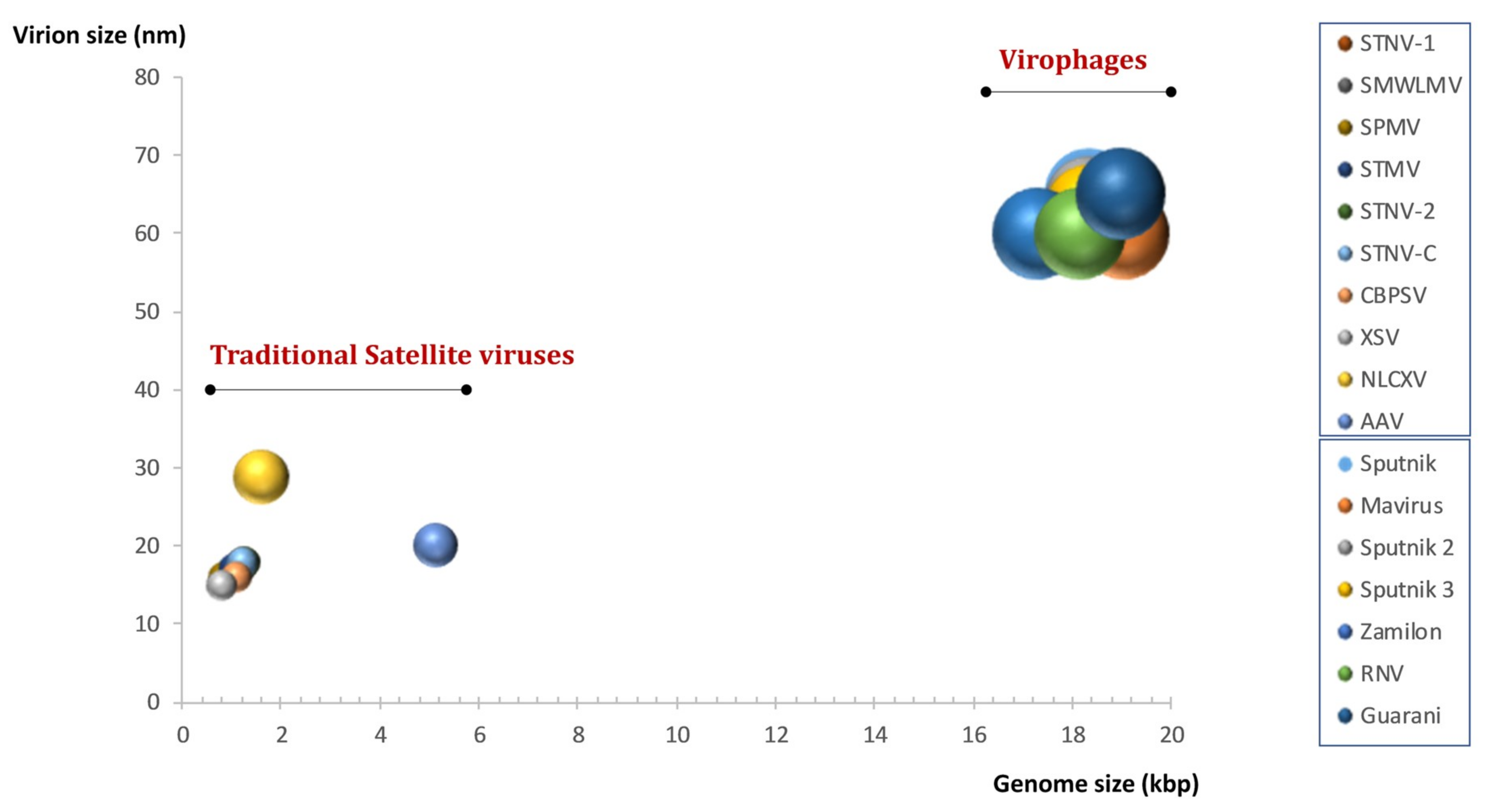
9. Definition and Classification
10. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Suttle, C.A. Marine viruses—Major players in the global ecosystem. Nat. Rev. Genet. 2007, 5, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Suttle, C.A. Viruses in the sea. Nature 2005, 437, 356–361. [Google Scholar] [CrossRef]
- Rohwer, F.; Thurber, R.V. Viruses manipulate the marine environment. Nature 2009, 459, 207–212. [Google Scholar] [CrossRef]
- Baltimore, D. Expression of Animal Virus genomes. Bacteriol. Rev. 1971, 35, 235–241. [Google Scholar]
- Koonin, E.V. Genome replication/expression strategies of positive-strand RNA viruses: A simple version of a combinatorial classification and prediction of new strategies. Virus Genes 1991, 5, 273–281. [Google Scholar] [CrossRef]
- Koonin, E.V.; Dolja, V.V. Virus world as an evolutionary network of viruses and capsidless selfish elements. Microbiol. Mol. Biol. Rev. 2014, 78, 278–303. [Google Scholar]
- Koonin, E.V.; Dolja, V.V. A virocentric perspective on the evolution of life. Curr. Opin. Virol. 2013, 3, 546–557. [Google Scholar] [CrossRef] [PubMed]
- Kassanis, B.; Nixon, H.L. Activation of one Tobacco Necrosis Virus by Another. J. Gen. Microbiol. 1961, 25, 459–471. [Google Scholar] [CrossRef]
- La Scola, B.; Audic, S.; Robert, C.; Jungang, L.; De Lamballerie, X.; Drancourt, M.; Birtles, R.; Claverie, J.M.R.D. A Giant Virus in Amoebae. Science 2003, 299, 2033. [Google Scholar] [CrossRef] [PubMed]
- Colson, P.; La Scola, B.; Raoult, D. Giant Viruses of Amoebae: A Journey Through Innovative Research and Paradigm Changes. Annu. Rev. Virol. 2017, 4, 61–85. [Google Scholar] [CrossRef] [PubMed]
- Raoult, D.; Audic, S.; Robert, C.; Abergel, C.; Renesto, P.; Ogata, H.; La Scola, B.; Suzan, M.; Claverie, J.M. The 1.2-Megabase Genome Sequence of Mimivirus. Science 2004, 306, 1344–1350. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.G.; Allen, M.J.; Wilson, W.H.; Suttle, C.A. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc. Natl. Acad. Sci. USA 2010, 107, 19508–19513. [Google Scholar] [CrossRef] [PubMed]
- Renesto, P.; Abergel, C.; Decloquement, P.; Moinier, D.; Azza, S.; Ogata, H.; Fourquet, P.; Gorvel, J.P.; Claverie, J.M. Mimivirus Giant Particles Incorporate a Large Fraction of Anonymous and Unique Gene Products. J. Virol. 2006, 80, 11678–11685. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.G.; Kelly, I.; Foster, L.J.; Suttle, C.A. The virion of Cafeteria roenbergensis virus (CroV) contains a complex suite of proteins for transcription and DNA repair. Virology 2014, 466, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Colson, P.; La Scola, B.; Levasseur, A.; Caetano-Anollés, G.; Raoult, D. Mimivirus: Leading the way in the discovery of giant viruses of amoebae. Nat. Rev. Genet. 2017, 15, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Fridmann-Sirkis, Y.; Milrot, E.; Mutsafi, Y.; Ben-Dor, S.; Levin, Y.; Savidor, A.; Kartvelishvily, E.; Minsky, A. Efficiency in Complexity: Composition and Dynamic Nature of Mimivirus Replication Factories. J. Virol. 2016, 90, 10039–10047. [Google Scholar] [CrossRef] [PubMed]
- Mutsafi, Y.; Zauberman, N.; Sabanay, I.; Minsky, A. Vaccinia-like cytoplasmic replication of the giant Mimivirus. Proc. Natl. Acad. Sci. USA 2010, 107, 5978–5982. [Google Scholar] [CrossRef]
- Suzan-Monti, M.; La Scola, B.; Barrassi, L.; Espinosa, L.; Raoult, D. Ultrastructural Characterization of the Giant Volcano-like Virus Factory of Acanthamoeba polyphaga Mimivirus. PLoS ONE 2007, 2, e328. [Google Scholar] [CrossRef]
- Forterre, P. Giant Viruses: Conflicts in Revisiting the Virus Concept. Intervirology 2010, 53, 362–378. [Google Scholar] [CrossRef]
- Forterre, P. Manipulation of cellular syntheses and the nature of viruses: The virocell concept. Comptes Rendus Chim. 2011, 14, 392–399. [Google Scholar] [CrossRef]
- Forterre, P.; Gaïa, M. Giant viruses and the origin of modern eukaryotes. Curr. Opin. Microbiol. 2016, 31, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Aherfi, S.; Colson, P.; La Scola, B.; Raoult, D. Giant Viruses of Amoebas: An Update. Front. Microbiol. 2016, 7, 12406. [Google Scholar] [CrossRef] [PubMed]
- Colson, P.; Ominami, Y.; Hisada, A.; La Scola, B.; Raoult, D. Giant mimiviruses escape many canonical criteria of the virus definition. Clin. Microbiol. Infect. 2018, 2, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Colson, P.; Pontarotti, P.; Raoult, D. Mimivirus inaugurated in the 21st century the beginning of a reclassification of viruses. Curr. Opin. Microbiol. 2016, 31, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, Y.G.; Xiao, C.; Sun, S.; Raoult, D.; Rossmann, M.; McPherson, A. Atomic force microscopy investigation of the giant mimivirus. Virology 2010, 404, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Klose, T.; Kuznetsov, Y.G.; Xiao, C.; Sun, S.; McPherson, A.; Rossmann, M.G. The Three-Dimensional Structure of Mimivirus. Intervirology 2010, 53, 268–273. [Google Scholar] [CrossRef]
- Xiao, C.; Kuznetsov, Y.G.; Sun, S.; Hafenstein, S.L.; Kostyuchenko, A.V.; Chipman, P.R.; Suzan-Monti, M.; Raoult, D.; McPherson, A.; Rossmann, M.G. Structural Studies of the Giant Mimivirus. PLoS Biol. 2009, 7, e1000092. [Google Scholar] [CrossRef]
- Legendre, M.; Santini, S.; Rico, A.; Abergel, C.; Claverie, J.M. Breaking the 1000-gene barrier for Mimivirus using ultra-deep genome and transcriptome sequencing. Virol. J. 2011, 8, 99. [Google Scholar] [CrossRef]
- La Scola, B.; de Lamballerie, X.N.; Claverie, J.M.; Drancourt, M.; Raoult, D. Genus Mimivirus in Virus Taxonomy. In Virus Taxonomy, 1st ed.; Fauquet, M., Mayo, M.A., Maniloff, J., Desselberger, U., Ball, L.A., Eds.; Elsevier Acad Press: Amsterdam, The Netherlands, 2005; pp. 275–276. [Google Scholar]
- La Scola, B.; Desnues, C.; Pagnier, I.; Robert, C.; Barrassi, L.; Fournous, G.; Merchat, M.; Suzan-Monti, M.; Forterre, P.; Koonin, E.; et al. The virophage as a unique parasite of the giant mimivirus. Nature 2008, 455, 100–104. [Google Scholar] [CrossRef]
- Fischer, M.G. Sputnik and Mavirus: More than just satellite viruses. Nat. Rev. Microbiol. 2012, 10, 88. [Google Scholar] [CrossRef]
- Raoult, D.; Forterre, P. Redefining viruses: Lessons from Mimivirus. Nat. Rev. Genet. 2008, 6, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Raoult, D. How the virophage compels the need to readdress the classification of microbes. Virology 2015, 477, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Claverie, J.; Abergel, C. Mimivirus and its Virophage. Annu. Rev. Genet. 2009, 43, 49–66. [Google Scholar] [CrossRef] [PubMed]
- Bekliz, M.; Colson, P.; La Scola, B. The expanding family of virophages. Viruses 2016, 8, 317. [Google Scholar] [CrossRef] [PubMed]
- Pearson, H. “Virophage” suggests viruses are alive. Nature 2008, 7205, 677. [Google Scholar] [CrossRef] [PubMed]
- Serrano, V.; Paulo, S.; Toscano, E.; Sávio, S. Genomic Signatures Among Acanthamoeba polyphaga Entoorganisms Unveil Evidence of Coevolution. J. Mol. Evol. 2018, 87, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Desnues, C.; La Scola, B.; Yutin, N.; Fournous, G.; Robert, C.; Azza, S.; Jardot, P.; Monteil, S.; Campocasso, A.; Koonin, E.V.; et al. Provirophages and transpovirons as the diverse mobilome of giant viruses. Proc. Natl. Acad. Sci. USA 2012, 109, 18078–18083. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Gallot-Lavallée, L.; Maumus, F. Provirophages in the Bigelowiella genome bear testimony to past encounters with giant viruses. Proc. Natl. Acad. Sci. USA 2015, 112, E5318–E5326. [Google Scholar] [CrossRef]
- Fischer, M.G.; Hackl, T. Host genome integration and giant virus-induced reactivation of the virophage mavirus. Nature 2016, 540, 288–291. [Google Scholar] [CrossRef]
- Levasseur, A.; Bekliz, M.; Chabriere, E.; Pontarotti, P.; La Scola, B.; Raoult, D. MIMIVIRE is a defence system in mimivirus that confers resistance to virophage. Nature 2016, 531, 249–252. [Google Scholar] [CrossRef]
- Koonin, E.V.; Krupovic, M. The depths of virus exaptation. Curr. Opin. Virol. 2018, 31, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Pagnier, I.; Reteno, D.G.I.; Saadi, H.; Boughalmi, M.; Gaia, M.; Slimani, M.; Ngounga, T.; Bekliz, M.; Colson, P.; Raoult, D.; et al. A Decade of Improvements in Mimiviridae and Marseilleviridae Isolation from Amoeba. Intervirology 2013, 56, 354–363. [Google Scholar] [CrossRef]
- Khalil, J.Y.B.; Robert, S.; Reteno, D.G.; Andreani, J.; Raoult, D.; La Scola, B. High-Throughput Isolation of Giant Viruses in Liquid Medium Using Automated Flow Cytometry and Fluorescence Staining. Front. Microbiol. 2016, 7, 1124. [Google Scholar] [CrossRef] [PubMed]
- Khalil, J.Y.B.; Andreani, J.; La Scola, B. Updating strategies for isolating and discovering giant viruses. Curr. Opin. Microbiol. 2016, 31, 80–87. [Google Scholar] [CrossRef]
- Boughalmi, M.; Saadi, H.; Pagnier, I.; Colson, P.; Fournous, G.; Raoult, D.; La Scola, B. High-throughput isolation of giant viruses of the Mimiviridae and Marseilleviridae families in the Tunisian environment. Env. Microbiol. 2012, 15, 2000–2007. [Google Scholar] [CrossRef] [PubMed]
- BouKhalil, J.Y.; Langlois, T.; Andreani, J.; Sorraing, J. Flow Cytometry Sorting to Separate Viable Giant Viruses from Amoeba. Front. Cell Infect. Microbiol. 2017, 6, 1–10. [Google Scholar]
- Kerepesi, C.; Grolmusz, V. The “Giant Virus Finder” discovers an abundance of giant viruses in the Antarctic dry valleys. Arch. Virol. 2017, 162, 1671–1676. [Google Scholar] [CrossRef] [PubMed]
- Mihara, T.; Koyano, H.; Hingamp, P.; Grimsley, N.; Goto, S.; Ogata, H. Taxon Richness of “Megaviridae” Exceeds those of Bacteria and Archaea in the Ocean. Microbes Environ. 2018, 33, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Andrade, A.C.D.S.P.; Arantes, T.S.; Rodrigues, R.A.L.; Machado, T.B.; Dornas, F.P.; Landell, M.F.; Furst, C.; Borges, L.G.A.; Dutra, L.A.L.; Almeida, G.; et al. Ubiquitous giants: A plethora of giant viruses found in Brazil and Antarctica. Virol. J. 2018, 15, 22. [Google Scholar] [CrossRef]
- Claverie, J.; Abergel, C. Mimiviridae: An Expanding Family of Highly Diverse Large dsDNA Viruses Infecting a Wide Phylogenetic. Viruses 2018, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Rolland, C.; Andreani, J.; Louazani, A.C.; Aherfi, S.; Francis, R.; Rodrigues, R.; Silva, L.S.; Sahmi, D.; Mougari, S.; Chelkha, N.; et al. Discovery and Further Studies on Giant Viruses at the IHU Mediterranee Infection That Modified the Perception of the Virosphere. Viruses 2019, 11, 312. [Google Scholar] [CrossRef] [PubMed]
- Yoosuf, N.; Yutin, N.; Colson, P.; Shabalina, S.A.; Pagnier, I.; Robert, C.; Azza, S.; Klose, T.; Wong, J.; Rossmann, M.G.; et al. Related Giant Viruses in Distant Locations and Different Close to the Megavirus Lineage. Genome Biol. Evol. 2012, 4, 1324–1330. [Google Scholar] [CrossRef] [PubMed]
- Arslan, D.; Legendre, M.; Seltzer, V.; Abergel, C.; Claverie, J.-M. Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae. Proc. Natl. Acad. Sci. USA 2011, 108, 17486–17491. [Google Scholar] [CrossRef] [PubMed]
- Yau, S.; Lauro, F.M.; DeMaere, M.Z.; Brown, M.V.; Thomas, T.; Raftery, M.J.; Andrews-Pfannkoch, C.; Lewis, M.; Hoffman, J.M.; Gibson, J.A.; et al. Virophage control of antarctic algal host–virus dynamics. Proc. Natl. Acad. Sci. USA 2011, 108, 6163–6168. [Google Scholar] [CrossRef] [PubMed]
- Schulz, F.; Yutin, N.; Ivanova, N.N.; Ortega, D.R.; Lee, T.K.; Vierheilig, J.; Daims, H.; Horn, M.; Wagner, M.; Jensen, G.J.; et al. Giant viruses with an expanded complement of translation system components. Science 2017, 356, 82–85. [Google Scholar] [CrossRef] [PubMed]
- Schulz, F.; Alteio, L.; Goudeau, D.; Ryan, E.M.; Yu, F.B.; Malmstrom, R.R.; Blanchard, J.; Woyke, T. Hidden diversity of soil giant viruses. Nat. Commun. 2018, 9, 4881. [Google Scholar] [CrossRef] [PubMed]
- Deeg, C.M.; Chow, C.E.T.; Suttle, A.C. The kinetoplastid-infecting Bodo saltans virus (BsV), a window into the most abundant giant viruses in the sea. eLife 2018, 7, e33014. [Google Scholar] [CrossRef]
- Abrahao, J.; Silva, L.; Silva, L.S.; Khalil, J.Y.B.; Rodrigues, R.; Arantes, T.; Assis, F.; Boratto, P.; Andrade, M.; Kroon, E.G.; et al. Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat. Commun. 2018, 9, 749. [Google Scholar] [CrossRef]
- Rodrigues, R.A.L.; Mougari, S.; Colson, P.; La Scola, B.; Abrahão, J.S. “Tupanvirus”, a new genus in the family Mimiviridae. Arch. Virol. 2018, 164, 325–331. [Google Scholar] [CrossRef]
- Rodrigues, R.A.L.; Arantes, T.S.; Oliveira, G.P.; Dos Santos Silva, L.K.; Abrahão, J.S. The Complex Nature of Tupanviruses. Adv. Virus Res. 2019, 103, 135–166. [Google Scholar]
- Santini, S.; Jeudy, S.; Bartoli, J.; Poirot, O.; Lescot, M.; Abergel, C.; Barbe, V.; Wommack, K.E.; Noordeloos, A.A.M.; Brussaard, C.P.D.; et al. Genome of Phaeocystis globosa virus PgV-16T highlights the common ancestry of the largest known DNA viruses infecting eukaryotes. Proc. Natl. Acad. Sci. USA 2013, 110, 10800–10805. [Google Scholar] [CrossRef] [PubMed]
- Schvarcz, C.R.; Steward, G.F. A giant virus infecting green algae encodes key fermentation genes. Virology 2018, 518, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Moniruzzaman, M.; Gann, E.R.; LeCleir, G.R.; Kang, Y.; Gobler, C.J.; Wilhelm, S.W. Diversity and dynamics of algal Megaviridae members during a harmful brown tide caused by the pelagophyte, Aureococcus anophagefferens. FEMS Microbiol. Ecol. 2016, 92, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sandaa, R.A.; Heldal, M.; Castberg, T.; Thyrhaug, R.; Bratbak, G. Isolation and Characterization of Two Viruses with Large Genome Size Infecting Chrysochromulina ericina (Prymnesiophyceae) and Pyramimonas orientalis (Prasinophyceae). Virology 2001, 290, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Yutin, N.; Colson, P.; Raoult, D.; Koonin, E.V. Mimiviridae: Clusters of orthologous genes, reconstruction of gene repertoire evolution and proposed expansion of the giant virus family. Virol. J. 2013, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Chipman, P.R.; Castberg, T.; Bratbak, G.; Baker, T.S. The Marine Algal Virus PpV01 Has an Icosahedral Capsid with T=219 Quasisymmetry. J. Virol. 2005, 79, 9236–9243. [Google Scholar] [CrossRef]
- Gallot-Lavallee, L.; Blanc, G.; Claverie, J.M. Comparative Genomics of Chrysochromulina Ericina Virus and Other Microalga-Infecting Large DNA Viruses Highlights Their Intricate Evolutionary Relationship with the Established Mimiviridae Family. J. Virol. 2017, 91, e00230-17. [Google Scholar] [CrossRef]
- Stough, J.M.A.; Yutin, N.; Chaban, Y.V.; Moniruzzaman, M.; Gann, E.R.; Pound, H.L.; Steffen, M.M.; Black, J.N.; Koonin, E.V.; Wilhelm, S.W.; et al. Genome and Environmental Activity of a Chrysochromulina parva Virus and Its Virophages. Front. Microbiol. 2019, 10, 1–12. [Google Scholar]
- Boyer, M.; Yutin, N.; Pagnier, I.; Barrassi, L.; Fournous, G.; Espinosa, L.; Robert, C.; Azza, S.; Sun, S.; Rossmann, M.G.; et al. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc. Natl. Acad. Sci. USA 2009, 106, 21848–21853. [Google Scholar] [CrossRef]
- Colson, P.; Pagnier, I.; Yoosuf, N. “Marseilleviridae”, a new family of giant viruses infecting amoebae. Arch. Virol. 2013, 158, 915–920. [Google Scholar] [CrossRef]
- Legendre, M.; Doutre, G.; Poirot, O.; Lescot, M.; Arslan, D.; Seltzer, V.; Bertaux, L.; Bruley, C. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 2013, 341, 281–287. [Google Scholar]
- Legendre, M.; Bartoli, J.; Shmakova, L.; Jeudy, S.; Labadie, K.; Adrait, A.; Lescot, M.; Poirot, O.; Bertaux, L.; Bruley, C.; et al. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc. Natl. Acad. Sci. USA 2014, 111, 4274–4279. [Google Scholar] [CrossRef] [PubMed]
- Reteno, D.G.; Benamar, S.; Khalil, J.B.; Andreani, J.; Armstrong, N.; Klose, T.; Rossmann, M.; Colson, P.; Raoult, D.; La Scola, B. Faustovirus, an Asfarvirus-Related New Lineage of Giant Viruses Infecting Amoebae. J. Virol. 2015, 89, 6585–6594. [Google Scholar] [CrossRef] [PubMed]
- Legendre, M.; Lartigue, A.; Bertaux, L.; Jeudy, S.; Bartoli, J.; Lescot, M.; Alempic, J.-M.; Ramus, C.; Bruley, C.; Labadie, K.; et al. In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. Proc. Natl. Acad. Sci. USA 2015, 112, E5327–E5335. [Google Scholar] [CrossRef] [PubMed]
- Bajrai, L.H.; Benamar, S.; Azhar, E.I.; Robert, C.; Levasseur, A.; Raoult, D.; Scola, B. La Kaumoebavirus, a New Virus That Clusters with. Viruses 2016, 8, 278. [Google Scholar] [CrossRef]
- Andreani, J.; Yaacoub, J.; Khalil, B.; Sevvana, M.; Benamar, S.; Pinto, F.; Di Bitam, I.; Colson, P.; Klose, T.; Rossmann, M.G.; et al. Pacmanvirus, a New Giant Icosahedral Virus at the Crossroads between. J. Virol. 2017, 91, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Andreani, J.; Aherfi, S.; Khalil, J.Y.B.; Di Pinto, F.; Bitam, I.; Raoult, D.; Colson, P.; La Scola, B. Cedratvirus, a Double-Cork Structured Giant Virus, is a Distant Relative of Pithoviruses. Viruses 2016, 8, 300. [Google Scholar] [CrossRef] [PubMed]
- Andreani, J.; Khalil, J.Y.B.; Baptiste, E.; Hasni, I.; Michelle, C.; Raoult, D.; Levasseur, A.; La Scola, B. Orpheovirus IHUMI-LCC2: A New Virus among the Giant Viruses. Front. Microbiol. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Iyer, L.M.; Aravind, L.; Koonin, E.V. Common Origin of Four Diverse Families of Large Eukaryotic DNA Viruses. J. Virol. 2001, 75, 11720–11734. [Google Scholar] [CrossRef]
- Yutin, N.; Wolf, Y.I.; Raoult, D.; Koonin, E.V. Eukaryotic large nucleo-cytoplasmic DNA viruses: Clusters of orthologous genes and reconstruction of viral genome evolution. Virol. J. 2009, 13, 1–13. [Google Scholar] [CrossRef]
- Yutin, N.; Koonin, E.V. Hidden evolutionary complexity of Nucleo-Cytoplasmic Large DNA viruses of eukaryotes. Virol. J. 2012, 9, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Colson, P. “Megavirales”, a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch. Virol. 2014, 158, 2517–2521. [Google Scholar] [CrossRef] [PubMed]
- Desnues, C.; Boyer, M.; Raoult, D. Sputnik, a Virophage Infecting the Viral Domain of Life; Elsevier BV: Amsterdam, The Netherlands, 2012; Volume 82, pp. 63–89. [Google Scholar]
- Sun, S.; La Scola, B.; Bowman, V.D.; Ryan, C.M.; Whitelegge, J.P.; Raoult, D.; Rossmann, M.G. Structural Studies of the Sputnik Virophage. J. Virol. 2010, 84, 894–897. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Sun, S.; Xiang, Y.; Wong, J.; Klose, T.; Raoult, D.; Rossmann, M.G. Structure of Sputnik, a virophage, at 3.5-Å resolution. Proc. Natl. Acad. Sci. USA 2012, 109, 18431–18436. [Google Scholar] [CrossRef]
- Fischer, M.G.; Suttle, C.A. A Virophage at the Origin of Large DNA Transposons. Science 2011, 332, 231–234. [Google Scholar] [CrossRef] [PubMed]
- Gaïa, M.; Pagnier, I.; Campocasso, A.; Fournous, G.; Raoult, D.; La Scola, B. Broad Spectrum of Mimiviridae Virophage Allows Its Isolation Using a Mimivirus Reporter. PLoS ONE 2013, 8, e61912. [Google Scholar] [CrossRef] [PubMed]
- Gaïa, M.; Benamar, S.; Boughalmi, M.; Pagnier, I.; Croce, O.; Colson, P.; Raoult, D.; La Scola, B. Zamilon, a Novel Virophage with Mimiviridae Host Specificity. PLoS ONE 2014, 9, e94923. [Google Scholar] [CrossRef]
- Borges, I.A.; De Assis, F.L.; Silva, L.K.D.S.; Abrahão, J. Rio Negro virophage: Sequencing of the near complete genome and transmission electron microscopy of viral factories and particles. Braz. J. Microbiol. 2018, 49, 260–261. [Google Scholar] [CrossRef]
- Michel, R.; Junglas, L.; Loch, S.; Wylezich, C.; Müller, K. Experimental co-infection of Saccamoeba lacustris with Mimivirus-like Giant virus and a small Satellite virus. Endocytobiosis Cell Res. Exp. 2018, 29, 1–6. [Google Scholar]
- Mougari, S.; Bekliz, M.; Abrahao, J.; Di Pinto, F.; Levasseur, A.; La Scola, B. Guarani Virophage, a New Sputnik-Like Isolate from a Brazilian Lake. Front. Microbiol. 2019, 10, 1–15. [Google Scholar] [CrossRef]
- Cohen, G.; Hoffart, L.; La Scola, B.; Raoult, D.; Drancourt, M. Ameba-associated Keratitis, France. Clin. Infect. Dis. 2009, 49, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Campos, R.K.; Boratto, P.V.; Assis, F.L.; Aguiar, E.R.; Silva, L.C.; Albarnaz, J.D.; Dornas, F.P.; Trindade, G.S.; Ferreira, P.P.; Marques, J.T.; et al. Samba virus: A novel mimivirus from a giant rain forest, the Brazilian Amazon. Virol. J. 2014, 11, 95. [Google Scholar] [CrossRef] [PubMed]
- Colson, P.; Gimenez, G.; Boyer, M.; Fournous, G.; Raoult, D. The Giant Cafeteria roenbergensis Virus That Infects a Widespread Marine Phagocytic Protist Is a New Member of the Fourth Domain of Life. PLoS ONE 2011, 6, e18935. [Google Scholar] [CrossRef] [PubMed]
- Michel, R.; Loch, S.; Dieter, K.; Junglas, L. Reisolation of Mimivirus-like giant viruses by using Saccamoeba sp. as bait resulted in loss of virophages. Endocytobiosis Cell Res. 2017, 28, 50–53. [Google Scholar]
- Yutin, N.; Shevchenko, S.; Kapitonov, V.; Krupovic, M.; Koonin, E.V. A novel group of diverse Polinton-like viruses discovered by metagenome analysis. BMC Biol. 2015, 13, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Chan, L.-K.; Egan, R.; Malmstrom, R.R.; McMahon, K.D.; Sullivan, M.B. Ecogenomics of virophages and their giant virus hosts assessed through time series metagenomics. Nat. Commun. 2017, 8, 858. [Google Scholar] [CrossRef] [PubMed]
- Bekliz, M.; Verneau, J.; Benamar, S.; Raoult, D.; La Scola, B.; Colson, P. A New Zamilon-like Virophage Partial Genome Assembled from a Bioreactor Metagenome. Front. Microbiol. 2015, 6, 403. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhang, W.; Yan, S.; Xiao, J.; Zhang, Y.; Li, B.; Pan, Y.; Wang, Y. Diversity of Virophages in Metagenomic Data Sets. J. Virol. 2013, 87, 4225–4236. [Google Scholar] [CrossRef]
- Zhou, J.; Sun, D.; Childers, A.; McDermott, T.R.; Wang, Y.; Liles, M.R. Three Novel Virophage Genomes Discovered from Yellowstone Lake Metagenomes. J. Virol. 2015, 89, 1278–1285. [Google Scholar] [CrossRef]
- Yutin, N.; Kapitonov, V.V.; Koonin, E.V. A new family of hybrid virophages from an animal gut metagenome. Biol. Direct 2015, 10, 19. [Google Scholar] [CrossRef]
- Gong, C.; Zhang, W.; Zhou, X.; Wang, H.; Sun, G.; Xiao, J.; Pan, Y.; Yan, S.; Wang, Y. Novel Virophages Discovered in a Freshwater Lake in China. Front. Microbiol. 2016, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Yoo, D.; Liu, W.T. Metagenomics Reveals a Novel Virophage Population in a Tibetan Mountain Lake. Microbes Environ. 2016, 31, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Kuhn, J.H.; Fischer, M.G. A classification system for virophages and satellite viruses. Arch. Virol. 2016, 161, 233–247. [Google Scholar] [CrossRef] [PubMed]
- Kapitonov, V.V.; Jurka, J. Self-synthesizing DNA transposons in eukaryotes. Proc. Natl. Acad. Sci. USA 2006, 103, 4540–4545. [Google Scholar] [CrossRef] [PubMed]
- Pritham, E.J.; Putliwala, T.; Feschotte, C. Mavericks, a novel class of giant transposable elements widespread in eukaryotes and related to DNA viruses. Gene 2007, 390, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Bamford, D.H.; Koonin, E.V. Conservation of major and minor jelly-roll capsid proteins in Polinton (Maverick) transposons suggests that they are bona fide viruses. Biol. Direct 2014, 9, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Yutin, N.; Raoult, D.; Koonin, E.V. Virophages, polintons, and transpovirons: A complex evolutionary network of diverse selfish genetic elements with different reproduction strategies. Virol. J. 2013, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Krupovič, M.; Yutin, N. Evolution of double-stranded DNA viruses of eukaryotes: From bacteriophages to transposons to giant viruses. Ann. N. Y. Acad. Sci. 2015, 1341, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Campbell, S.; Aswad, A.; Katzourakis, A. Disentangling the origins of virophages and polintons. Curr. Opin. Virol. 2017, 25, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Taylor, B.P.; Cortez, M.H.; Weitz, J.S. The virus of my virus is my friend: Ecological effects of virophage with alternative modes of coinfection. J. Theor. Biol. 2014, 354, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Desnues, C.; Raoult, D. Inside the Lifestyle of the Virophage. Intervirology 2010, 53, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Boyer, M.; Azza, S.; Barrassi, L.; Klose, T.; Campocasso, A.; Pagnier, I.; Fournous, G.; Borg, A.; Robert, C.; Zhang, X.; et al. Mimivirus shows dramatic genome reduction after intraamoebal culture. Proc. Natl. Acad. Sci. USA 2011, 108, 10296–10301. [Google Scholar] [CrossRef] [PubMed]
- Legendre, M.; Audic, S.; Poirot, O.; Hingamp, P.; Seltzer, V.; Byrne, D.; Lartigue, A.; Lescot, M.; Bernadac, A.; Poulain, J.; et al. mRNA deep sequencing reveals 75 new genes and a complex transcriptional landscape in Mimivirus. Genome Res. 2010, 20, 664–674. [Google Scholar] [CrossRef]
- Duponchel, S.; Fischer, M.G. Viva lavidaviruses! Five features of virophages that parasitize giant DNA viruses. PLOS Pathog. 2019, 15, e1007592. [Google Scholar] [CrossRef] [PubMed]
- Born, D.; Reuter, L.; Mersdorf, U.; Mueller, M.; Fischer, M.G.; Meinhart, A.; Reinstein, J. Capsid protein structure, self-assembly, and processing reveal morphogenesis of the marine virophage mavirus. Proc. Natl. Acad. Sci. USA 2018, 115, 7332–7337. [Google Scholar] [CrossRef] [PubMed]
- Andrade, A.C.S.P.; Rodrigues, R.A.L.; Oliveira, G.P.; Andrade, K.R.; Bonjardim, C.A.; La Scola, B.; Kroon, E.G.; Abrahão, J.S. Filling Knowledge Gaps for Mimivirus Entry, Uncoating, and Morphogenesis. J. Virol. 2017, 91, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Slimani, M.; Pagnier, I.; Raoult, D.; La Scola, B. Amoebae as Battlefields for Bacteria, Giant Viruses, and Virophages. J. Virol. 2013, 87, 4783–4785. [Google Scholar] [CrossRef] [PubMed]
- Marie, V.; Lin, J. Cannibalistic viruses in the aquatic environment: Role of virophages in manipulating microbial communities. Int. J. Environ. Sci. Technol. 2016, 13, 2097–2104. [Google Scholar] [CrossRef]
- Wodarz, D. Evolutionary dynamics of giant viruses and their virophages. Ecol. Evol. 2013, 3, 2103–2115. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.G. Virophages go nuclear in the marine alga Bigelowiella natans. Proc. Natl. Acad. Sci. USA 2015, 112, 11750–11751. [Google Scholar] [CrossRef] [PubMed]
- Berjón-Otero, M.; Koslová, A.; Fischer, M.G. The dual lifestyle of genome-integrating virophages in protists. Ann. N. Y. Acad. Sci. 2019, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Krupovic, M. Polintons, virophages and transpovirons: A tangled web linking viruses, transposons and immunity. Curr. Opin. Virol. 2017, 25, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.T.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef] [PubMed]
- Smargon, A.A.; Cox, D.B.T.; Pyzocha, N.K.; Slaymaker, I.M.; Gootenberg, J.S.; Abudayyeh, O.A.; Essletzbichler, P.; Shmakov, S.; Makarova, K.S.; Koonin, E.V. Cas13b is a Type VI-B CRISPR-associated RNA-Guided RNAse differentially regulated by accessory proteins Csx27 and Csx28. Mol. Cell 2017, 65, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Zhang, F. Prospects & Overviews Coupling immunity and programmed cell suicide in prokaryotes: Life-or-death choices. Bioessays 2016, 1600186, 1–9. [Google Scholar]
- Koonin, E.V. CRISPR: A new principle of genome engineering linked to conceptual shifts in evolutionary biology. Biol. Philos. 2019, 34, 9. [Google Scholar] [CrossRef]
- Krupovic, M.; Béguin, P.; Koonin, E.V. Casposons: Mobile genetic elements that gave rise to the CRISPR-Cas adaptation machinery. Curr. Opin. Microbiol. 2017, 38, 36–43. [Google Scholar] [CrossRef]
- Rose, J.A.; Berns, K.I.; Hoggan, M.D.; Koczot, F.J. Evidence for a Single-Stranded Adenovirus-Associated Virus Genome: Formation of a Dna Density Hybrid on Release of Viral DNA. Proc. Natl. Acad. Sci. USA 1969, 64, 863–869. [Google Scholar] [CrossRef]
- Muzyczka, N.; Bernst, K.I. Parvoviridae: The viruses and their replication. In Fields Virology; Wilkins: Philadelphia Fields, PA, USA, 2001. [Google Scholar]
- Cheung, A.K.; Hoggan, M.D.; Hauswirth, W.W.; Berns, I.K. Integration of the adeno-associated virus genome into cellular DNA in latently infected human Detroit 6 cells. J. Virol. 1980, 33, 739–748. [Google Scholar]
- Linden, R.M.; Winocour, E.; Berns, K.I. The recombination signals for adeno-associated virus site-specific integration. Proc. Natl. Acad. Sci. USA 1996, 93, 7966–7972. [Google Scholar] [CrossRef]
- Linden, R.M.; Ward, P.; Giraud, C.; Winocour, E.; Berns, K.I. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. USA 1996, 93, 11288–11294. [Google Scholar] [CrossRef] [PubMed]
- Mayor, H.D.; Drake, S.; Stahmann, J.; Mumford, D.M. Antibodies to adeno-associated satellite virus and herpes simplex in sera from cancer patients and normal adults. Am. J. Obstet. Gynecol. 1976, 126, 100–104. [Google Scholar] [CrossRef]
- Ward, P.; Elias, P.; Linden, R.M. Rescue of the Adeno-Associated Virus Genome from a Plasmid Vector: Evidence for Rescue by Replication. J. Virol. 2003, 77, 11480–11490. [Google Scholar] [CrossRef] [PubMed]
- Timpe, J.M.; Verrill, K.C.; Trempe, J.P. Effects of Adeno-Associated Virus on Adenovirus Replication and Gene Expression during Coinfection. J. Virol. 2006, 80, 7807–7815. [Google Scholar] [CrossRef] [PubMed]
- Meyers, C.; Mane, M.; Kokorina, N.; Alam, S.; Hermonat, P.L. Ubiquitous Human Adeno-Associated Virus Type 2 Autonomously Replicates in Differentiating Keratinocytes of a Normal Skin Model. Virology 2000, 272, 338–346. [Google Scholar] [CrossRef] [PubMed]
- Yalkinoglu, A.; Heilbronn, R.; Bürkle, A.; Schlehofer, J.; Harlard, Z.H. DNA Amplification of Adeno-associated Genotoxic Virus as a Response to Cellular Stress. CANCER Res. 1988, 48, 3123–3129. [Google Scholar] [PubMed]
- Chelkha, N.; Colson, P.; Levasseur, A.; La Scola, B. Deciphering the genomes of 16 Acanthamoeba species does not provide evidence of integration of known giant virus-associated mobile genetic elements. Virus Res. 2018, 251, 14–16. [Google Scholar] [CrossRef]
- Bekliz, M.; Levasseur, A.; La Scola, B.; Raoult, D. MIMIVIRE, un système de défense chez mimivirus qui illustre l’hypothèse de la Reine Rouge. Médecine/Sciences 2016, 32, 818–819. [Google Scholar] [CrossRef][Green Version]
- Dou, C.; Yu, M.; Gu, Y.; Cheng, W. Structural and mechanistic analyses reveal a unique Cas4-like protein in the mimivirus virophage resistance element system. iScience 2018, 3, 1–10. [Google Scholar] [CrossRef]
- Mougari, S.; Abrahao, J.; Oliveira, G.P.; Khalil, J.Y.B.; La Scola, B. Role of the R349 Gene and Its Repeats in the MIMIVIRE Defense System. Front. Microbiol. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Claverie, J.M.; Abergel, C. CRISPR-Cas-like system in giant viruses: Why MIMIVIRE is not likely to be an adaptive immune system. Virol. Sin. 2016, 31, 193–196. [Google Scholar] [CrossRef] [PubMed]
- Westra, E.R.; Semenova, E.; Datsenko, K.A.; Jackson, R.N.; Wiedenheft, B.; Severinov, K.; Brouns, S.J.J. Type I-E CRISPR-Cas Systems Discriminate Target from Non-Target DNA through Base Pairing-Independent PAM Recognition. PLoS Genet. 2013, 9, e1003742. [Google Scholar] [CrossRef] [PubMed]
- Nishimasu, H.; Cong, L.; Yan, W.X.; Ran, F.A.; Li, Y.; Kurabayashi, A.; Ishitani, R.; Zhang, F.; Nureki, O. Crystal structure of Staphylococcus aureus Cas9. Cell 2016, 162, 1113–1126. [Google Scholar] [CrossRef]
- Yamano, T.; Nishimasu, H.; Zetsche, B.; Hirano, H.; Slaymaker, I.M.; Li, Y.; Fedorova, I.; Nakane, T.; Makarova, K.S.; Koonin, E.V.; et al. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell 2016, 165, 949–962. [Google Scholar] [CrossRef] [PubMed]
- Deveau, H.; Garneau, J.E.; Moineau, S. CRISPR/Cas System and Its Role in Phage-Bacteria Interactions. Annu. Rev. Microbiol. 2010, 64, 475–493. [Google Scholar] [CrossRef] [PubMed]
- Sobhy, H. Virophages and Their Interactions with Giant Viruses and Host Cells. Proteomes 2018, 6, 23. [Google Scholar] [CrossRef]
- Gaia, M.; Colson, P.; Desnues, C.; La Scola, B. Virophage Concept, The; The eLS. John Wiley Sons: Chichester, UK, 2013; pp. 1–12. [Google Scholar]
- Krupovic, M. Virophages or satellite viruses? Nat. Rev. Microbiol. 2011, 9, 762–764. [Google Scholar] [CrossRef]
- Krupovič, M.; Cvirkaitė-Krupovič, V. Sputnik and Mavirus: Not more than satellite viruses. Nat. Rev. Genet. 2011, 10, 78. [Google Scholar] [CrossRef]
- Bawden, F.C.; Pirie, N.W. A Preliminary Description of Preparations of Some of the Viruses Causing Tobacco Necrosis. Br. J. Exp. Pathol. 1942, 23, 314–327. [Google Scholar]
- Kassanis, B. Properties and Behaviour of a Virus Depending for its Multiplication on Another. J. Gen. Microbiol. 1902, 27, 477–488. [Google Scholar] [CrossRef]
- Van, M.Y.J.; Emmelo, W.F. Total nucleotide sequence of a nearly full-size DNA copy of satellite tobacco necrosis virus RNA. J. Mol. Biol. 1980, 143, 273–287. [Google Scholar]
- Reichmann, E. The Satellite Tobacco Necrosis Virus: A Single Protein and Its Genetic Code. Proc. Natl. Acad. Sci. USA 1964, 52, 1009–1017. [Google Scholar] [CrossRef] [PubMed]
- Hoggan, M.D.; Blacklow, N.R.; Rowe, W.P. Studies of small DNA viruses found in various adenovirus preparations: Physical, biological, and immunological characteristics. Proc. Natl. Acad. Sci. USA 1966, 55, 1467–1474. [Google Scholar] [CrossRef] [PubMed]
- Hastie, E.; Samulski, R.J. Adeno-Associated Virus at 50: A Golden Anniversary of Discovery, Research, and Gene Therapy Success—A Personal Perspective. Hum. Gene Ther. 2015, 26, 257–265. [Google Scholar] [CrossRef]
- Buller, R.M.L.; Janik, J.E.; Sebring, E.D.; Rose, J.A. Herpes Simplex Virus Types 1 and 2 Completely Help Adenovirus-Associated Virus Replication. J. Virol. 1981, 40, 241–247. [Google Scholar] [PubMed]
- McPherson, R.A.; Rosenthal, L.J.; Rose, J.A. Human cytomegalovirus completely helps adeno-associated virus replication. Virology 1985, 147, 217–222. [Google Scholar] [CrossRef]
- Walz, C.; Deprez, A.; Dupressoir, T.; Du, M.; Schlehofer, R. Interaction of human papillomavirus type 16 and adeno-associated virus type 2 co-infecting human cervical epithelium. J. Gen. Virol. 1997, 78, 1441–1452. [Google Scholar] [CrossRef][Green Version]
- Wang, Z.; Deng, X.; Zou, W.; Engelhardt, J.F.; Yan, Z. Human Bocavirus 1 Is a Novel Helper for. J. Virol. 2017, 91, 1–18. [Google Scholar] [CrossRef]
- Carter, B.J.; Laughlin, C.A.; De La Maza, L.M.; Myers, M. Adeno-associated virus autointerference. Virology 1979, 92, 449–462. [Google Scholar] [CrossRef]
- Casto, B.C.; Atchison, R.W.; Hammon, W.M. Studies on the relationship between adeno-associated virus type I (AAV-1) and adenoviruses. I. Replication of AAV-1 in certain cell cultures and its effect on helper adenovirus. Virology 1967, 32, 52–59. [Google Scholar] [CrossRef]
- Laughlin, A.; Myers, W.; Risin, L. Defective-Interfering Particles of the Human Parvovirus Adeno-Associated Virus. Virology 1979, 94, 162–174. [Google Scholar] [CrossRef]
- Desnues, C.; Raoult, D. Virophages question the existence of satellites. Nat. Rev. Genet. 2012, 10, 234. [Google Scholar] [CrossRef] [PubMed]
| Virophage | Year of Description | Place of Isolation | Associated Giant Virus | Capsid Size (nm) | Genome Features | GenBank/EBI Accession No. | References | ||
|---|---|---|---|---|---|---|---|---|---|
| Size (bp) | Number of ORFs | G+C (%) | |||||||
| Sputnik | 2008 | France | Mamavirus | 50–74 | 18,343 | 21 | 27 | EU606015 | [30] |
| Mavirus | 2010 | USA | Cafeteria roenbergensis virus | 60 | 19,063 | 20 | 30 | NC_015230 | [87] |
| Sputnik 2 | 2012 | France | Lentillevirus | 50–74 | 18,338 | 20 | 27 | NC_023846 | [38] |
| Sputnik 3 | 2013 | France | APMV 1 | 50–74 | 18,338 | 20 | 27 | NC_023847 | [88] |
| PgVV | 2013 | North Sea | Phaeocystis globosa virus | N.d. | 19,527 | 16 | 36 | NC_021333 | [62] |
| Zamilon | 2014 | Tunisia | Mont1 | 60 | 17,276 | 20 | 30 | NC_022990 | [89] |
| RNV | 2014 | Brazil | Sambavirus | 50–70 | 18,145 | 20 | 27 | MG676470 | [90] |
| Platanovirus saccamoebae virophage | 2018 | Germany | KSL5 virus | 50 | N.d. | N.d. | N.d. | N.d. | [91] |
| Guarani | 2019 | Brazil | APMV 1 | 50–74 | 18,967 | 22 | 26 | LS999520 | [92] |
| 3 CpV-PLV virophages | 2019 | USA | CpV-BQ2 | N.d. | 21,750- 22,879 | 19–23 | 30–39 | MH920636/ MH919296/ MH919297 | [69] |
| Virophage | Year of Description | Origin of the Metagenome | Associated Giant Virus | Genome Features | Genbenk/EBI Accession No. | References | ||
|---|---|---|---|---|---|---|---|---|
| Size (bp) | Number of ORFs | G+C (%) | ||||||
| OLV | 2011 | Antarctica | Organic lake phycodnavirus | 26,421 | 24 | 37 | HQ704801 | [55] |
| YSLV1 | 2013 | USA | N.d. | 27,849 | 26 | 33 | KC556924 | [100] |
| YLSV2 | 2013 | USA | N.d. | 23,184 | 21 | 34 | KC556925 | [100] |
| YLSV3 | 2013 | USA | N.d. | 27,05 | 23 | 35 | KC556926 | [100] |
| YLSV4 | 2013 | USA | N.d. | 28,306 | 34 | 37 | KC556922 | [100] |
| ALM | 2013 | Antarctica | N.d. | 17,767 | 22 | 27 | KC556923 | [100] |
| YLSV5 | 2014 | USA | N.d. | 29,767 | 32 | 51 | KM502589 | [101] |
| YLSV6 | 2014 | USA | N.d. | 24,837 | 29 | 27 | KM502590 | [101] |
| YLSV7 | 2014 | USA | N.d. | 23,193 | 26 | 27 | KM502591 | [101] |
| 3 RVPs (incompletes) | 2015 | USA | Mimiviridae | Up to 26, 209 | Up to 21 | - | - | [102] |
| Zamilon 2 (incomplete) | 2015 | USA | Mimiviridae | 6716 | 15 | 32 | - | [99] |
| QLV | 2016 | China | N.d. | 23,379 | 25 | 33 | KJ854379 | [104] |
| DSLV1 | 2016 | China | N.d. | 28,788 | 28 | 43 | KT894027 | [103] |
| 17 Freshwater virophages (10 incompletes) | 2017 | USA | Mimiviridae | 13,800–25,800 | 13–25 | - | - | [98] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mougari, S.; Sahmi-Bounsiar, D.; Levasseur, A.; Colson, P.; La Scola, B. Virophages of Giant Viruses: An Update at Eleven. Viruses 2019, 11, 733. https://doi.org/10.3390/v11080733
Mougari S, Sahmi-Bounsiar D, Levasseur A, Colson P, La Scola B. Virophages of Giant Viruses: An Update at Eleven. Viruses. 2019; 11(8):733. https://doi.org/10.3390/v11080733
Chicago/Turabian StyleMougari, Said, Dehia Sahmi-Bounsiar, Anthony Levasseur, Philippe Colson, and Bernard La Scola. 2019. "Virophages of Giant Viruses: An Update at Eleven" Viruses 11, no. 8: 733. https://doi.org/10.3390/v11080733
APA StyleMougari, S., Sahmi-Bounsiar, D., Levasseur, A., Colson, P., & La Scola, B. (2019). Virophages of Giant Viruses: An Update at Eleven. Viruses, 11(8), 733. https://doi.org/10.3390/v11080733





