Crystal Structure of the Japanese Encephalitis Virus Capsid Protein
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Construction
2.2. Protein Expression and Purification
2.3. Mass-Spectrometry Analysis
2.4. Protein Crystallization
2.5. Diffraction Experiment, Data Processing, and Model Building
2.6. Data Deposition
3. Results and Discussion
3.1. The JEV Capsid Protein N-terminus is Prone to Proteolytic Cleavage
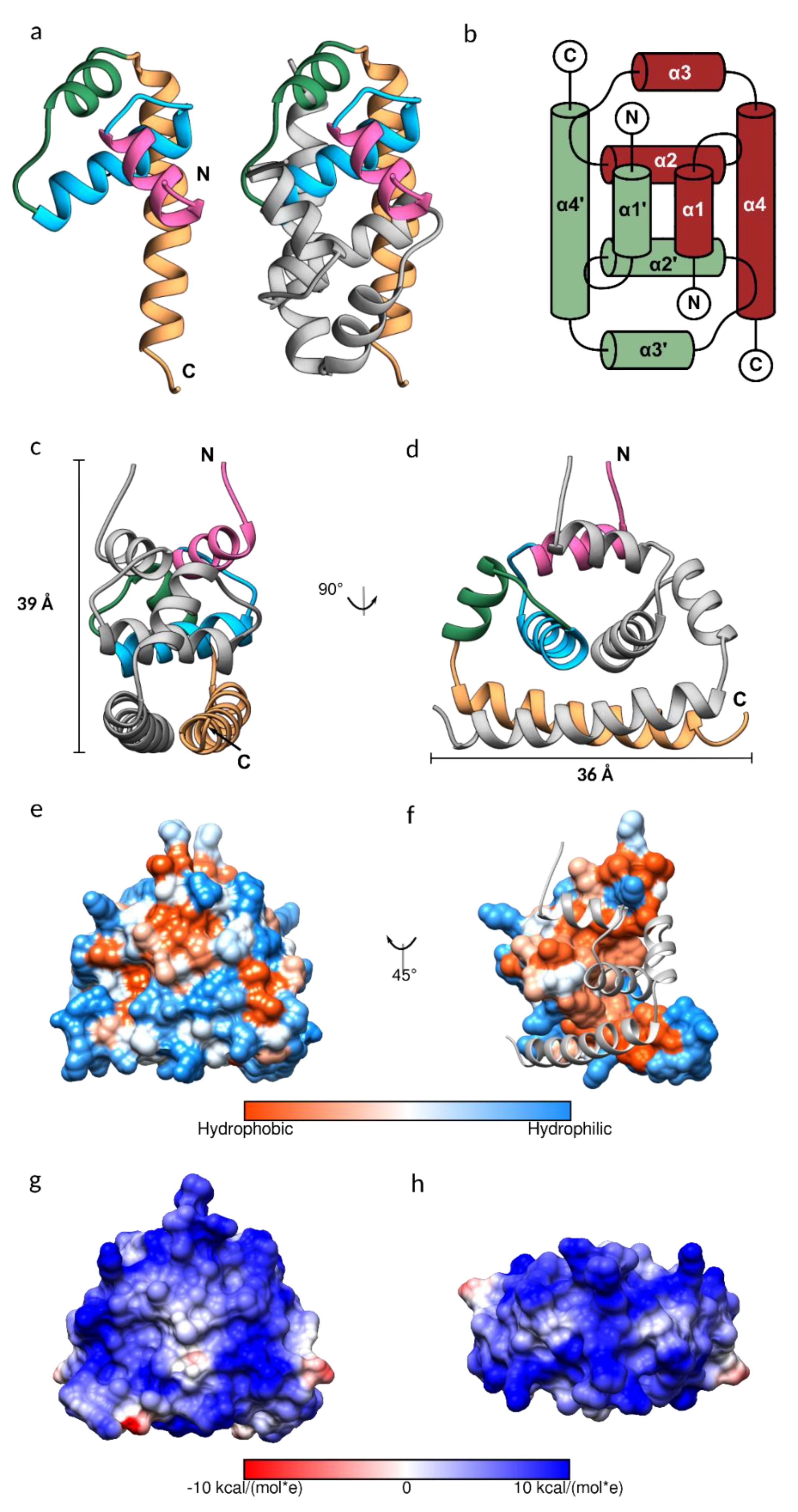
3.2. JEV Capsid Protein Structure
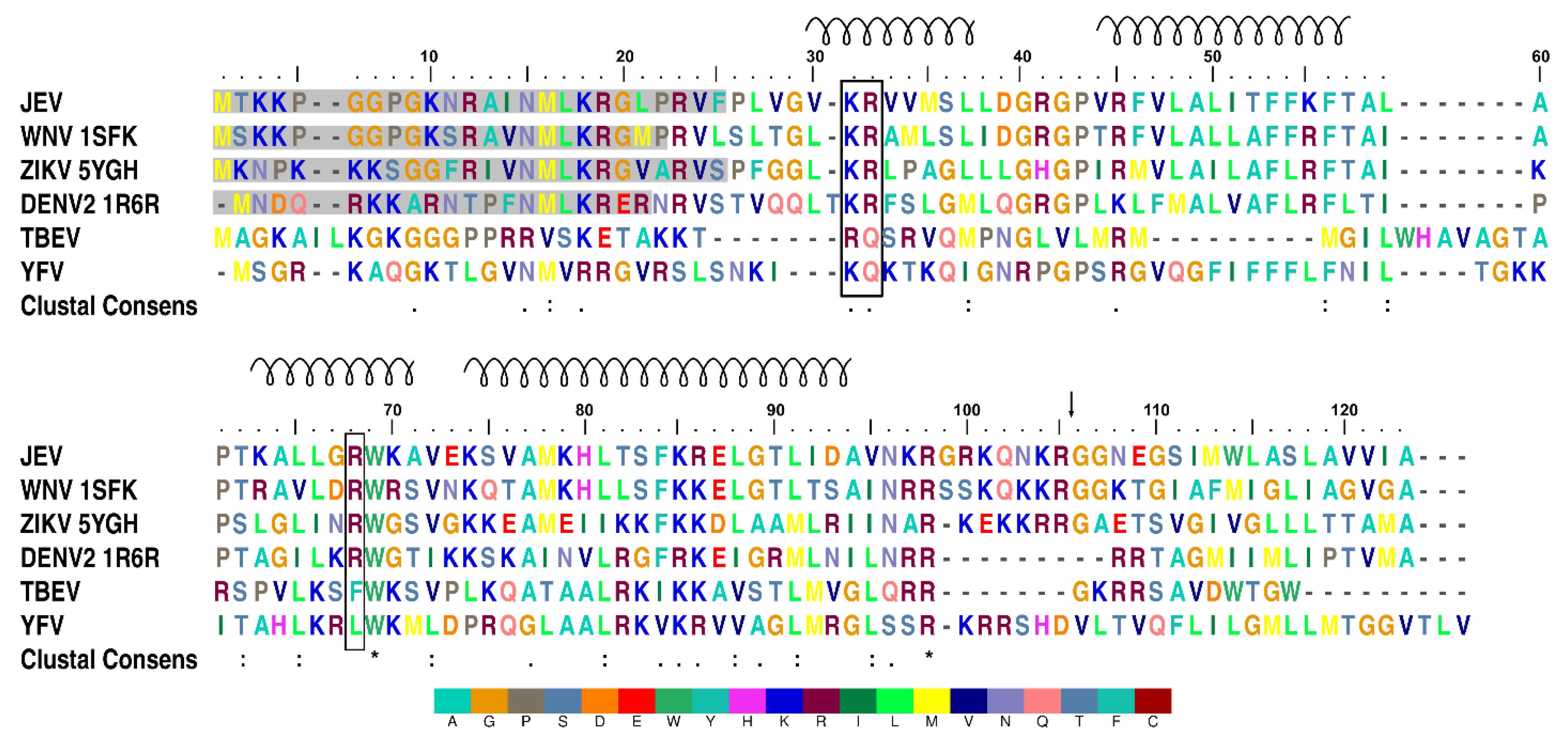
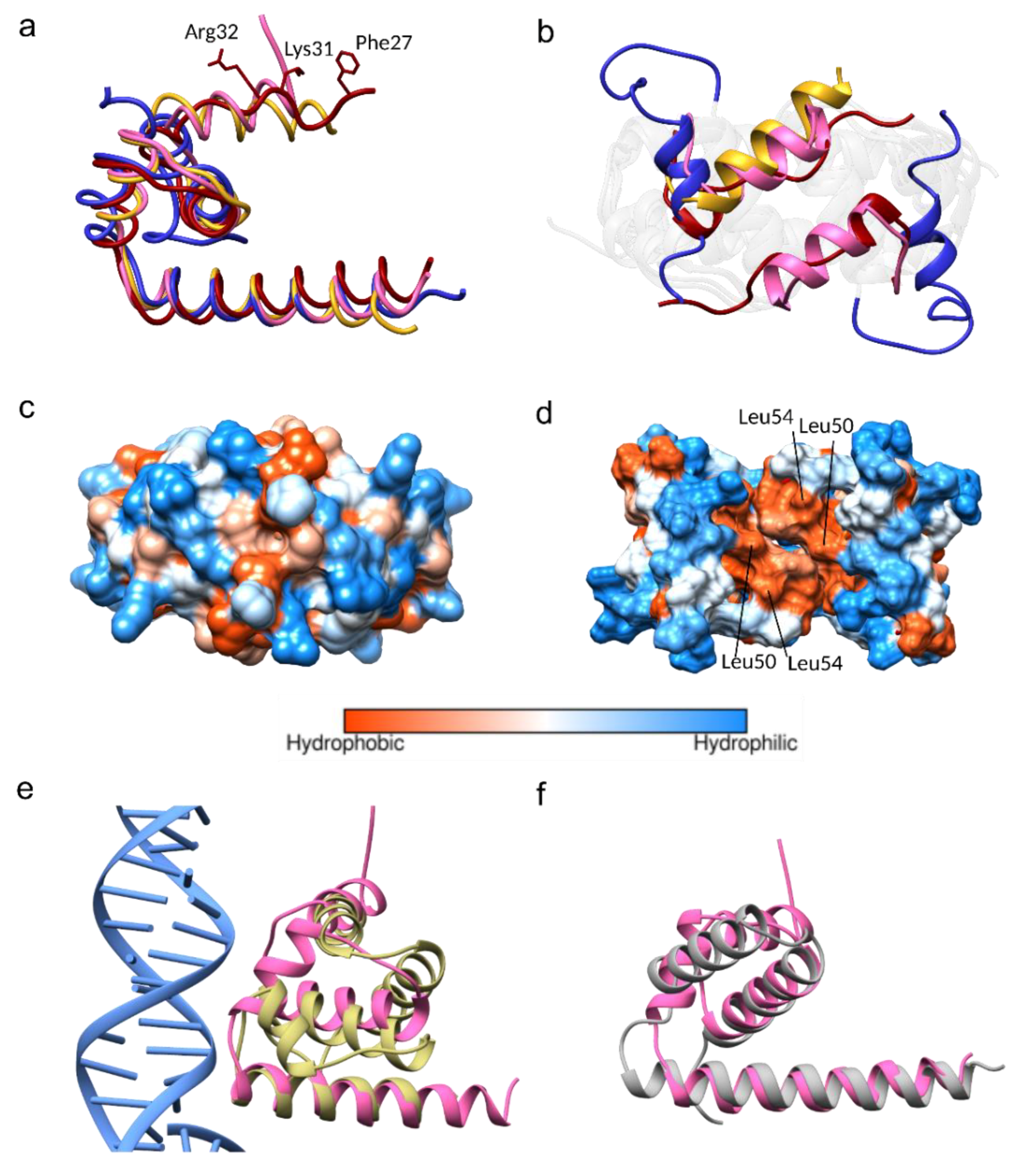
3.3. Conformational Plasticity of α-helix-1
3.4. Homologous Protein Superposition
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Turtle, L.; Solomon, T. Japanese encephalitis—the prospects for new treatments. Nat. Rev. Neurol. 2018, 14, 298–313. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, R.J.; Zhang, W.; Rossmann, M.G.; Pletnev, S.V.; Corver, J.; Lenches, E.; Jones, C.T.; Mukhopadhyay, S.; Chipman, P.R.; Strauss, E.G.; et al. Structure of dengue virus: Implications for flavivirus organization, maturation, and fusion. Cell 2002, 108, 717–725. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Kuhn, R.J.; Rossmann, M.G. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 2005, 3, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Perera, R.; Kuhn, R.J. Structural proteomics of dengue virus. Curr. Opin. Microbiol. 2008, 11, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, S.H.; Zhu, L.; Nian, Q.G.; Yuan, S.; Gao, Q.; Hu, Z.; Ye, Q.; Li, X.F.; Xie, D.Y.; et al. Near-atomic structure of Japanese encephalitis virus reveals critical determinants of virulence and stability. Nat. Commun. 2017, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Welsch, S.; Miller, S.; Romero-Brey, I.; Merz, A.; Bleck, C.K.; Walther, P.; Fuller, S.D.; Antony, C.; Krijnse-Locker, J.; Bartenschlager, R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 2009, 5, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Byk, L.A.; Gamarnik, A.V. Properties and Functions of the Dengue Virus Capsid Protein. Annu. Rev. Virol. 2016, 3, 263–281. [Google Scholar] [CrossRef]
- Oliveira, E.R.; Mohana-Borges, R.; de Alencastro, R.B.; Horta, B.A. The flavivirus capsid protein: Structure, function and perspectives towards drug design. Virus Res. 2017, 227, 115–123. [Google Scholar] [CrossRef]
- Aljanabi, S.M.; Martinez, I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997, 25, 4692–4693. [Google Scholar] [CrossRef]
- Samsa, M.M.; Mondotte, J.A.; Caramelo, J.J.; Gamarnik, A.V. Uncoupling cis-Acting RNA elements from coding sequences revealed a requirement of the N-terminal region of dengue virus capsid protein in virus particle formation. J. Virol. 2012, 86, 1046–1058. [Google Scholar] [CrossRef]
- Ivanyi-Nagy, R.; Lavergne, J.P.; Gabus, C.; Ficheux, D.; Darlix, J.L. RNA chaperoning and intrinsic disorder in the core proteins of Flaviviridae. Nucleic Acids Res. 2008, 36, 712–725. [Google Scholar] [CrossRef] [PubMed]
- Pong, W.L.; Huang, Z.S.; Teoh, P.G.; Wang, C.C.; Wu, H.N. RNA binding property and RNA chaperone activity of dengue virus core protein and other viral RNA-interacting proteins. FEBS Lett. 2011, 585, 2575–2581. [Google Scholar] [CrossRef] [PubMed]
- Khromykh, A.A.; Westaway, E.G. RNA binding properties of core protein of the flavivirus Kunjin. Arch. Virol. 1996, 141, 685–699. [Google Scholar] [CrossRef] [PubMed]
- Bulich, R.; Aaskov, J.G. Nuclear localization of dengue 2 virus core protein detected with monoclonal antibodies. J. Gen. Virol. 1992, 73, 2999–3003. [Google Scholar] [CrossRef] [PubMed]
- Shang, Z.; Song, H.; Shi, Y.; Qi, J.; Gao, G.F. Crystal Structure of the Capsid Protein from Zika Virus. J. Mol. Biol. 2018, 430, 948–962. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.H.; Syu, W.J.; Huang, K.J.; Lei, H.Y.; Yao, C.W.; King, C.C.; Hu, S.T. Intracellular localization and determination of a nuclear localization signal of the core protein of dengue virus. J. Gen. Virol. 2002, 83, 3093–3102. [Google Scholar] [CrossRef] [PubMed]
- Schlick, P.; Taucher, C.; Schittl, B.; Tran, J.L.; Kofler, R.M.; Schueler, W.; von Gabain, A.; Meinke, A.; Mandl, C.W. Helices alpha2 and alpha3 of West Nile virus capsid protein are dispensable for assembly of infectious virions. J. Virol. 2009, 83, 5581–5591. [Google Scholar] [CrossRef] [PubMed]
- Markoff, L.; Falgout, B.; Chang, A. A conserved internal hydrophobic domain mediates the stable membrane integration of the dengue virus capsid protein. Virology 1997, 233, 105–117. [Google Scholar] [CrossRef]
- Samsa, M.M.; Mondotte, J.A.; Iglesias, N.G.; Assuncao-Miranda, I.; Barbosa-Lima, G.; Da Poian, A.T.; Bozza, P.T.; Gamarnik, A.V. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 2009, 5, e1000632. [Google Scholar] [CrossRef]
- Luca, V.C.; AbiMansour, J.; Nelson, C.A.; Fremont, D.H. Crystal structure of the Japanese encephalitis virus envelope protein. J. Virol. 2012, 86, 2337–2346. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, X.; Na, R.; Li, L.; Warkentin, E.; Witt, J.; Lu, X.; Yu, Y.; Wei, Y.; Peng, G.; et al. The structure differences of Japanese encephalitis virus SA14 and SA14-14-2 E proteins elucidate the virulence attenuation mechanism. Protein Cell 2019, 10, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Weinert, T.; Olieric, V.; Waltersperger, S.; Panepucci, E.; Chen, L.; Zhang, H.; Zhou, D.; Rose, J.; Ebihara, A.; Kuramitsu, S.; et al. Fast native-SAD phasing for routine macromolecular structure determination. Nat. Methods 2015, 12, 131–133. [Google Scholar] [CrossRef] [PubMed]
- Crystal Structure of Catalytic Domain of Japanese Encephalitis Virus NS3 Helicase/Nucleoside Triphosphatase at a Resolution 1.8 Å. Available online: http://www.rcsb.org/structure/2Z83 (accessed on 23 June 2019).
- Surana, P.; Satchidanandam, V.; Nair, D.T. RNA-dependent RNA polymerase of Japanese encephalitis virus binds the initiator nucleotide GTP to form a mechanistically important pre-initiation state. Nucleic Acids Res. 2014, 42, 2758–2773. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Gong, P. Crystal Structure of the full-length Japanese encephalitis virus NS5 reveals a conserved methyltransferase-polymerase interface. PLoS Pathog. 2013, 9, e1003549. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, T.; Konishi, E. Potential chemotherapeutic targets for Japanese encephalitis: Current status of antiviral drug development and future challenges. Expert Opin. Ther. Targets 2015, 19, 1379–1395. [Google Scholar] [CrossRef] [PubMed]
- Eyer, L.; Nencka, R.; de Clercq, E.; Seley-Radtke, K.; Ruzek, D. Nucleoside analogs as a rich source of antiviral agents active against arthropod-borne flaviviruses. Antivir. Chem. Chemother. 2018, 26, 2040206618761299. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, M.; Umeyama, H.; Iwadate, M.; Tanahashi, T.; Yano, Y.; Enomoto, M.; Tamori, A.; Kawada, N.; Murakami, Y. Development of novel hepatitis B virus capsid inhibitor using in silico screening. Biochem. Biophys. Res. Commun. 2015, 463, 1165–1175. [Google Scholar] [CrossRef] [PubMed]
- Makarov, V.A.; Braun, H.; Richter, M.; Riabova, O.B.; Kirchmair, J.; Kazakova, E.S.; Seidel, N.; Wutzler, P.; Schmidtke, M. Pyrazolopyrimidines: Potent Inhibitors Targeting the Capsid of Rhino- and Enteroviruses. ChemMedChem 2015, 10, 1629–1634. [Google Scholar] [CrossRef]
- Tang, C.; Loeliger, E.; Kinde, I.; Kyere, S.; Mayo, K.; Barklis, E.; Sun, Y.; Huang, M.; Summers, M.F. Antiviral inhibition of the HIV-1 capsid protein. J. Mol. Biol. 2003, 327, 1013–1020. [Google Scholar] [CrossRef]
- Poonsiri, T.; Wright, G.S.; Diamond, M.S.; Turtle, L.; Solomon, T.; Antonyuk, S.V. Structural study of the C-terminal domain of nonstructural protein 1 from Japanese encephalitis virus. Journal of Virology 2018, 92, e01868-17. [Google Scholar] [CrossRef]
- Shiryaev, S.A.; Kozlov, I.A.; Ratnikov, B.I.; Smith, J.W.; Lebl, M.; Strongin, A.Y. Cleavage preference distinguishes the two-component NS2B–NS3 serine proteinases of Dengue and West Nile viruses. Biochem. J. 2007, 401, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Winter, G. xia2: An expert system for macromolecular crystallography data reduction. J. Appl. Cryst. 2010, 43, 186–190. [Google Scholar] [CrossRef]
- Cowtan, K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta. Crystallogr. D. Biol. Crystallogr. 2006, 62, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- Murshudov, G.N.; Skubak, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta. Crystallogr. D. Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta. Crystallogr. D. Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Ma, L.; Jones, C.T.; Groesch, T.D.; Kuhn, R.J.; Post, C.B. Solution structure of dengue virus capsid protein reveals another fold. Proc. Natl. Acad. Sci. USA 2004, 101, 3414–3419. [Google Scholar] [CrossRef]
- Dokland, T.; Walsh, M.; Mackenzie, J.M.; Khromykh, A.A.; Ee, K.H.; Wang, S. West Nile virus core protein; tetramer structure and ribbon formation. Structure 2004, 12, 1157–1163. [Google Scholar] [CrossRef]
- Mori, Y.; Yamashita, T.; Tanaka, Y.; Tsuda, Y.; Abe, T.; Moriishi, K.; Matsuura, Y. Processing of capsid protein by cathepsin L plays a crucial role in replication of Japanese encephalitis virus in neural and macrophage cells. J. Virol. 2007, 81, 8477–8487. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kiermayr, S.; Kofler, R.M.; Mandl, C.W.; Messner, P.; Heinz, F.X. Isolation of capsid protein dimers from the tick-borne encephalitis flavivirus and in vitro assembly of capsid-like particles. J. Virol. 2004, 78, 8078–8084. [Google Scholar] [CrossRef]
- Hakoshima, T. Leucine Zippers. In Encyclopedia of Life Sciences; John Wiley & Sons, Ltd.: Chichester, UK, 2005. [Google Scholar] [CrossRef]
- Zhang, Y.; Corver, J.; Chipman, P.R.; Zhang, W.; Pletnev, S.V.; Sedlak, D.; Baker, T.S.; Strauss, J.H.; Kuhn, R.J.; Rossmann, M.G. Structures of immature flavivirus particles. EMBO J. 2003, 22, 2604–2613. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ge, P.; Yu, X.; Brannan, J.M.; Bi, G.; Zhang, Q.; Schein, S.; Zhou, Z.H. Cryo-EM structure of the mature dengue virus at 3.5-A resolution. Nat. Struct. Mol. Biol. 2013, 20, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Prasad, V.M.; Miller, A.S.; Klose, T.; Sirohi, D.; Buda, G.; Jiang, W.; Kuhn, R.J.; Rossmann, M.G. Structure of the immature Zika virus at 9 A resolution. Nat. Struct. Mol. Biol. 2017, 24, 184–186. [Google Scholar] [CrossRef] [PubMed]
- Martins, I.C.; Gomes-Neto, F.; Faustino, A.F.; Carvalho, F.A.; Carneiro, F.A.; Bozza, P.T.; Mohana-Borges, R.; Castanho, M.A.; Almeida, F.C.; Santos, N.C.; et al. The disordered N-terminal region of dengue virus capsid protein contains a lipid-droplet-binding motif. Biochem. J. 2012, 444, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.T.; Ma, L.; Burgner, J.W.; Groesch, T.D.; Post, C.B.; Kuhn, R.J. Flavivirus capsid is a dimeric alpha-helical protein. J. Virol. 2003, 77, 7143–7149. [Google Scholar] [CrossRef] [PubMed]
- Byrd, C.M.; Dai, D.; Grosenbach, D.W.; Berhanu, A.; Jones, K.F.; Cardwell, K.B.; Schneider, C.; Wineinger, K.A.; Page, J.M.; Harver, C.; et al. A Novel Inhibitor of Dengue Virus Replication That Targets the Capsid Protein. Antimicrob. Agents Chemother. 2013, 57, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Holm, L.; Rosenstrom, P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010, 38, W545–W549. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poonsiri, T.; Wright, G.S.A.; Solomon, T.; Antonyuk, S.V. Crystal Structure of the Japanese Encephalitis Virus Capsid Protein. Viruses 2019, 11, 623. https://doi.org/10.3390/v11070623
Poonsiri T, Wright GSA, Solomon T, Antonyuk SV. Crystal Structure of the Japanese Encephalitis Virus Capsid Protein. Viruses. 2019; 11(7):623. https://doi.org/10.3390/v11070623
Chicago/Turabian StylePoonsiri, Thanalai, Gareth S. A. Wright, Tom Solomon, and Svetlana V. Antonyuk. 2019. "Crystal Structure of the Japanese Encephalitis Virus Capsid Protein" Viruses 11, no. 7: 623. https://doi.org/10.3390/v11070623
APA StylePoonsiri, T., Wright, G. S. A., Solomon, T., & Antonyuk, S. V. (2019). Crystal Structure of the Japanese Encephalitis Virus Capsid Protein. Viruses, 11(7), 623. https://doi.org/10.3390/v11070623





