Hypsugopoxvirus: A Novel Poxvirus Isolated from Hypsugo savii in Italy
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Virological Analysis
2.3. Molecular Analysis
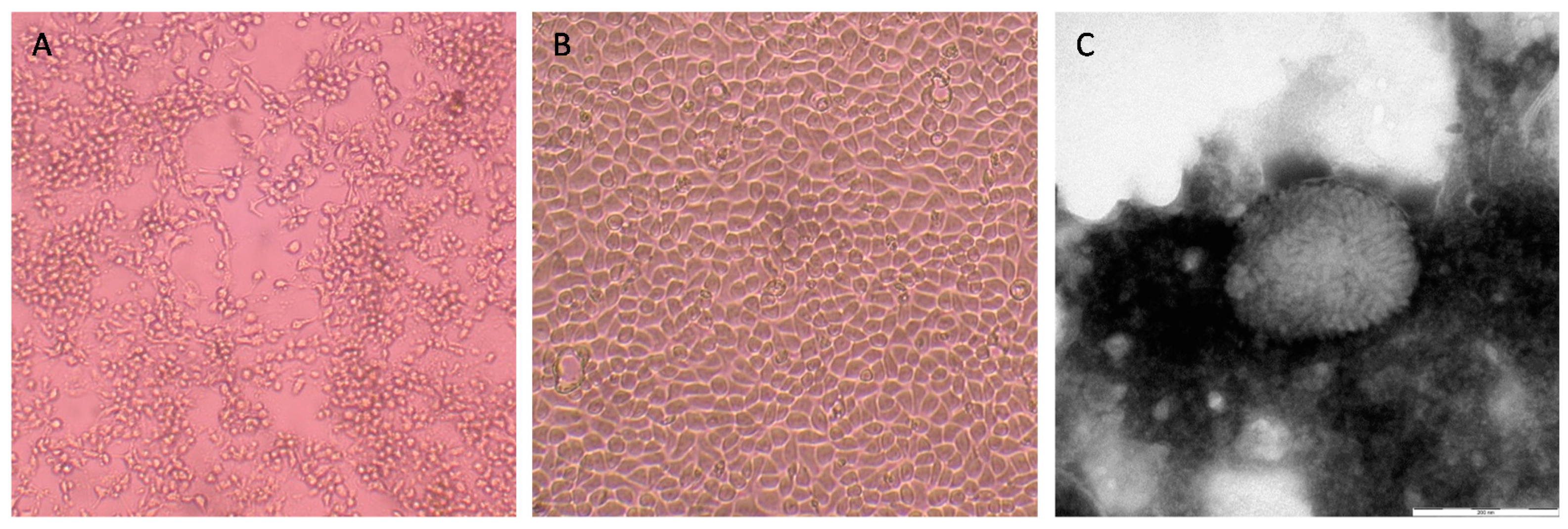
3. Results
3.1. Clinical Case
3.2. Virus Isolation and Identification
3.3. Genome Characterization
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Adams, M.J.; Lefkowitz, E.J.; King, A.M.Q.; Harrach, B.; Harrison, R.L.; Knowles, N.J.; Kropinski, A.M.; Krupovic, M.; Kuhn, J.H.; Mushegian, A.R.; et al. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2016). Arch. Virol. 2016, 161, 2921–2949. [Google Scholar] [CrossRef] [PubMed]
- Shchelkunov, S.N. An increasing danger of zoonotic orthopoxvirus infections. PLoS Pathog. 2013, 9, e1003756. [Google Scholar] [CrossRef] [PubMed]
- Calisher, C.H.; Childs, J.E.; Field, H.E.; Holmes, K.V.; Schountz, T. Bats: Important reservoir hosts of emerging virus. Clin. Microbiol. 2006, 19, 531–545. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.F.; Walker, P.J.; Poon, L.L. Mass extinctions, biodiversity and mitochondrial function: Are bats “special” as reservoirs for emerging viruses? Curr. Opin. Virol. 2011, 1, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.L.; Schountz, T.; Wang, L.F. Antiviral immune responses of bats: A review. Zoonoses Public Health 2013, 60, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Luis, A.D.; Hayman, D.T.; O’Shea, T.J.; Cryan, P.M.; Gilbert, A.T.; Pulliam, J.R.; Mills, J.N.; Timonin, M.E.; Willis, C.K.; Cunningham, A.A.; et al. A comparison of bats and rodents as reservoirs of zoonotic viruses: Are bats special? Proc. Biol. Sci. 2013, 280, 20122753. [Google Scholar] [CrossRef] [PubMed]
- Olival, K.J.; Hosseini, P.R.; Zambrana-Torrelio, C.; Ross, N.; Bogich, T.L.; Daszak, P. Host and viral traits predict zoonotic spillover from mammals. Nature 2017, 546, 646–650. [Google Scholar] [CrossRef] [PubMed]
- Baker, K.S.; Murcia, P.R. Poxviruses in bats … so what? Viruses 2014, 6, 1564–1577. [Google Scholar] [CrossRef] [PubMed]
- Emerson, G.L.; Nordhausen, R.; Garner, M.M.; Huckabee, J.R.; Johnson, S.; Wohrle, R.D.; Davidson, W.B.; Wilkins, K.; Li, Y.; Doty, J.B.; et al. Novel poxvirus in big brown bats, northwestern United States. Emerg. Infect. Dis. 2013, 19, 1002–1004. [Google Scholar] [CrossRef]
- Tu, S.L.; Nakazawa, Y.; Gao, J.; Wilkins, K.; Gallardo-Romero, N.; Li, Y.; Emerson, G.L.; Carroll, D.S.; Upton, C. Characterization of Eptesipoxvirus, a novel poxvirus from a microchiropteran bat. Virus Genes 2017, 53, 856–867. [Google Scholar] [CrossRef]
- Baker, K.S.; Leggett, R.M.; Bexfield, N.H.; Alston, M.; Daly, G.; Todd, S.; Tachedjian, M.; Holmes, C.E.; Crameri, S.; Wang, L.F.; et al. Metagenomic study of the viruses of African straw-coloured fruit bats: Detection of a chiropteran poxvirus and isolation of a novel adenovirus. Virology 2013, 441, 95–106. [Google Scholar] [CrossRef] [PubMed]
- O’Dea, M.A.; Tu, S.L.; Pang, S.; de Ridder, T.; Jackson, B.; Upton, C. Genomic characterization of a novel poxvirus from a flying fox: Evidence for a new genus? J. Gen. Virol. 2016, 97, 2363–2375. [Google Scholar] [CrossRef] [PubMed]
- McLelland, D.J.; Reardon, T.; Bourne, S.; Dickason, C.; Kessell, A.; Boardman, W. Outbreak of Skin Nodules associated with Riouxgolvania beveridgei (Nematoda: Muspiceida) in the Southern Bentwing Bat (Miniopterus schreibersii bassanii), South Australia. J. Wildl. Dis. 2013, 49, 1009–1013. [Google Scholar] [CrossRef] [PubMed]
- Lelli, D.; Moreno, A.; Lavazza, A.; Bresaola, M.; Canelli, E.; Boniotti, M.B.; Cordioli, P. Identification of Mammalian Orthoreovirus type 3 in Italian bats. Zoonosis Public Health 2013, 60, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Lelli, D.; Papetti, A.; Sabelli, C.; Rosti, E.; Moreno, A.; Boniotti, M.B. Detection of coronaviruses in bats of various species in Italy. Viruses 2013, 5, 2679–2689. [Google Scholar] [CrossRef] [PubMed]
- Lelli, D.; Prosperi, A.; Moreno, A.; Chiapponi, C.; Gibellini, A.M.; De Benedictis, P.; Leopardi, S.; Sozzi, E.; Lavazza, A. Isolation of a novel Rhabdovirus from an insectivorous bat (Pipistrellus kuhlii) in Italy. Virol. J. 2018, 15, 37. [Google Scholar] [CrossRef] [PubMed]
- Lavazza, A.; Pascucci, S.; Gelmetti, D. Rod-shaped virus-like particles in intestinal contents of three avian species. Vet. Rec. 1990, 126, 581. [Google Scholar]
- Da Silva, M.; Upton, C. Bioinformatics for analysis of poxvirus genomes. Methods Mol. Biol. 2012, 890, 233–258. [Google Scholar]
- Tcherepanov, V.; Ehlers, A.; Upton, C. Genome Annotation Transfer Utility (GATU): Rapid annotation of viral genomes using a closely related reference genome. BMC Genom. 2006, 7, 150. [Google Scholar] [CrossRef]
- Balboni, A.; Gallina, L.; Palladini, A.; Prosperi, S.; Battilani, M. A real-time PCR assay for bat SARS-like coronavirus detection and its application to Italian greater horseshoe bat faecal sample surveys. Sci. World J. 2012, 2012, 989514. [Google Scholar] [CrossRef]
- De Benedictis, P.; Marciano, S.; Scaravelli, D.; Priori, P.; Zecchin, B.; Capua, I.; Monne, I.; Cattoli, G. Alpha and lineage C betaCoV infections in Italian bats. Virus Genes 2014, 48, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.; Lelli, D.; de Sabato, L.; Zaccaria, G.; Boni, A.; Sozzi, E.; Prosperi, A.; Lavazza, A.; Cella, E.; Castrucci, M.R.; et al. Detection and full genome characterization of two beta CoV viruses related to Middle East respiratory syndrome from bats in Italy. Virol. J. 2017, 14, 239. [Google Scholar] [CrossRef] [PubMed]
- De Sabato, L.; Lelli, D.; Faccin, F.; Canziani, S.; Di Bartolo, I.; Vaccari, G.; Moreno, A. Full genome characterization of two novel Alpha-coronavirus species from Italian bats. Virus Res. 2018, 260, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, F.; Edenborough, K.M.; Toffoli, R.; Culasso, P.; Zoppi, S.; Dondo, A.; Robetto, S.; Rosati, S.; Lander, A.; Kurth, A.; et al. Coronavirus and paramyxovirus in bats from Northwest Italy. BMC Vet. Res. 2017, 13, 396. [Google Scholar] [CrossRef]
- Leopardi, S.; Priori, P.; Zecchin, B.; Poglayen, G.; Trevisiol, K.; Lelli, D.; Zoppi, S.; Scicluna, M.T.; D’Avino, N.; Schiavon, E.; et al. Active and passive surveillance for bat lyssaviruses in Italy revealed serological evidence for their circulation in three bat species. Epidemiol. Infect. 2018, 4, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Amoroso, M.G.; Russo, D.; Lanave, G.; Cistrone, L.; Pratelli, A.; Martella, V.; Galiero, G.; Decaro, N.; Fusco, G. Detection and phylogenetic characterization of astroviruses in insectivorous bats from Central-Southern Italy. Zoonoses Public Health 2018, 65, 702–710. [Google Scholar] [CrossRef] [PubMed]
- Lopesode, S.; Lacerda, J.P.; Fonseca, I.E.; Castro, D.P.; Forattini, O.P.; Rabello, E.X. Cotia virus: A new agent isolated from sentinel mice in Sao Paulo, Brazil. Am. J. Trop. Med. Hyg. 1965, 14, 156–157. [Google Scholar] [CrossRef] [PubMed]
- Afonso, P.P.; Silva, P.M.; Schnellrath, L.C.; Jesus, D.M.; Hu, J.; Yang, Y.; Renne, R.; Attias, M.; Condit, R.C.; Moussatche, N.; et al. Biological characterization and next-generation genome sequencing of the unclassified Cotia virus SPAn232 (Poxviridae). J. Virol. 2012, 86, 5039–5054. [Google Scholar] [CrossRef] [PubMed]
| Poxvirus Strain | Host | Sample Source | Origin | Collection Date | Clinical/Post-Mortem Findings | Laboratory Outcomes | Ref. |
|---|---|---|---|---|---|---|---|
| Hypsugopox virus (HYPV) Id lab: IZSLER 251170-23/2017 | Hypsugo savii | Pool of viscera (heart and lungs) | Europe (Italy) | 2017 | Humerus fracture and osteomalacia, calcium deficiency | CC, EM, nFGS (166,600 nt), GA (161 genes) | This study |
| Pteropox virus (PTPV) | Pteropus scapulatus | Wing membrane | North Western Australia (Kimberley region) | 2015 | Multiple nodules on the wing membranes | PGS (133,492 nt), GA (143 genes) | [12] |
| Eptesipox virus (EPTV) strain “Washington“ | Eptesicus fuscus | Elbow joint | America (WA, USA) | 2011 | Necro-suppurative osteomyelitis in multiple joints | CC, EM, FGS (176,688 nt), GA (191 genes) | [9,10] |
| Eidolon helvum poxvirus 1 (EHPV1) | Eidolon helvum | Throat swabs | Africa (Ghana) | 2009 | Apparently healthy bats | PGS | [11] |
| NA | Miniopterus schreibersii bassanii | Skin biopsies | South Australia (Naracoorte) | 2009 | Nodular cutaneous lesions | EM | [13] |
| % Similarity | Query Cover % | Poxvirus Strain | GenBank Accession No. | Host | Ref. |
|---|---|---|---|---|---|
| 85 | 75 | Eptesipoxvirus strain “Washington” | KY747497 | Eptesicus fuscus | [3,4] |
| Gene Name | Putative Product Identity | Start | Stop | +/− | Size | % Id. to EPTV | Orthologs |
|---|---|---|---|---|---|---|---|
| HYPV-1 | Hypothetical protein | 87 | 557 | − | 471 | 58 | EPTV-001 |
| HYPV-2 | Serpin 2 | 1037 | 1552 | − | 516 | 42.5 | EPTV-002 |
| HYPV-3 | Hypothetical protein | 1581 | 2261 | − | 681 | 82.4 | EPTV-003 |
| HYPV-4 | IL-1 receptor-like protein | 2309 | 3316 | − | 1008 | 65.1 | EPTV-004 |
| HYPV-5 | Hypothetical protein | 3356 | 3835 | − | 480 | 88.8 | EPTV-005 |
| HYPV-6 | Tyrosine protein kinase-like protein | 3872 | 4774 | − | 903 | 91.7 | EPTV-006 |
| HYPV-7 | ER-localized apoptosis regulator | 4842 | 5522 | − | 681 | 63.6 | EPTV-007 |
| HYPV-8 | Hypothetical protein | 7002 | 7481 | − | 480 | 80.5 | EPTV-008 |
| HYPV-9 | Ankyrin repeat-containing protein, host range | 8141 | 9826 | − | 1686 | 63.0 | EPTV-010 |
| HYPV-10 | Monoglyceride lipase | 11,053 | 11,913 | − | 861 | 93.1 | EPTV-014 |
| HYPV-11 | Secreted EGF-like growth factor | 12,340 | 12,588 | − | 249 | 62.4 | EPTV-015 |
| HYPV-12 | Anti-apoptotic factor | 12,594 | 13,100 | − | 507 | 65.7 | EPTV-016 |
| HYPV-13 | dUTPase | 13,144 | 13,569 | − | 426 | 87.2 | EPTV-017 |
| HYPV-14 | IFN-inducible protein | 13,597 | 14,004 | − | 408 | 83.7 | EPTV-018 |
| HYPV-15 | Ribonucleotide reductase small subunit | 14,060 | 15,034 | − | 975 | 93.8 | EPTV-019 |
| HYPV-16 | F5L membrane protein | 15,075 | 16,139 | − | 1065 | 68.4 | EPTV-020 |
| HYPV-17 | Cytoplasmic protein | 16,687 | 16,869 | − | 183 | 71.4 | EPTV-023 |
| HYPV-18 | S–S bond formation pathway protein | 17,361 | 18,008 | − | 648 | 92.6 | EPTV-025 |
| HYPV-19 | Ser|Thr protein kinase | 17,998 | 19,314 | − | 1317 | 94.7 | EPTV-026 |
| HYPV-20 | RhoA signaling inhibitor, virus release protein | 19,334 | 20,626 | − | 1293 | 88.0 | EPTV-07 |
| HYPV-21 | EEV maturation protein | 20,659 | 22,602 | − | 1944 | 89.0 | EPTV-028 |
| HYPV-22 | Palmitylated EEV membrane glycoprotein | 22,640 | 23,755 | − | 1116 | 98.9 | EPTV-029 |
| HYPV-23 | Hypothetical protein | 23,781 | 24,008 | − | 228 | 67.1 | EPTV-031 |
| HYPV-24 | Hypothetical protein | 24,050 | 24,250 | − | 201 | 97.0 | Unique to HYPV |
| HYPV-25 | Hypothetical protein | 24,471 | 24,917 | − | 447 | 92.6 | Unique to HYPV |
| HYPV-26 | Conserved non-functional serine recombinase | 24,992 | 25,654 | − | 663 | 78.8 | EPTV-033 |
| HYPV-27 | DNA-binding phosphoprotein | 25,714 | 26,052 | + | 339 | 86.7 | EPTV-034 |
| HYPV-28 | Poly (A) polymerase catalytic subunit | 26,046 | 27,461 | − | 1416 | 92.6 | EPTV-035 |
| HYPV-29 | IEV morphogenesis | 27,478 | 29,676 | − | 2199 | 93.3 | EPTV-036 |
| HYPV-30 | RNA polymerase subunit | 29,733 | 30,455 | − | 723 | 93.8 | EPTV-038 |
| HYPV-31 | IMV protein, virion morphogenesis | 30,760 | 32,463 | + | 1704 | 95.8 | EPTV-039 |
| HYPV-32 | ER-localized membrane protein, virion core protein | 32,490 | 33,302 | + | 813 | 95.6 | EPTV-040 |
| HYPV-33 | DNA polymerase | 33,299 | 36,319 | − | 3021 | 93.8 | EPTV-041 |
| HYPV-34 | Sulfhydryl oxidase (FAD-linked) | 36,352 | 36,642 | + | 291 | 96.9 | EPTV-042 |
| HYPV-35 | Virion core protein | 36,645 | 37,055 | − | 411 | 87.9 | EPTV-043 |
| HYPV-36 | Virulence, modulates Raf|MEK|ERK pathway | 37,039 | 39,117 | − | 2079 | 91.9 | EPTV-044 |
| HYPV-37 | Nonessential glutaredoxin | 39,173 | 39,487 | − | 315 | 91.3 | EPTV-045 |
| HYPV-38 | DNA-binding core protein | 39,613 | 40,545 | − | 933 | 90.3 | EPTV-046 |
| HYPV-39 | IMV membrane protein | 40,546 | 40,767 | − | 222 | 83.6 | EPTV-047 |
| HYPV-40 | ssDNA-binding phosphoprotein | 40,768 | 41,577 | − | 810 | 87.5 | EPTV-048 |
| HYPV-41 | Ribonucleotide reductase large subunit | 41,640 | 43,925 | − | 2286 | 95.2 | EPTV-049 |
| HYPV-42 | IMV protein (VP13) | 43,966 | 44,202 | − | 237 | 88.5 | EPTV-050 |
| HYPV-43 | Telomere-binding protein | 44,220 | 45,371 | − | 1152 | 90.9 | EPTV-051 |
| HYPV-44 | Viral core cysteine proteinase | 45,364 | 46,650 | − | 1287 | 94.6 | EPTV-052 |
| HYPV-45 | RNA-helicase, DExH-NPH-II | 46,656 | 48,686 | + | 2031 | 94.3 | EPTV-053 |
| HYPV-46 | Insulin metalloproteinase-like protein | 48,678 | 50,465 | − | 1788 | 92.3 | EPTV-054 |
| HYPV-47 | Entry|fusion complex component | 50,462 | 50,794 | − | 333 | 97.3 | EPTV-055 |
| HYPV-48 | Late transcription elongation factor (VLTF) | 50,788 | 51,456 | + | 669 | 90.5 | EPTV-056 |
| HYPV-49 | Thioredoxin-like protein | 51,423 | 51,800 | − | 378 | 89.6 | EPTV-057 |
| HYPV-50 | FEN1-like nuclease | 51,803 | 53,140 | + | 1338 | 87.0 | EPTV-058 |
| HYPV-51 | RNA polymerase subunit | 53,142 | 53,333 | + | 192 | 96.8 | EPTV-059 |
| HYPV-52 | NLPc|P60 superfamily protein | 53,337 | 53,870 | + | 534 | 87.7 | EPTV-060 |
| HYPV-53 | Virion structural phosphoprotein, early morphogenesis | 53,836 | 54,933 | − | 1098 | 91.3 | EPTV-061 |
| HYPV-54 | Late transcription factor | 54,962 | 55,744 | + | 783 | 98.5 | EPTV-062 |
| HYPV-55 | Myristylated entry|fusion protein | 55,760 | 56,782 | + | 1023 | 93.8 | EPTV-063 |
| HYPV-56 | Myristylated IMV envelope protein | 56,783 | 57,532 | + | 750 | 96.4 | EPTV-064 |
| HYPV-57 | Crescent membrane|immature virion protein | 57,558 | 57,833 | + | 276 | 84.6 | EPTV-065 |
| HYPV-58 | Internal virion protein | 57,825 | 58,790 | − | 966 | 92.1 | EPTV-066 |
| HYPV-59 | DNA-binding virion protein | 58,815 | 59,573 | + | 759 | 98.4 | EPTV-067 |
| HYPV-60 | IMV protein, entry|fusion | 59,588 | 59,992 | + | 405 | 94.0 | EPTV-068 |
| HYPV-61 | IMV membrane protein, virion morphogenesis | 59,934 | 60,380 | + | 447 | 95.9 | EPTV-069 |
| HYPV-62 | Thymidine kinase | 60,402 | 60,932 | + | 531 | 93.8 | EPTV-070 |
| HYPV-63 | Type I IFN inhibitor | 61,026 | 61,625 | + | 600 | 73.6 | EPTV-071 |
| HYPV-64 | Poly (A) polymerase small subunit | 61,692 | 62,693 | + | 1002 | 94.3 | EPTV-072 |
| HYPV-65 | RNA polymerase subunit (RPO22) | 62,608 | 63,165 | + | 558 | 96.8 | EPTV-073 |
| HYPV-66 | IMV membrane protein, entry|fusion | 63,170 | 63,580 | − | 411 | 94.1 | EPTV-074 |
| HYPV-67 | RNA polymerase subunit (RPO147) | 63,688 | 67,545 | + | 3858 | 98.5 | EPTV-075 |
| HYPV-68 | Tyr|Ser kinase, virus assembly, IFN-gamma inhibitor | 67,542 | 68,060 | − | 519 | 97.7 | EPTV-076 |
| HYPV-69 | Entry|fusion IMV protein | 68,074 | 68,646 | + | 573 | 98.9 | EPTV-077 |
| HYPV-70 | IMV heparin-binding surface protein | 68,654 | 69,667 | − | 1014 | 90.6 | EPTV-078 |
| HYPV-71 | RNA polymerase-associated protein (RAP94) | 69,671 | 72,058 | − | 2388 | 97.5 | EPTV-079 |
| HYPV-72 | Late transcription factor | 72,228 | 72,872 | + | 645 | 71.9 | EPTV-080 |
| HYPV-73 | DNA topoisomerase type I | 72,894 | 73,829 | + | 936 | 93.9 | EPTV-081 |
| HYPV-74 | Crescent membrane|immature virion protein | 73,868 | 74,314 | + | 447 | 88.6 | EPTV-082 |
| HYPV-75 | mRNA capping enzyme large subunit | 74,355 | 76,889 | + | 2535 | 95.7 | EPTV-083 |
| HYPV-76 | Virion core protein | 76,851 | 77,288 | − | 438 | 89.8 | EPTV-084 |
| HYPV-77 | Virion core protein | 77,287 | 78,030 | + | 744 | 83.0 | EPTV-085 |
| HYPV-78 | Uracil DNA glycosylase, DNA pol processivity factor | 78,027 | 78,683 | + | 657 | 96.8 | EPTV-086 |
| HYPV-79 | NTPase, DNA primase | 78,717 | 81,080 | + | 2364 | 96.6 | EPTV-087 |
| HYPV-80 | Early transcription factor small subunit (VETF-s) | 81,077 | 82,984 | + | 1908 | 99.1 | EPTV-088 |
| HYPV-81 | RNA polymerase subunit | 83,017 | 83,532 | + | 516 | 90.1 | EPTV-089 |
| HYPV-82 | Carbonic anhydrase, GAG-binding MV membrane protein | 83,464 | 84,339 | − | 876 | 82.5 | EPTV-090 |
| HYPV-83 | mRNA decapping enzyme | 84,397 | 85,068 | + | 672 | 85.7 | EPTV-091 |
| HYPV-84 | mRNA decapping enzyme | 85,043 | 85,822 | + | 780 | 92.0 | EPTV-092 |
| HYPV-85 | ATPase, NPH1 | 85,796 | 87,703 | − | 1908 | 98.1 | EPTV-093 |
| HYPV-86 | mRNA capping enzyme small subunit | 87,746 | 88,609 | − | 864 | 96.5 | EPTV-094 |
| HYPV-87 | Trimeric virion coat protein | 88,643 | 90,295 | − | 1653 | 94.6 | EPTV-095 |
| HYPV-88 | Late transcription factor (VLTF-2) | 90,321 | 90,776 | − | 456 | 93.4 | EPTV-096 |
| HYPV-89 | Late transcription factor (VLTF-3) | 90,805 | 91,479 | − | 675 | 100.0 | EPTV-097 |
| HYPV-90 | S-S bond formation pathway protein | 91,476 | 91,706 | − | 231 | 93.4 | EPTV-098 |
| HYPV-91 | P4b precursor | 91,726 | 93,726 | − | 2001 | 93.3 | EPTV-099 |
| HYPV-92 | RNA polymerase subunit (RPO19) | 94,462 | 94,992 | + | 531 | 87.5 | EPTV-101 |
| HYPV-93 | Virion morphogenesis core protein | 94,989 | 96,107 | − | 1119 | 94.1 | EPTV-102 |
| HYPV-94 | Early transcription factor large subunit (VETF-L) | 96,131 | 98,275 | − | 2145 | 97.8 | EPTV-103 |
| HYPV-95 | Intermediate transcription factor (VITF-3s) | 98,338 | 99,213 | + | 876 | 94.2 | EPTV-104 |
| HYPV-96 | IMV membrane protein, early morphogenesis | 99,223 | 99,459 | − | 237 | 92.5 | EPTV-105 |
| HYPV-97 | P4a precursor | 99,460 | 102,192 | − | 2733 | 90.5 | EPTV-106 |
| HYPV-98 | Viral membrane formation | 102,207 | 103,142 | + | 936 | 96.1 | EPTV-107 |
| HYPV-99 | Virion core and cleavage processing protein | 103,139 | 103,705 | − | 567 | 76.7 | EPTV-108 |
| HYPV-100 | IMV membrane protein, virion maturation | 103,799 | 104,002 | − | 204 | 71.6 | EPTV-109 |
| HYPV-101 | IMV membrane protein, essential | 104,067 | 104,348 | − | 282 | 96.8 | EPTV-110 |
| HYPV-102 | IMV membrane protein, non-essential | 104,365 | 104,526 | − | 162 | 98.1 | EPTV-111 |
| HYPV-103 | Core protein | 104,516 | 104,809 | − | 294 | 95.9 | EPTV-112 |
| HYPV-104 | Myristylated protein, essential for entry | 104,793 | 105,935 | − | 1143 | 91.8 | EPTV-113 |
| HYPV-105 | IMV membrane protein | 105,936 | 106,526 | − | 591 | 96.9 | EPTV-114 |
| HYPV-106 | DNA helicase, transcript release factor | 106,541 | 107,995 | + | 1455 | 90.1 | EPTV-115 |
| HYPV-107 | Zn finger-like protein, late morphogenesis | 107,967 | 108,188 | − | 222 | 93.2 | EPTV-116 |
| HYPV-108 | IMV membrane protein, entry|fusion | 108,189 | 108,533 | − | 345 | 95.6 | EPTV-117 |
| HYPV-109 | DNA polymerase processivity factor | 108,532 | 109,809 | + | 1278 | 89.4 | EPTV-118 |
| HYPV-110 | Holliday junction resolvase | 109,793 | 110,338 | + | 546 | 91.3 | EPTV-119 |
| HYPV-111 | Intermediate transcription factor (VITF-3L) | 110,335 | 111,495 | + | 1161 | 91.5 | EPTV-120 |
| HYPV-112 | RNA polymerase subunit (RPO132) | 111,492 | 115,010 | + | 3519 | 97.7 | EPTV-121 |
| HYPV-113 | A-type inclusion protein | 114,996 | 117,869 | − | 2874 | 77.4 | EPTV-122 |
| HYPV-114 | P4c precursor | 117,926 | 119,800 | − | 1875 | 82.1 | EPTV-123 |
| HYPV-115 | IMV membrane protein, fusion | 119,856 | 120,206 | − | 351 | 86.2 | EPTV-124 |
| HYPV-116 | IMV membrane protein, entry | 120,207 | 120,623 | − | 417 | 94.2 | EPTV-125 |
| HYPV-117 | RNA polymerase subunit (RPO35) | 120,637 | 121,539 | − | 903 | 94.7 | EPTV-126 |
| HYPV-118 | IMV protein | 121,523 | 121,750 | − | 228 | 92.0 | EPTV-127 |
| HYPV-119 | Hypothetical protein | 121,953 | 122,435 | + | 483 | 80.7 | EPTV-128 |
| HYPV-120 | ATPase|DNA packaging protein | 122,465 | 123,235 | − | 771 | 95.3 | EPTV-129 |
| HYPV-121 | C-type lectin-like EEV membrane phosphoglycoprotein | 123,371 | 123,922 | + | 552 | 77.2 | EPTV-130 |
| HYPV-122 | C-type lectin like IEV|EEV membrane glycoprotein | 123,970 | 124,470 | + | 501 | 91.0 | EPTV-131 |
| HYPV-123 | MHC class II antigen presentation inhibitor | 124,509 | 125,033 | + | 525 | 81.0 | EPTV-132 |
| HYPV-124 | Concanavalin-like precursor | 125,073 | 125,918 | + | 846 | 78.0 | EPTV-133 |
| HYPV-125 | EEV glycoprotein | 125,953 | 126,681 | + | 729 | 68.7 | EPTV-134 |
| HYPV-126 | Hypothetical protein | 126,724 | 127,554 | + | 831 | 79.4 | EPTV-135 |
| HYPV-127 | Hypothetical protein | 127,578 | 127,817 | + | 240 | 68.2 | EPTV-136 |
| HYPV-128 | Truncated CD47-like protein, integral membrane protein | 127,814 | 128,404 | − | 591 | 80.3 | EPTV-137 |
| HYPV-129 | Myristylated protein | 128,422 | 128,829 | + | 408 | 73.3 | EPTV-138 |
| HYPV-130 | Hypothetical protein | 128,826 | 129,587 | + | 762 | 78.3 | EPTV-139 |
| HYPV-131 | Chemokine binding protein | 129,575 | 130,438 | − | 864 | 69.0 | EPTV-140 |
| HYPV-132 | Profilin-like protein, ATI-localized | 130,558 | 130,959 | + | 402 | 98.5 | EPTV-141 |
| HYPV-133 | Hypothetical protein | 130,956 | 131,339 | − | 384 | 76.9 | EPTV-142 |
| HYPV-134 | 3 beta-hydroxysteroid dehydrogenase|delta 5->4 isomerase | 131,948 | 133,015 | + | 1068 | 84.2 | EPTV-144 |
| HYPV-135 | Thymidylate kinase | 133,646 | 134,233 | + | 588 | 85.2 | EPTV-147 |
| HYPV-136 | DNA ligase-like protein | 134,265 | 135,944 | + | 1680 | 87.2 | EPTV-148 |
| HYPV-137 | A52R-like family protein | 137,441 | 138,046 | + | 606 | 77.9 | EPTV-150 |
| HYPV-138 | Hypothetical protein | 138,641 | 139,717 | + | 1077 | 65.2 | EPTV-151 |
| HYPV-139 | Toll|IL-1 receptor-like protein, IL-1, NFkB signaling inhibitor | 139,781 | 140,428 | + | 648 | 89.8 | EPTV-153 |
| HYPV-140 | Hypothetical protein | 140,538 | 140,954 | − | 417 | 70.5 | EPTV-154 |
| HYPV-141 | BTB kelch-domain protein | 141,056 | 142,690 | + | 1635 | 76.8 | EPTV-155 |
| HYPV-142 | Hemagglutinin | 142,737 | 143,477 | + | 741 | 70.2 | EPTV-156 |
| HYPV-143 | Ser|Thr protein kinase | 143,532 | 144,467 | + | 936 | 91.0 | EPTV-157 |
| HYPV-144 | IL-1 receptor antagonist | 144,505 | 145,470 | + | 966 | 71.5 | EPTV-158 |
| HYPV-145 | RING finger protein, host range | 145,503 | 146,378 | + | 876 | 68.4 | EPTV-159 |
| HYPV-146 | Partial schlafen-like protein | 146,424 | 147,011 | + | 588 | 81.1 | EPTV-160 |
| HYPV-147 | EEV type-1 membrane glycoprotein | 147,108 | 147,815 | + | 708 | 75.3 | EPTV-161 |
| HYPV-148 | Anti-apoptotic Bcl-2-like protein | 147,851 | 148,294 | + | 444 | 73.5 | EPTV-162 |
| HYPV-149 | Serpin 1 | 149,551 | 150,555 | + | 1005 | 76.9 | EPTV-164 |
| HYPV-150 | Hypothetical protein | 150,662 | 151,132 | + | 471 | 69.4 | EPTV-165 |
| HYPV-151 | Tyrosine protein kinase-like protein | 151,173 | 152,036 | + | 864 | 77.7 | EPTV-166 |
| HYPV-152 | IL-1 beta-receptor | 152,065 | 153,093 | + | 1029 | 71.9 | EPTV-167 |
| HYPV-153 | Ankyrin repeat protein | 153,125 | 155,104 | + | 1980 | 68.2 | EPTV-168 |
| HYPV-154 | Ankyrin repeat protein | 157,276 | 157,827 | + | 552 | 60.6 | EPTV-169 |
| HYPV-155 | Alpha-amanitin target protein | 158,306 | 159,013 | + | 708 | 82.3 | EPTV-170 |
| HYPV-156 | NFkB inhibitor | 159,072 | 159,752 | + | 681 | 81.1 | EPTV-171 |
| HYPV-157 | Endothelin precursor | 159,796 | 160,020 | + | 225 | 64.9 | EPTV-172 |
| HYPV-158 | NFkB inhibitor | 160,059 | 160,712 | + | 654 | 65.0 | EPTV-173 |
| HYPV-159 | Secreted complement binding protein C3b|C4b | 160,747 | 161,529 | + | 783 | 74.6 | EPTV-174 |
| HYPV-160 | IL-18 binding protein | 162,096 | 162,521 | + | 426 | 83.2 | EPTV-175 |
| HYPV-161 | Ankyrin repeat protein | 164,706 | 166,571 | + | 1866 | 60.3 | EPTV-179 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lelli, D.; Lavazza, A.; Prosperi, A.; Sozzi, E.; Faccin, F.; Baioni, L.; Trogu, T.; Cavallari, G.L.; Mauri, M.; Gibellini, A.M.; et al. Hypsugopoxvirus: A Novel Poxvirus Isolated from Hypsugo savii in Italy. Viruses 2019, 11, 568. https://doi.org/10.3390/v11060568
Lelli D, Lavazza A, Prosperi A, Sozzi E, Faccin F, Baioni L, Trogu T, Cavallari GL, Mauri M, Gibellini AM, et al. Hypsugopoxvirus: A Novel Poxvirus Isolated from Hypsugo savii in Italy. Viruses. 2019; 11(6):568. https://doi.org/10.3390/v11060568
Chicago/Turabian StyleLelli, Davide, Antonio Lavazza, Alice Prosperi, Enrica Sozzi, Francesca Faccin, Laura Baioni, Tiziana Trogu, Gian Luca Cavallari, Matteo Mauri, Anna Maria Gibellini, and et al. 2019. "Hypsugopoxvirus: A Novel Poxvirus Isolated from Hypsugo savii in Italy" Viruses 11, no. 6: 568. https://doi.org/10.3390/v11060568
APA StyleLelli, D., Lavazza, A., Prosperi, A., Sozzi, E., Faccin, F., Baioni, L., Trogu, T., Cavallari, G. L., Mauri, M., Gibellini, A. M., Chiapponi, C., & Moreno, A. (2019). Hypsugopoxvirus: A Novel Poxvirus Isolated from Hypsugo savii in Italy. Viruses, 11(6), 568. https://doi.org/10.3390/v11060568







