Influenza A in Bovine Species: A Narrative Literature Review
Abstract
1. Introduction
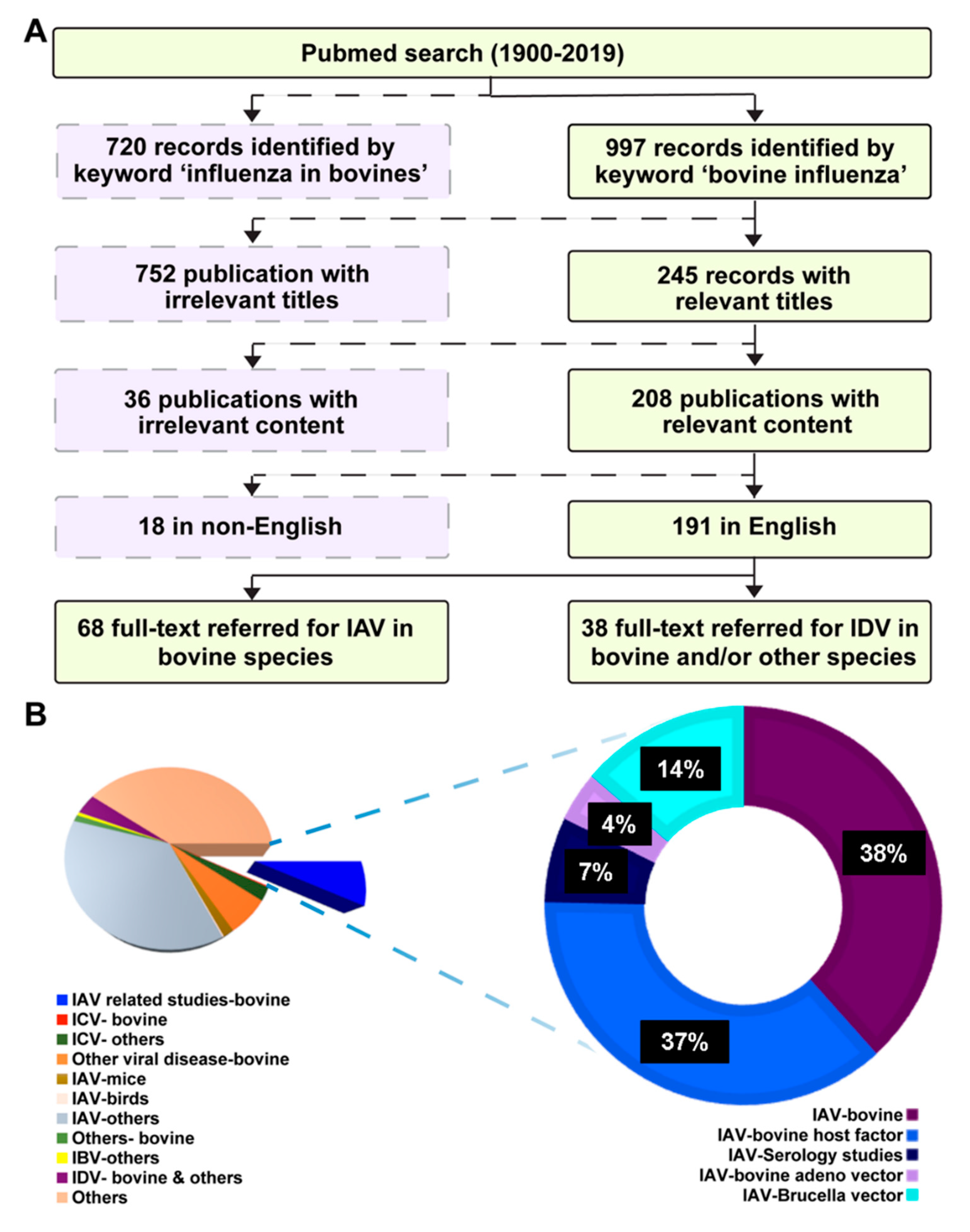
2. Literature Search Strategy
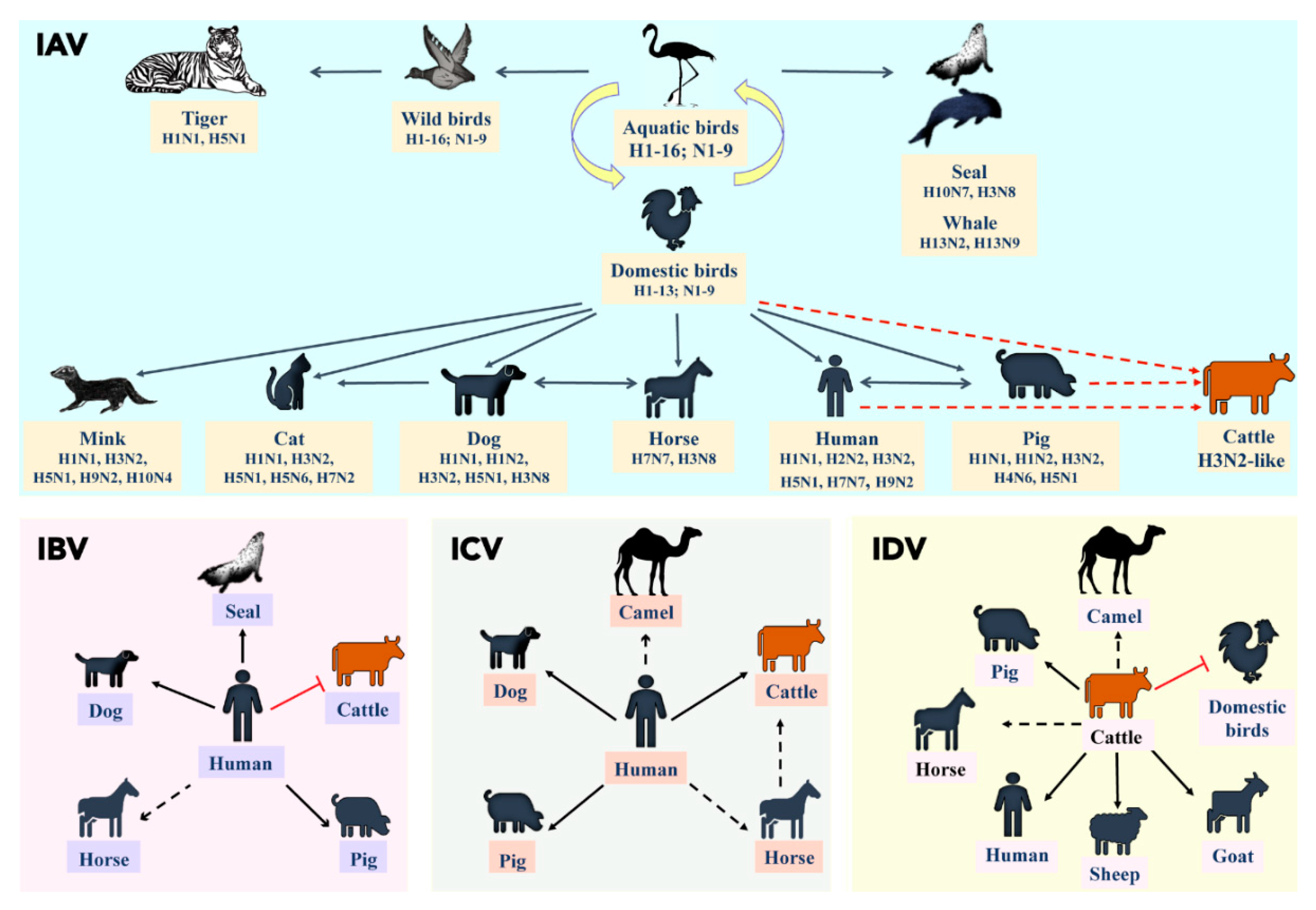
3. A Brief Overview of Influenza Viral Ecology
4. Role of Influenza in Bovine Respiratory Diseases and Evidence of Zoonosis/Reverse Zoonosis
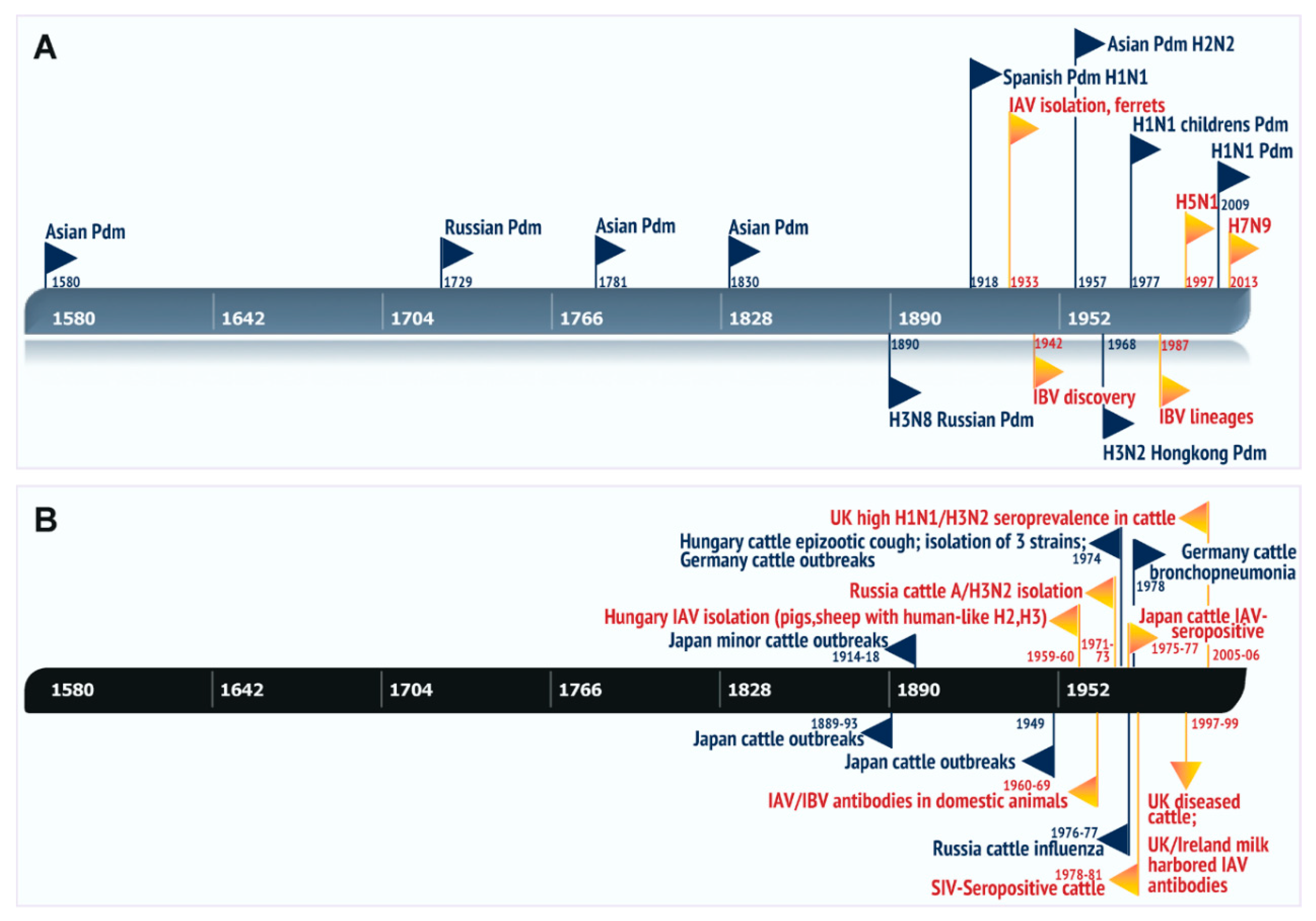
5. Natural Cases of Influenza A in Bovines
6. Experimental Infections of Influenza A in Ruminant Species
7. Seroprevalence Studies of Influenza A in Bovine Species
8. Bovine Cell Cultures for Influenza Studies in Vitro
9. Host Restriction Factors in Bovines: Sensitivity of Influenza to Cellular/Serum Factors
9.1. Bovine Colostrum/Milk
9.2. Bovine Lactoferrin
9.3. Bovine Serum Inhibitors: Conglutinin, Collectin
9.4. Bovine Lung Factors
9.5. Bovine Antivirals
10. Influenza Vaccine Studies in Bovines and Use of Bovine Viral Vectors
11. Summary
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Potter, C.W. A history of influenza. J. Appl. Microbiol. 2001, 91, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Aramini, J.M.; Ma, L.-C.; Krug, R.M.; Arnold, E. Structures of influenza A proteins and insights into antiviral drug targets. Nat. Struct. Mol. Boil. 2010, 17, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Richardson, J.C.; Akkina, R.K. NS2 protein of influenza virus is found in purified virus and phosphorylated in infected cells. Arch. Virol. 1991, 116, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, E.C.; Charles, P.D.; Hester, S.S.; Thomas, B.; Trudgian, D.; Martinez-Alonso, M.; Fodor, E. Conserved and host-specific features of influenza virion architecture. Nat. Commun. 2014, 5, 4816. [Google Scholar] [CrossRef] [PubMed]
- Paragas, J.; Talon, J.; O’Neill, R.E.; Anderson, D.K.; García-Sastre, A.; Palese, P. Influenza B and C virus NEP (NS2) proteins possess nuclear export activities. J. Virol. 2001, 75, 7375–7383. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.; Li, Y.; Rivailler, P.; Conrardy, C.; Castillo, D.A.; Chen, L.M.; Recuenco, S.; Ellison, J.A.; Davis, C.T.; York, I.A.; et al. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. USA 2012, 109, 4269–4274. [Google Scholar] [CrossRef] [PubMed]
- Campos, A.C.A.; Goes, L.G.B.; Moreira-Soto, A.; de Carvalho, C.; Ambar, G.; Sander, A.L.; Fischer, C.; Ruckert da Rosa, A.; Cardoso de Oliveira, D.; Kataoka, A.P.G.; et al. Bat influenza A(HL18NL11) virus in Fruit Bats, Brazil. Emerg. Infect. Dis. 2019, 25, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Palese, P. Influenza: Old and new threats. Nat. Med. 2004, 10, S82–S87. [Google Scholar] [CrossRef] [PubMed]
- Berhane, Y.; Ojkic, D.; Neufeld, J.; Leith, M.; Hisanaga, T.; Kehler, H.; Ferencz, A.; Wojcinski, H.; Cottam-Birt, C.; Suderman, M.; et al. Molecular characterization of pandemic H1N1 influenza viruses isolated from turkeys and pathogenicity of a human pH1N1 isolate in turkeys. Avian Dis. 2010, 54, 1275–1285. [Google Scholar] [CrossRef]
- Campagnolo, E.R.; Rankin, J.T.; Daverio, S.A.; Hunt, E.A.; Lute, J.R.; Tewari, D.; Acland, H.M.; Ostrowski, S.R.; Moll, M.E.; Urdaneta, V.V.; et al. Fatal pandemic (H1N1) 2009 influenza A virus infection in a Pennsylvania domestic cat. Zoonoses Public Health 2011, 58, 500–507. [Google Scholar] [CrossRef]
- Dundon, W.G.; De Benedictis, P.; Viale, E.; Capua, I. Serologic evidence of pandemic (H1N1) 2009 infection in dogs, Italy. Emerg. Infect. Dis. 2010, 16, 2019–2021. [Google Scholar] [CrossRef] [PubMed]
- Pasma, T.; Joseph, T. Pandemic (H1N1) 2009 infection in swine herds, Manitoba, Canada. Emerg. Infect. Dis. 2010, 16, 706–708. [Google Scholar] [CrossRef] [PubMed]
- Sponseller, B.A.; Strait, E.; Jergens, A.; Trujillo, J.; Harmon, K.; Koster, L.; Jenkins-Moore, M.; Killian, M.; Swenson, S.; Bender, H.; et al. Influenza A pandemic (H1N1) 2009 virus infection in domestic cat. Emerg. Infect. Dis. 2010, 16, 534–537. [Google Scholar] [CrossRef] [PubMed]
- Welsh, M.D.; Baird, P.M.; Guelbenzu-Gonzalo, M.P.; Hanna, A.; Reid, S.M.; Essen, S.; Russell, C.; Thomas, S.; Barrass, L.; McNeilly, F.; et al. Initial incursion of pandemic (H1N1) 2009 influenza A virus into European pigs. Vet. Rec. 2010, 166, 642–645. [Google Scholar] [CrossRef] [PubMed]
- Callan, R.J.; Early, G.; Kida, H.; Hinshaw, V.S. The appearance of H3 influenza viruses in seals. J. Gen. Virol. 1995, 76, 199–203. [Google Scholar] [PubMed]
- Hinshaw, V.S.; Bean, W.J.; Geraci, J.; Fiorelli, P.; Early, G.; Webster, R.G. Characterization of two influenza A viruses from a pilot whale. J. Virol. 1986, 58, 655–656. [Google Scholar]
- Lang, G.; Gagnon, A.; Geraci, J.R. Isolation of an influenza A virus from seals. Arch. Virol. 1981, 68, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Vladimirtseva, E.A.; Sadykhova, F.; Iamnikova, S.S.; L’Vov, D.K.; Zhdanov, V.M. Physicochemical properties of the RNA and proteins of an influenza virus H1N3 isolated from an ill child and antigenically analogous to A/whale/TO/19/76. Vopr. Virusol. 1985, 30, 163–166. [Google Scholar]
- Taubenberger, J.K.; Kash, J.C. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 2010, 7, 440–451. [Google Scholar] [CrossRef]
- Alexander, P.E.; De, P.; Rave, S. Is H9N2 avian influenza virus a pandemic potential? Can. J. Infect. Dis. Med. Microbiol. 2009, 20, e35–e36. [Google Scholar] [CrossRef]
- Taubenberger, J.K.; Morens, D.M. Pandemic influenza—Including a risk assessment of H5N1. Rev. Sci. Tech. 2009, 28, 187–202. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Sorrell, E.M.; Song, H.; Hossain, M.J.; Ramirez-Nieto, G.; Monne, I.; Stevens, J.; Cattoli, G.; Capua, I.; Chen, L.M.; et al. Replication and transmission of H9N2 influenza viruses in ferrets: Evaluation of pandemic potential. PLoS ONE 2008, 3, e2923. [Google Scholar] [CrossRef] [PubMed]
- Cauldwell, A.V.; Long, J.S.; Moncorge, O.; Barclay, W.S. Viral determinants of influenza A virus host range. J. Gen. Virol. 2014, 95, 1193–1210. [Google Scholar] [CrossRef] [PubMed]
- Manz, B.; Schwemmle, M.; Brunotte, L. Adaptation of avian influenza A virus polymerase in mammals to overcome the host species barrier. J. Virol. 2013, 87, 7200–7209. [Google Scholar] [CrossRef] [PubMed]
- Busuioc, C.; Popovici, M.; Ionescu, V.; Stoicescu, A.; Scheau, A.; Cazacu, E. Incidence and level of influenza and adenovirus antibodies in various species of domestic animals. Stud. Cercet. Inframicrobiol. 1969, 20, 191–195. [Google Scholar] [PubMed]
- Cilli, V.; Davoli, R. Reaction of bovines to experimental infection with human influenza virus. Nuovi Ann. Ig. Microbiol. 1954, 5, 106–109. [Google Scholar] [PubMed]
- Smetanin, M.A.; Molibog, E.V.; Borovik, R.V.; Zakstel’skaia, L.; Isachenko, V.A. Experimental inoculation of calves with influenza virus A/csf/Udmurtiia/116/73. Vopr. Virusol. 1977, 3, 285–287. [Google Scholar]
- Yoon, S.W.; Webby, R.J.; Webster, R.G. Evolution and ecology of influenza A viruses. Curr. Top. Microbiol. Immunol. 2014, 385, 359–375. [Google Scholar]
- Shope, R.E. Swine influenza: Iii. filtration experiments and etiology. J. Exp. Med. 1931, 54, 373–385. [Google Scholar] [CrossRef]
- Webby, R.J.; Swenson, S.L.; Krauss, S.L.; Gerrish, P.J.; Goyal, S.M.; Webster, R.G. Evolution of swine H3N2 influenza viruses in the United States. J. Virol. 2000, 74, 8243–8251. [Google Scholar] [CrossRef]
- Zhou, N.N.; Senne, D.A.; Landgraf, J.S.; Swenson, S.L.; Erickson, G.; Rossow, K.; Liu, L.; Yoon, K.; Krauss, S.; Webster, R.G. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J. Virol. 1999, 73, 8851–8856. [Google Scholar] [PubMed]
- Vincent, A.; Awada, L.; Brown, I.; Chen, H.; Claes, F.; Dauphin, G.; Donis, R.; Culhane, M.; Hamilton, K.; Lewis, N.; et al. Review of influenza A virus in swine worldwide: A call for increased surveillance and research. Zoonoses Public Health 2014, 61, 4–17. [Google Scholar] [CrossRef] [PubMed]
- Webster, R.G.; Thomas, T.L. Efficacy of equine influenza vaccines for protection against A/Equine/Jilin/89 (H3N8)—A new equine influenza virus. Vaccine 1993, 11, 987–993. [Google Scholar] [CrossRef]
- Hamed, M.I.; Amen, O.A.K.; Rateb, H.Z. Detection of avian influenza virus H5N1 in horses at Assiut Governorate, Egypt. J. Adv. Vet. Res. 2014, 4, 161–165. [Google Scholar]
- Feng, K.H.; Gonzalez, G.; Deng, L.; Yu, H.; Tse, V.L.; Huang, L.; Huang, K.; Wasik, B.R.; Zhou, B.; Wentworth, D.E.; et al. Equine and canine influenza H3N8 viruses show minimal biological differences despite phylogenetic divergence. J. Virol. 2015, 89, 6860. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Hughes, J.; Murcia, P.R. Origins and evolutionary dynamics of H3N2 canine influenza virus. J. Virol. 2015, 89, 5406. [Google Scholar] [CrossRef]
- Bodewes, R.; Bestebroer, T.M.; van der Vries, E.; Verhagen, J.H.; Herfst, S.; Koopmans, M.P.; Fouchier, R.A.; Pfankuche, V.M.; Wohlsein, P.; Siebert, U.; et al. Avian Influenza A(H10N7) virus-associated mass deaths among harbor seals. Emerg. Infect. Dis. 2015, 21, 720–722. [Google Scholar] [CrossRef] [PubMed]
- Berg, M.; Englund, L.; Abusugra, I.A.; Klingeborn, B.; Linne, T. Close relationship between mink influenza (H10N4) and concomitantly circulating avian influenza viruses. Arch. Virol. 1990, 113, 61–71. [Google Scholar] [CrossRef]
- Englund, L. Studies on influenza viruses H10N4 and H10N7 of avian origin in mink. Vet. Microbiol. 2000, 74, 101–107. [Google Scholar] [CrossRef]
- Englund, L.; Klingeborn, B.; Mejerland, T. Avian influenza A virus causing an outbreak of contagious interstitial pneumonia in mink. Acta Vet. Scand. 1986, 27, 497–504. [Google Scholar]
- Feldmann, H.; Kretzschmar, E.; Klingeborn, B.; Rott, R.; Klenk, H.D.; Garten, W. The structure of serotype H10 hemagglutinin of influenza A virus: Comparison of an apathogenic avian and a mammalian strain pathogenic for mink. Virology 1988, 165, 428–437. [Google Scholar] [CrossRef]
- Peng, L.; Chen, C.; Kai-yi, H.; Feng-xia, Z.; Yan-li, Z.; Zong-shuai, L.; Xing-xiao, Z.; Shi-jin, J.; Zhi-jing, X. Molecular characterization of H9N2 influenza virus isolated from mink and its pathogenesis in mink. Vet. Microbiol. 2015, 176, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, D.; Allard, V.; Doyon, J.F.; Bellehumeur, C.; Spearman, J.G.; Harel, J.; Gagnon, C.A. Emerg.ence of a new swine H3N2 and pandemic (H1N1) 2009 influenza A virus reassortant in two Canadian animal populations, mink and swine. J. Clin. Microbiol. 2011, 49, 4386–4390. [Google Scholar] [CrossRef]
- Yoon, K.J.; Schwartz, K.; Sun, D.; Zhang, J.; Hildebrandt, H. Naturally occurring Influenza A virus subtype H1N2 infection in a Midwest United States mink (Mustela vison) ranch. J. Vet. Diagn Investig. 2012, 24, 388–391. [Google Scholar] [CrossRef] [PubMed]
- Fatkhuddinova, M.F.; Kir’ianova, A.I.; Isachenko, V.A.; Zakstel’skaia, L. Isolation and identification of the A-Hong Kong (H3N2) virus in respiratory diseases of cattle. Vopr. Virusol. 1973, 18, 474–478. [Google Scholar]
- Tanyi, J.; Romvary, J.; Aldasy, P.; Mathe, Z. Isolation of influenza a virus strains from cattle. Preliminary report. Acta Vet. Acad. Sci. Hung. 1974, 24, 341–343. [Google Scholar] [PubMed]
- Lopez, J.W.; Woods, G.T. Influenza virus in ruminants: A review. Res. Commun. Chem. Pathol. Pharmacol. 1984, 45, 445–462. [Google Scholar] [PubMed]
- Campbell, C.H.; Easterday, B.C.; Webster, R.G. Strains of Hong Kong influenza virus in calves. J. Infect. Dis. 1977, 135, 678–680. [Google Scholar] [CrossRef]
- Bodewes, R.; Morick, D.; de Mutsert, G.; Osinga, N.; Bestebroer, T.; van der Vliet, S.; Smits, S.L.; Kuiken, T.; Rimmelzwaan, G.F.; Fouchier, R.A.; et al. Recurring influenza B virus infections in seals. Emerg. Infect. Dis. 2013, 19, 511–512. [Google Scholar] [CrossRef]
- Osterhaus, A.D.; Rimmelzwaan, G.F.; Martina, B.E.; Bestebroer, T.M.; Fouchier, R.A. Influenza B virus in seals. Science 2000, 288, 1051–1053. [Google Scholar] [CrossRef]
- Paules, C.; Subbarao, K. Influenza. Lancet 2017, 390, 697–708. [Google Scholar] [CrossRef]
- Takatsy, G.Y.; Farkas, E.; Romvary, J. Susceptibility of the domestic pig to influenza B virus. Nature 1969, 222, 184–185. [Google Scholar] [CrossRef] [PubMed]
- Ran, Z.; Shen, H.; Lang, Y.; Kolb, E.A.; Turan, N.; Zhu, L.; Ma, J.; Bawa, B.; Liu, Q.; Liu, H.; et al. Domestic pigs are susceptible to infection with influenza B viruses. J. Virol. 2015, 89, 4818–4826. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.P.; New, A.E.; Taylor, J.F.; Chiang, H.S. Influenza virus isolations from dogs during a human epidemic in Taiwan. Int. J. Zoonoses 1976, 3, 61–64. [Google Scholar] [PubMed]
- Ditchfield, J.; Macpherson, L.W. Upper respiratory disease in thoroughbred horses: Studies of its viral etiology in the Toronto area, 1960 to 1963. Can. J. Comp. Med. Vet. Sci. 1965, 29, 18–22. [Google Scholar] [PubMed]
- Kawano, J.; Onta, T.; Kida, H.; Yanagawa, R. Distribution of antibodies in animals against influenza B and C viruses. Jpn. J. Vet. Res. 1978, 26, 74–80. [Google Scholar]
- Anton, A.; Marcos, M.A.; Codoner, F.M.; de Molina, P.; Martinez, A.; Cardenosa, N.; Godoy, P.; Torner, N.; Martinez, M.J.; Ramon, S.; et al. Influenza C virus surveillance during the first influenza A (H1N1) 2009 pandemic wave in Catalonia, Spain. Diagn. Microbiol. Infect. Dis. 2011, 69, 419–427. [Google Scholar] [CrossRef]
- Brown, I.H.; Harris, P.A.; Alexander, D.J. Serological studies of influenza viruses in pigs in Great Britain 1991–2. Epidemiol. Infect. 1995, 114, 511–520. [Google Scholar] [CrossRef]
- Kimura, H.; Abiko, C.; Peng, G.; Muraki, Y.; Sugawara, K.; Hongo, S.; Kitame, F.; Mizuta, K.; Numazaki, Y.; Suzuki, H.; et al. Interspecies transmission of influenza C virus between humans and pigs. Virus Res. 1997, 48, 71–79. [Google Scholar] [CrossRef]
- Guo, Y.J.; Jin, F.G.; Wang, P.; Wang, M.; Zhu, J.M. Isolation of influenza C virus from pigs and experimental infection of pigs with influenza C virus. J. Gen. Virol. 1983, 64, 177–182. [Google Scholar]
- Nedland, H.; Wollman, J.; Sreenivasan, C.; Quast, M.; Singrey, A.; Fawcett, L.; Christopher-Hennings, J.; Nelson, E.; Kaushik, R.S.; Wang, D.; et al. Serological evidence for the co-circulation of two lineages of influenza D viruses in equine populations of the Midwest United States. Zoonoses Public Health 2018, 65, e148–e154. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Porter, E.; Lohman, M.; Lu, N.; Peddireddi, L.; Hanzlicek, G.; Marthaler, D.; Liu, X.; Bai, J. Influenza C virus in cattle with respiratory disease, United States, 2016–2018. Emerg. Infect. Dis. 2018, 24, 1926–1929. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Hill, J.E.; Fernando, C.; Alexander, T.W.; Timsit, E.; van der Meer, F.; Huang, Y. Respiratory viruses identified in western Canadian beef cattle by metagenomic sequencing and their association with bovine respiratory disease. Transbound. Emerg. Dis. 2019, 66, 1379–1386. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, L.; Eckard, L.; Epperson, W.B.; Long, L.P.; Smith, D.; Huston, C.; Genova, S.; Webby, R.; Wan, X.F. Influenza D virus infection in Mississippi beef cattle. Virology 2015, 486, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Hause, B.M.; Ducatez, M.; Collin, E.A.; Ran, Z.; Liu, R.; Sheng, Z.; Armien, A.; Kaplan, B.; Chakravarty, S.; Hoppe, A.D.; et al. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog. 2013, 9, e1003176. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Ferguson, L.; Smith, D.R.; Woolums, A.R.; Epperson, W.B.; Wan, X.F. Serological evidence for high prevalence of Influenza D Viruses in Cattle, Nebraska, United States, 2003–2004. Virology 2017, 501, 88–91. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Endoh, M.; Kobayashi, T.; Takenaka-Uema, A.; Chambers, J.K.; Uchida, K.; Nishihara, M.; Hause, B.; Horimoto, T. Influenza D virus infection in herd of cattle, Japan. Emerg. Infect. Dis. 2016, 22, 1517–1519. [Google Scholar] [CrossRef]
- Snoeck, C.J.; Oliva, J.; Pauly, M.; Losch, S.; Wildschutz, F.; Muller, C.P.; Hubschen, J.M.; Ducatez, M.F. Influenza D virus circulation in cattle and swine, Luxembourg, 2012–2016. Emerg. Infect. Dis. 2018, 24, 1388–1389. [Google Scholar] [CrossRef]
- Ferguson, L.; Luo, K.; Olivier, A.K.; Cunningham, F.L.; Blackmon, S.; Hanson-Dorr, K.; Sun, H.; Baroch, J.; Lutman, M.W.; Quade, B.; et al. Influenza D Virus Infection in Feral Swine Populations, United States. Emerg. Infect. Dis. 2018, 24, 1020–1028. [Google Scholar] [CrossRef]
- Quast, M.; Sreenivasan, C.; Sexton, G.; Nedland, H.; Singrey, A.; Fawcett, L.; Miller, G.; Lauer, D.; Voss, S.; Pollock, S.; et al. Serological evidence for the presence of influenza D virus in small ruminants. Vet. Microbiol. 2015, 180, 281–285. [Google Scholar] [CrossRef]
- White, S.K.; Ma, W.; McDaniel, C.J.; Gray, G.C.; Lednicky, J.A. Serologic evidence of exposure to influenza D virus among persons with occupational contact with cattle. J. Clin. Virol. 2016, 81, 31–33. [Google Scholar] [CrossRef] [PubMed]
- Borkenhagen, L.K.; Mallinson, K.A.; Tsao, R.W.; Ha, S.J.; Lim, W.H.; Toh, T.H.; Anderson, B.D.; Fieldhouse, J.K.; Philo, S.E.; Chong, K.S.; et al. Surveillance for respiratory and diarrheal pathogens at the human-pig interface in Sarawak, Malaysia. PLoS ONE 2018, 13, e0201295. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.Y.; Zemke, J.; Philo, S.E.; Bailey, E.S.; Yondon, M.; Gray, G.C. Aerosol sampling in a hospital emergency room setting: A complementary surveillance method for the detection of respiratory viruses. Front. Public Health 2018, 6, 174. [Google Scholar] [CrossRef] [PubMed]
- Bollongino, R.; Burger, J.; Powell, A.; Mashkour, M.; Vigne, J.D.; Thomas, M.G. Modern taurine cattle descended from small number of near-eastern founders. Mol. Biol. Evol. 2012, 29, 2101–2104. [Google Scholar] [CrossRef] [PubMed]
- Giuffra, E.; Kijas, J.M.; Amarger, V.; Carlborg, O.; Jeon, J.T.; Andersson, L. The origin of the domestic pig: Independent domestication and subsequent introgression. Genetics 2000, 154, 1785–1791. [Google Scholar]
- Saito, K. An outbreak of cattle influenza in Japan in the fall of 1949. J. Am. Vet. Med. Assoc. 1951, 118, 316–319. [Google Scholar]
- Reisinger, R.C. Parainfluenza 3 virus in cattle. Ann. N. Y. Acad. Sci. 1962, 101, 576–582. [Google Scholar] [CrossRef]
- Hoerlein, A.B.; Mansfield, M.E.; Abinanti, F.R.; Huebner, R.J. Studies of shipping fever of cattle. I. Para-influenza 3 virus antibodies in feeder calves. J. Am. Vet. Med. Assoc. 1959, 135, 153–160. [Google Scholar]
- Madin, S.H.; McKercher, D.G.; York, C.J. Isolation of the infectious bovine rhinotracheitis virus. Science 1956, 124, 721–722. [Google Scholar] [CrossRef]
- Noice, F.; Schipper, I.A. Isolation of mucosal disease virus by tissue cultures in mixture 199, Morgan, Morton and Parker. Proc. Soc. Exp. Biol. Med. 1959, 100, 84–86. [Google Scholar] [CrossRef]
- Paccaud, M.F.; Jacquier, C. A respiratory syncytial virus of bovine origin. Arch. Gesamte Virusforsch. 1970, 30, 327–342. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, W.R. The bovine viral diarrhea-mucosal disease complex. Adv. Vet. Sci. 1963, 8, 1–47. [Google Scholar] [PubMed]
- Schiott, C.R.; Jensen, C.H. A mouse pathogenic strain of parainfluenza virus type 3 isolated from cattle. Acta Pathol. Microbiol. Scand. 1962, 56, 479–480. [Google Scholar] [CrossRef] [PubMed]
- USDA, Feedlot 2011. Part IV: Health and Health Management on U.S. Feedlots with a Capacity of 1000 or More Head. 2011. Available online: https://www.aphis.usda.gov/animal_health/nahms/feedlot/downloads/feedlot2011/Feed11_dr_PartIV.pdf (accessed on 17 June 2019).
- Mitra, N.; Cernicchiaro, N.; Torres, S.; Li, F.; Hause, B.M. Metagenomic characterization of the virome associated with bovine respiratory disease in feedlot cattle identified novel viruses and suggests an etiologic role for influenza D virus. J. Gen. Virol. 2016, 97, 1771–1784. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Porter, E.P.; Lohman, M.; Lu, N.; Peddireddi, L.; Hanzlicek, G.; Marthaler, D.; Liu, X.; Bai, J. Complete genome sequence of an influenza C virus strain identified from a sick calf in the United States. Microbiol. Resour Announc. 2018, 7, e00828-18. [Google Scholar] [CrossRef]
- Hause, B.M.; Collin, E.A.; Liu, R.; Huang, B.; Sheng, Z.; Lu, W.; Wang, D.; Nelson, E.A.; Li, F. Characterization of a novel influenza virus in cattle and Swine: Proposal for a new genus in the Orthomyxoviridae family. MBio 2014, 5, e00031-14. [Google Scholar] [CrossRef] [PubMed]
- Romvary, J.; Takatsy, G.; Barb, K.; Farkas, E. Isolation of influenza virus strains from animals. Nature 1962, 193, 907–908. [Google Scholar] [CrossRef]
- Isachenko, V.A.; Molibog, E.V.; Zakstel’skaia, L. Antigenic characteristics of the influenza viruses isolated from domestic animals and birds in the USSR. Vopr. Virusol. 1973, 18, 700–705. [Google Scholar]
- Izmailov, I.A.; Atamas, V.A.; Kniazeva, N.I.; Petrova, M.S.; Kaimakan, P.V. Outbreak of influenza among cattle. Veterinariia 1973, 49, 69–70. [Google Scholar]
- Koval’chuk-Ivaniuk, T.V.; Stel’makh, S.G.; Shablovskaia, E.A.; Repko, A.N.; Zlonkevich Ia, D. Results of an examination of cattle for influenza. Vopr. Virusol. 1977, 4, 418–421. [Google Scholar]
- Smetain, M.A.; Gaffarov Kh, Z.; Vakhrameev, V.I.; Egorchev, I.V. Study of cattle influenza. Veterinariia 1976, 12, 35–36. [Google Scholar]
- Wagner, K.; Becker, W.; Bromel, J. Influenza of cattle. Enzootic bronchopneumonia of cattle. Tierarztl. Prax. 1978, 6, 51–61. [Google Scholar] [PubMed]
- Sabisch, G. Results of pathological-anatomical examination of lung material for bovine influenza. Berl. Munch. Tierarztl. Wochenschr. 1977, 90, 414–416. [Google Scholar] [PubMed]
- Gunning, R.F.; Pritchard, G.C. Unexplained sporadic milk drop in dairy cows. Vet. Rec. 1997, 140, 488. [Google Scholar] [PubMed]
- Brown, I.H.; Crawshaw, T.R.; Harris, P.A.; Alexander, D.J. Detection of antibodies to influenza A virus in cattle in association with respiratory disease and reduced milk yield. Vet. Rec. 1998, 143, 637–638. [Google Scholar] [PubMed]
- Crawshaw, T.R.; Brown, I. Bovine influenza. Vet. Rec. 1998, 143, 372. [Google Scholar] [PubMed]
- Gunning, R.F.; Brown, I.H.; Crawshaw, T.R. Evidence of influenza A virus infection in dairy cows with sporadic milk drop syndrome. Vet. Rec. 1999, 145, 556–557. [Google Scholar] [CrossRef] [PubMed]
- Graham, D.A.; Calvert, V.; McLaren, E. Retrospective analysis of serum and nasal mucus from cattle in Northern Ireland for evidence of infection with influenza A virus. Vet. Rec. 2002, 150, 201–204. [Google Scholar] [CrossRef] [PubMed]
- Crawshaw, T.R.; Brown, I.H.; Essen, S.C.; Young, S.C. Significant rising antibody titres to influenza A are associated with an acute reduction in milk yield in cattle. Vet. J. 2008, 178, 98–102. [Google Scholar] [CrossRef]
- Szeredi, L.; Janosi, S.; Palfi, V. Microbiol.ogical and pathological examination of fatal calf pneumonia cases induced by bacterial and viral respiratory pathogens. Acta Vet. Hung. 2010, 58, 341–356. [Google Scholar] [CrossRef]
- Simpson, V.R. Wild animals as reservoirs of infectious diseases in the UK. Vet. J. 2002, 163, 128–146. [Google Scholar] [CrossRef] [PubMed]
- Graves, I.L.; Pyakural, S.; Sousa, V.O. Susceptibility of a yak to influenza A viruses and presence of H3N2 antibodies in animals in Nepal and India. Bull. World Health Organ. 1974, 51, 173–177. [Google Scholar] [PubMed]
- Boyle, D.B.; Coupar, B.E.; Parsonson, I.M.; Bagust, T.J.; Both, G.W. Responses of cattle, sheep and poultry to a recombinant vaccinia virus expressing a swine influenza haemagglutinin. Res. Vet. Sci. 1986, 41, 40–44. [Google Scholar] [CrossRef]
- Lopez, J.W.; Woods, G.T. Response of calves to exposure with swine influenza virus. Am. J. Vet. Res. 1987, 48, 1264–1268. [Google Scholar] [PubMed]
- Kalthoff, D.; Hoffmann, B.; Harder, T.; Durban, M.; Beer, M. Experimental infection of cattle with highly pathogenic avian influenza virus (H5N1). Emerg. Infect. Dis. 2008, 14, 1132–1134. [Google Scholar] [CrossRef]
- Phuong do, Q.; Dung, N.T.; Jorgensen, P.H.; Van, D.T.; Tung, D.D.; Christensen, J.P. Virulence of H5N1 influenza virus in Cattle Egrets (Bubulcus ibis). J. Wildl. Dis. 2011, 47, 314–320. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Francis, M.E.; King, M.L.; Kelvin, A.A. Back to the future for influenza preimmunity-looking back at influenza virus History to infer the outcome of future infections. Viruses 2019, 11, 122. [Google Scholar] [CrossRef]
- Kilbourne, E.D. Influenza pandemics of the 20th century. Emerg. Infect. Dis. 2006, 12, 9–14. [Google Scholar] [CrossRef]
- Nakamura, R.M.; Easterday, B.C. Serological studies of influenza in animals. Bull. World Health Organ. 1967, 37, 559–567. [Google Scholar]
- Onta, T.; Kida, H.; Kawano, J.; Matsuoka, Y.; Yanagawa, R. Dis.tribution of antibodies against various influenza A viruses in animals. Nihon Juigaku Zasshi 1978, 40, 451–454. [Google Scholar] [CrossRef]
- Saunders-Hastings, P.R.; Krewski, D. Reviewing the history of pandemic influenza: Understanding patterns of emergence and transmission. Pathogens 2016, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.J.; Bahl, J.; Vijaykrishna, D.; Zhang, J.; Poon, L.L.; Chen, H.; Webster, R.G.; Peiris, J.S.; Guan, Y. Dating the emergence of pandemic influenza viruses. Proc. Natl. Acad. Sci. USA 2009, 106, 11709–11712. [Google Scholar] [CrossRef] [PubMed]
- Jones-Lang, K.; Ernst-Larson, M.; Lee, B.; Goyal, S.M.; Bey, R. Prevalence of influenza A virus (H1N1) antibodies in bovine sera. New Microbiol. 1998, 21, 153–160. [Google Scholar] [PubMed]
- Mitchell, C.A.; Walker, R.V.; Bannister, G.L. Studies relating to the formation of neutralizing antibody following the propagation of influenza and Newcastle disease virus in the bovine mammary gland. Can. J. Microbiol. 1956, 2, 322–328. [Google Scholar] [CrossRef] [PubMed]
- Moustafa, M.E.-S.; Basem, M.A.; Ahmed, A.E.-S.; Youssef, I.Y. Isolation of H5 highly pathogenic avian influenza virus from cattle Egret (Bubulcus ibis) near affected broiler chicken flocks in Egypt. J. Virol. Sci. 2017, 2, 62. [Google Scholar]
- Lin, C.; Holland, R.E., Jr.; McCoy, M.H.; Donofrio-Newman, J.; Vickers, M.L.; Chambers, T.M. Infectivity of equine H3N8 influenza virus in bovine cells and calves. Influenza Other Respir. Viruses 2010, 4, 357–361. [Google Scholar] [CrossRef] [PubMed]
- Bronitki, A.; Demetrescu, R.; Malian, A. Incidence of anti-influenzal and anti-adenoviral antibodies in domestic and wild animals. Stud. Cercet. Inframicrobiol. 1965, 16, 217–220. [Google Scholar]
- Fyson, R.E.; Westwood, J.C.; Brunner, A.H. An immunoprecipitin study of the incidence of influenza A antibodies in animal sera in the Ottawa area. Can. J. Microbiol. 1975, 21, 1089–1101. [Google Scholar] [CrossRef] [PubMed]
- Chapman, M.S.; Lamont, P.H.; Harkness, J.W. Serological evidence of continuing infection of swine in Great Britain with an influenza A virus (H3N2). J. Hyg. 1978, 80, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Adair, B.M.; McFerran, J.B.; McKillop, E.R.; McCullough, S.J. Survey for antibodies to respiratory viruses in two groups of sheep in Northern Ireland. Vet. Rec. 1984, 115, 403–406. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.W.; Woods, G.T. Epidemiological study of swine influenza virus as a component of the respiratory disease complex of feeder calves. Res. Commun. Chem. Pathol. Pharmacol. 1986, 51, 417–420. [Google Scholar] [PubMed]
- Lopez, J.W. A single radial hemolysis technique for the measurement of influenza virus antibody in cattle serum. Res. Commun. Chem. Pathol. Pharmacol. 1986, 51, 273–276. [Google Scholar] [PubMed]
- El-Sayed, A.; Prince, A.; Fawzy, A.; Nadra, E.; Abdou, M.I.; Omar, L.; Fayed, A.; Salem, M. Sero-prevalence of avian influenza in animals and human in Egypt. Pak. J. Biol. Sci. 2013, 16, 524–529. [Google Scholar] [CrossRef] [PubMed]
- Samina, I.; Zakay-Rones, Z.; Peleg, B.A. Homologous and heterologous antibody response of cattle and sheep after vaccination with foot and mouth disease and influenza viruses. Vaccine 1998, 16, 551–557. [Google Scholar] [CrossRef]
- Knight, C.A. The nucleic acid and carbohydrate of influenza virus. J. Exp. Med. 1947, 85, 99–116. [Google Scholar] [CrossRef] [PubMed]
- Ada, G.L.; Perry, B.T. Infectivity and nucleic acid content of influenza virus. Nature 1955, 175, 209–210. [Google Scholar] [CrossRef] [PubMed]
- Rafelson, M.E., Jr. Studies on the metabolism of virus-infected tissues. III. The effect of influenza virus on ribonucleic acid fractions of chick chorio-allantoic membranes. Arch. Biochem. Biophys. 1960, 90, 68–72. [Google Scholar] [CrossRef]
- Davies, P.; Barry, R.D. Nucleic acid of influenza virus. Nature 1966, 211, 384–387. [Google Scholar] [CrossRef]
- Deusberg, P.H.; Robinson, W.S. On the structure and replication of influenza virus. J. Mol. Biol. 1967, 25, 383–405. [Google Scholar] [CrossRef]
- Pons, M.W. Studies on influenza virus ribonucleic acid. Virology 1967, 31, 523–531. [Google Scholar] [CrossRef]
- Duesberg, P.H. The RNA of influenza virus. Proc. Natl. Acad. Sci. USA 1968, 59, 930–937. [Google Scholar] [CrossRef] [PubMed]
- Compans, R.W.; Dimmock, N.J. An electron microscopic study of single-cycle infection of chick embryo fibroblasts by influenza virus. Virology 1969, 39, 499–515. [Google Scholar] [CrossRef]
- Duesberg, P.H. Distinct subunits of the ribonucleoprotein of influenza virus. J. Mol. Biol. 1969, 42, 485–499. [Google Scholar] [CrossRef]
- Nayak, D.P. Influenza virus: Structure, replication and defectiveness. Fed. Proc. 1969, 28, 1858–1866. [Google Scholar] [PubMed]
- Skehel, J.J.; Burke, D.C. Ribonucleic acid synthesis in chick embryo cells infected with fowl-plague virus. J. Virol. 1969, 3, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Bishop, D.H.; Obijeski, J.F.; Simpson, R.W. Transcription of the influenza ribonucleic acid genome by a virion polymerase. II. Nature of the in vitro polymerase product. J. Virol. 1971, 8, 74–80. [Google Scholar]
- Bishop, D.H.; Obijeski, J.F.; Simpson, R.W. Transcription of the influenza ribonucleic acid genome by a virion polymerase. I. Optimal conditions for in vitro activity of the ribonucleic acid-dependent ribonucleic acid polymerase. J. Virol. 1971, 8, 66–73. [Google Scholar]
- Penhoet, E.; Miller, H.; Doyle, M.; Blatti, S. RNA-dependent RNA polymerase activity in influenza virions. Proc. Natl. Acad. Sci. USA 1971, 68, 1369–1371. [Google Scholar] [CrossRef]
- Skehel, J.J. RNA-dependent RNA polymerase activity of the influenza virus. Virology 1971, 45, 793–796. [Google Scholar] [CrossRef]
- Schwarz, R. Purification and function of RNA dependent RNA polymerase of an influenza virus. Hoppe Seylers Z Physiol. Chem. 1972, 353, 1569. [Google Scholar]
- Hastie, N.D.; Mahy, B.W. RNA-dependent RNA polymerase in nuclei of cells infected with influenza virus. J. Virol. 1973, 12, 951–961. [Google Scholar] [PubMed]
- Hirst, G.K. Genetic recombination with Newcastle disease virus, polioviruses, and influenza. Cold Spring Harb. Symp. Quant. Biol. 1962, 27, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Choppin, P.W. Replication of influenza virus in a continuous cell line: High yield of infective virus from cells inoculated at high multiplicity. Virology 1969, 39, 130–134. [Google Scholar] [CrossRef]
- Choppin, P.W.; Pons, M.W. The RNAs of infective and incomplete influenza virions grown in MDBK and HeLa cells. Virology 1970, 42, 603–610. [Google Scholar] [CrossRef]
- Leiderman, E.; Mogabgab, W.J. Antigenicity of influenza vaccine from bovine cell cultures. Appl. Microbiol. 1969, 18, 596–600. [Google Scholar] [PubMed]
- Niven, J.S.; Armstrong, J.A.; Balfour, B.M.; Klemperer, H.G.; Tyrrell, D.A. Cellular changes accompanying the growth of influenza virus in bovine cell cultures. J. Pathol. Bacteriol. 1962, 84, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Ramos, B.A.; De Torres, R.A. B influenza virus proliferation in bovine etal kidney cell cultures, formation of incomplete virus. Rev. Latinoam. Microbiol. 1970, 12, 123–130. [Google Scholar]
- Enami, M.; Fukuda, R.; Ishihama, A. Transcription and replication of eight RNA segments of influenza virus. Virology 1985, 142, 68–77. [Google Scholar] [CrossRef]
- Compans, R.W.; Content, J.; Duesberg, P.H. Structure of the ribonucleoprotein of influenza virus. J. Virol. 1972, 10, 795–800. [Google Scholar]
- Compans, R.W.; Klenk, H.D.; Caliguiri, L.A.; Choppin, P.W. Influenza virus proteins. I. Analysis of polypeptides of the virion and identification of spike glycoproteins. Virology 1970, 42, 880–889. [Google Scholar] [CrossRef]
- Compans, R.W.; Caliguiri, L.A. Isolation and properties of an RNA polymerase from influenza virus-infected cells. J. Virol. 1973, 11, 441–448. [Google Scholar] [PubMed]
- Content, J.; Duesberg, P.H. Base sequence differences among the ribonucleic acids of influenza virus. J. Mol. Biol. 1971, 62, 273–285. [Google Scholar] [CrossRef]
- Lazarowitz, S.G.; Compans, R.W.; Choppin, P.W. Influenza virus structural and nonstructural proteins in infected cells and their plasma membranes. Virology 1971, 46, 830–843. [Google Scholar] [CrossRef]
- Schulman, J.L.; Palese, P. Virulence factors of influenza A viruses: WSN virus neuraminidase required for plaque production in MDBK cells. J. Virol. 1977, 24, 170–176. [Google Scholar] [PubMed]
- Beare, A.S.; Keast, K. Plaque formation by influenza B viruses in primary calf kidney cell monolayes. J. Gen. Virol. 1971, 13, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Lisok, T.P.; Sominina, A.A. Improved methods of influenza virus propagation. I. Enhancement of virus reproduction in cell cultures. Acta Virol. 1977, 21, 234–240. [Google Scholar] [PubMed]
- Sominina, A.A.; Lisok, T.P.; Rumel, N.B. Improved methods of influenza virus propagation. II. Characteristics of cell culture and allantoic virus preparations. Acta Virol. 1977, 21, 241–245. [Google Scholar]
- Gavrilov, V.I.; Asher, D.M.; Vyalushkina, S.D.; Ratushkina, L.S.; Zmieva, R.G.; Tumyan, B.G. Persistent infection of continuous line of pig kidney cells with a variant of the WSN strain of influenza A 0 virus. Proc. Soc. Exp. Biol. Med. 1972, 140, 109–117. [Google Scholar] [CrossRef]
- Krug, R.M. Cytoplasmic and nucleoplasmic viral RNPs in influenza virus-infected MDCK cells. Virology 1972, 50, 103–113. [Google Scholar] [CrossRef]
- Krug, R.M.; Etkind, P.R. Cytoplasmic and nuclear virus-specific proteins in influenza virus-infected MDCK cells. Virology 1973, 56, 334–348. [Google Scholar] [CrossRef]
- Urabe, M.; Tanaka, T.; Tobita, K. MDBK cells which survived infection with a mutant of influenza virus A/WSN and subsequently received many passages contained viral M and NS genes in full length in the absence of virus production. Arch. Virol. 1993, 130, 457–462. [Google Scholar] [CrossRef] [PubMed]
- De, B.K.; Nayak, D.P. Defective interfering influenza viruses and host cells: Establishment and maintenance of persistent influenza virus infection in MDBK and HeLa cells. J. Virol. 1980, 36, 847–859. [Google Scholar] [PubMed]
- Nayak, D.P. Defective interfering influenza viruses. Annu. Rev. Microbiol. 1980, 34, 619–644. [Google Scholar] [CrossRef]
- Von Magnus, P. Incomplete forms of influenza virus. Adv. Virus Res. 1954, 2, 59–79. [Google Scholar] [PubMed]
- Lazarowitz, S.G.; Compans, R.W.; Choppin, P.W. Proteolytic cleavage of the hemagglutinin polypeptide of influenza virus. Function of the uncleaved polypeptide HA. Virology 1973, 52, 199–212. [Google Scholar] [CrossRef]
- Lazarowitz, S.G.; Goldberg, A.R.; Choppin, P.W. Proteolytic cleavage by plasmin of the HA polypeptide of influenza virus: Host cell activation of serum plasminogen. Virology 1973, 56, 172–180. [Google Scholar] [CrossRef]
- Beare, A.S.; Keast, K.A. Influenza virus plaque formation in different species of cell monolayers. J. Gen. Virol. 1974, 22, 347–354. [Google Scholar] [CrossRef]
- Crecelius, D.M.; Deom, C.M.; Schulze, I.T. Biological properties of a hemagglutinin mutant of influenza virus selected by host cells. Virology 1984, 139, 164–177. [Google Scholar] [CrossRef]
- Deom, C.M.; Schulze, I.T. Oligosaccharide composition of an influenza virus hemagglutinin with host-determined binding properties. J. Biol. Chem. 1985, 260, 14771–14774. [Google Scholar]
- Mir-Shekari, S.Y.; Ashford, D.A.; Harvey, D.J.; Dwek, R.A.; Schulze, I.T. The glycosylation of the influenza A virus hemagglutinin by mammalian cells. A site-specific study. J. Biol. Chem. 1997, 272, 4027–4036. [Google Scholar] [CrossRef]
- Schulze, I.T. Effects of glycosylation on the properties and functions of influenza virus hemagglutinin. J. Infect. Dis. 1997, 176 (Suppl. 1), S24–S28. [Google Scholar] [CrossRef] [PubMed]
- Murphy, B.R.; Chalhub, E.G.; Nusinoff, S.R.; Chanock, R.M. Temperature-sensitive mutants of influenza virus. II. Attenuation of ts recombinants for man. J. Infect. Dis. 1972, 126, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Kitayama, T.; Togo, Y.; Hornick, R.B.; Friedwald, W.T. Low-temperature-adapted influenza A2/AA/6/60 virus vaccine in man. Infect. Immun. 1973, 7, 119–122. [Google Scholar] [PubMed]
- Sugiura, A.; Tobita, K.; Kilbourne, E.D. Isolation and preliminary characterization of temperature-sensitive mutants of influenza virus. J. Virol. 1972, 10, 639–647. [Google Scholar] [PubMed]
- Sokolova, N.N.; Irzhanov, S.D. Use of organ cultures of animal trachea and lung for studying respiratory viruses. Vopr. Virusol. 1976, 2, 220–222. [Google Scholar]
- Reed, S.E. The interaction of mycoplasmas and influenza viruses in tracheal organ cultures. J. Infect. Dis. 1971, 124, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Gorev, N.E. The effect of species and organ-specific features of tissue systems on the reactogenic and immunogenic properties of influenza virus. Acta Virol. 1975, 19, 41–46. [Google Scholar]
- Schaffer, F.L.; Soergel, M.E.; Straube, D.C. Survival of airborne influenza virus: Effects of propagating host, relative humidity, and composition of spray fluids. Arch. Virol. 1976, 51, 263–273. [Google Scholar] [CrossRef]
- Sreenivasan, C.C.; Jandhyala, S.S.; Luo, S.; Hause, B.M.; Thomas, M.; Knudsen, D.E.B.; Leslie-Steen, P.; Clement, T.; Reedy, S.E.; Chambers, T.M.; et al. Phylogenetic analysis and characterization of a sporadic isolate of equine influenza A H3N8 from an unvaccinated horse in 2015. Viruses 2018, 10, 31. [Google Scholar] [CrossRef]
- Gagov, I.; Pancheva-Golovinska, S. Studies on acitivity of some cellular enzymes and their connection with cytopathogenic effect at cultivation of the influenza virus in tissue cultures. Izv. Mikrobiol. Inst. (Sofiia) 1967, 19, 235–239. [Google Scholar]
- Gagov, I. Experiments in the adaptation of 2 strains of the influenza virus in a model monolayer cell culture of calf kidney. Izv. Mikrobiol. Inst. (Sofiia) 1969, 20, 191–201. [Google Scholar] [PubMed]
- De Sousa, C.P.; Belyavin, G. Some aspects of plaque formation by human influenza viruses. I. Cell and media factors affecting a modified plaque technique. Arch. Gesamte Virusforsch. 1970, 32, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Gagov, I.; Manolova, N. Adaptation to influenza virus A2 (Hong Kong) 68 in calf kidney cell cultures. Izv. Mikrobiol. Inst. (Sofiia) 1972, 23, 123–128. [Google Scholar] [PubMed]
- Lenard, J.; Compans, R.W. Polypeptide composition of incomplete influenza virus grown in MDBK cells. Virology 1975, 65, 418–426. [Google Scholar] [CrossRef]
- Lee, Y.S.; Seong, B.L. Nucleotides in the panhandle structure of the influenza B virus virion RNA are involved in the specificity between influenza A and B viruses. J. Gen. Virol. 1998, 79, 673–681. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lenard, J.; Tsai, D.K.; Compans, R.W.; Landsberger, F.R. Observations on the membrane organization of standard and incomplete influenza grown in MDBK cells. Virology 1976, 71, 389–394. [Google Scholar] [CrossRef]
- Ransohoff, R.M.; Maroney, P.A.; Nayak, D.P.; Chambers, T.M.; Nilsen, T.W. Effect of human alpha A interferon on influenza virus replication in MDBK cells. J. Virol. 1985, 56, 1049–1052. [Google Scholar]
- Lanni, F.; Lanni, Y.T.; Beard, J.W. Inhibitory effect of cow’s milk on influenza virus hemagglutination. Proc. Soc. Exp. Biol. Med. 1949, 72, 227–232. [Google Scholar] [CrossRef]
- Perdijk, O.; Van Splunter, M.; Savelkoul, H.F.J.; Brugman, S.; Van Neerven, R.J.J. Cow’s milk and immune function in the respiratory tract: Potential mechanisms. Front. Immunol. 2018, 9, 143. [Google Scholar] [CrossRef]
- Alisky, J. Bovine and human-derived passive immunization could help slow a future avian influenza pandemic. Med. Hypotheses 2009, 72, 74–75. [Google Scholar] [CrossRef]
- Curtain, C.C.; Pye, J. A mucoprotein from bovine submaxillary glands with restricted inhibitory action against influenza virus haemagglutination. Aust. J. Exp. Biol. Med. Sci. 1955, 33, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Lycke, E. A factor in bovine amniotic fluid inhibiting influenza virus hemagglutination. Arch. Gesamte Virusforsch. 1954, 5, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Ng, W.C.; Wong, V.; Muller, B.; Rawlin, G.; Brown, L.E. Prevention and treatment of influenza with hyperimmune bovine colostrum antibody. PLoS ONE 2010, 5, e13622. [Google Scholar] [CrossRef] [PubMed]
- Uchida, K.; Hiruta, N.; Yamaguchi, H.; Yamashita, K.; Fujimura, K.; Yasui, H. Augmentation of cellular immunity and protection against influenza virus infection by bovine late colostrum in mice. Nutrition 2012, 28, 442–446. [Google Scholar] [CrossRef] [PubMed]
- Wong, E.B.; Mallet, J.F.; Duarte, J.; Matar, C.; Ritz, B.W. Bovine colostrum enhances natural killer cell activity and immune response in a mouse model of influenza infection and mediates intestinal immunity through toll-like receptors 2 and 4. Nutr. Res. 2014, 34, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.L.; Kim, H.J.; Chang, D.Y.; Kim, H.J. The effect of dietary intake of the acidic protein fraction of bovine colostrum on influenza A (H1N1) virus infection. J. Microbiol. 2013, 51, 389–393. [Google Scholar] [CrossRef]
- Benson, K.F.; Carter, S.G.; Patterson, K.M.; Patel, D.; Jensen, G.S. A novel extract from bovine colostrum whey supports anti-bacterial and anti-viral innate immune functions in vitro and in vivo: I. Enhanced immune activity in vitro translates to improved microbial clearance in animal infection models. Prev. Med. 2012, 54, S116–S123. [Google Scholar] [CrossRef]
- Ammendolia, M.G.; Agamennone, M.; Pietrantoni, A.; Lannutti, F.; Siciliano, R.A.; De Giulio, B.; Amici, C.; Superti, F. Bovine lactoferrin-derived peptides as novel broad-spectrum inhibitors of influenza virus. Pathog. Glob. Health 2012, 106, 12–19. [Google Scholar] [CrossRef]
- Pietrantoni, A.; Ammendolia, M.G.; Superti, F. Bovine lactoferrin: Involvement of metal saturation and carbohydrates in the inhibition of influenza virus infection. Biochem. Cell. Biol. 2012, 90, 442–448. [Google Scholar] [CrossRef]
- Pietrantoni, A.; Dofrelli, E.; Tinari, A.; Ammendolia, M.G.; Puzelli, S.; Fabiani, C.; Donatelli, I.; Superti, F. Bovine lactoferrin inhibits influenza A virus induced programmed cell death in vitro. Biometals 2010, 23, 465–475. [Google Scholar] [CrossRef]
- Shin, K.; Wakabayashi, H.; Yamauchi, K.; Teraguchi, S.; Tamura, Y.; Kurokawa, M.; Shiraki, K. Effects of orally administered bovine lactoferrin and lactoperoxidase on influenza virus infection in mice. J. Med. Microbiol. 2005, 54, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Scala, M.C.; Sala, M.; Pietrantoni, A.; Spensiero, A.; Di Micco, S.; Agamennone, M.; Bertamino, A.; Novellino, E.; Bifulco, G.; Gomez-Monterrey, I.M.; et al. Lactoferrin-derived peptides active towards influenza: Identification of three potent tetrapeptide inhibitors. Sci. Rep. 2017, 7, 10593. [Google Scholar] [CrossRef] [PubMed]
- Superti, F.; Agamennone, M.; Pietrantoni, A.; Ammendolia, M.G. Bovine lactoferrin prevents influenza A virus infection by interfering with the fusogenic function of viral hemagglutinin. Viruses 2019, 11, 51. [Google Scholar] [CrossRef]
- Yamauchi, K.; Wakabayashi, H.; Shin, K.; Takase, M. Bovine lactoferrin: Benefits and mechanism of action against infections. Biochem. Cell. Biol. 2006, 84, 291–296. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, C.P.; Bal, A. Sensitivity of A2-Hong Kong and pre-1968 A2 influenza virus strains to normal serum inhibitors. Acta Virol. 1971, 15, 367–373. [Google Scholar] [PubMed]
- Beardmore, W.B.; Jones, K.V.; Clark, T.D.; Hebeka, E.K. Induction of an inhibitor of influenza virus hemagglutination by treatment of serum with periodate. Appl. Microbiol. 1968, 16, 563–568. [Google Scholar] [PubMed]
- Saiatov, M.; Shuratov, I.; Beisembaeva, R.U.; Akhmetova, N.A.; Asanova, S.E. Quantitative indices and physico-chemical properties of non-specific inhibitors of influenza A virus hemagglutination in the sera of different species of animals and birds. Vopr. Virusol. 1977, 3, 299–304. [Google Scholar]
- Kawai, T.; Kase, T.; Suzuki, Y.; Eda, S.; Sakamoto, T.; Ohtani, K.; Wakamiya, N. Anti-influenza A virus activities of mannan-binding lectins and bovine conglutinin. J. Vet. Med. Sci. 2007, 69, 221–224. [Google Scholar] [CrossRef][Green Version]
- Anders, E.M.; Hartley, C.A.; Jackson, D.C. Bovine and mouse serum beta inhibitors of influenza A viruses are mannose-binding lectins. Proc. Natl. Acad. Sci. USA 1990, 87, 4485–4489. [Google Scholar] [CrossRef]
- Eda, S.; Suzuki, Y.; Kase, T.; Kawai, T.; Ohtani, K.; Sakamoto, T.; Kurimura, T.; Wakamiya, N. Recombinant bovine conglutinin, lacking the N-terminal and collagenous domains, has less conglutination activity but is able to inhibit haemagglutination by influenza A virus. Biochem. J. 1996, 316, 43–48. [Google Scholar] [CrossRef]
- Hartley, C.A.; Jackson, D.C.; Anders, E.M. Two distinct serum mannose-binding lectins function as beta inhibitors of influenza virus: Identification of bovine serum beta inhibitor as conglutinin. J. Virol. 1992, 66, 4358–4363. [Google Scholar] [PubMed]
- Hartshorn, K.L.; Sastry, K.; Brown, D.; White, M.R.; Okarma, T.B.; Lee, Y.M.; Tauber, A.I. Conglutinin acts as an opsonin for influenza A viruses. J. Immunol. 1993, 151, 6265–6273. [Google Scholar] [PubMed]
- Wakamiya, N.; Okuno, Y.; Sasao, F.; Ueda, S.; Yoshimatsu, K.; Naiki, M.; Kurimura, T. Isolation and characterization of conglutinin as an influenza A virus inhibitor. Biochem. Biophys. Res. Commun. 1992, 187, 1270–1278. [Google Scholar] [CrossRef]
- Suzuki, Y.; Eda, S.; Kawai, T.; Ohtani, K.; Kase, T.; Sakamoto, T.; Wakamiya, N. Characterization of recombinant bovine conglutinin expressed in a mammalian cell. Biochem. Biophys. Res. Commun. 1997, 238, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Reading, P.C.; Holmskov, U.; Anders, E.M. Antiviral activity of bovine collectins against rotaviruses. J. Gen. Virol. 1998, 79, 2255–2263. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hartshorn, K.L.; White, M.R.; Smith, K.; Sorensen, G.; Kuroki, Y.; Holmskov, U.; Head, J.; Crouch, E.C. Increasing antiviral activity of surfactant protein d trimers by introducing residues from bovine serum collectins: Dissociation of mannan-binding and antiviral activity. Scand. J. Immunol. 2010, 72, 22–30. [Google Scholar] [CrossRef]
- Dec, M.; Wernicki, A. Conglutinin, CL-43 and CL-46--three bovine collectins. Pol. J. Vet. Sci. 2006, 9, 265–275. [Google Scholar]
- Zhirnov, O.P.; Matrosovich, T.Y.; Matrosovich, M.N.; Klenk, H.D. Aprotinin, a protease inhibitor, suppresses proteolytic activation of pandemic H1N1v influenza virus. Antivir. Chem. Chemother. 2011, 21, 169–174. [Google Scholar] [CrossRef]
- Zhirnov, O.P.; Klenk, H.D.; Wright, P.F. Aprotinin and similar protease inhibitors as drugs against influenza. Antivir. Res. 2011, 92, 27–36. [Google Scholar] [CrossRef]
- Kimoto, T.; Mizuno, D.; Takei, T.; Kunimi, T.; Ono, S.; Sakai, S.; Kido, H. Intranasal influenza vaccination using a new synthetic mucosal adjuvant SF-10: Induction of potent local and systemic immunity with balanced Th1 and Th2 responses. Influenza Other Respir. Viruses 2013, 7, 1218–1226. [Google Scholar] [CrossRef]
- Glasky, A.J.; Simon, L.; Holper, J.C. Interferons: Selectivity and specificity of action in cell-free systems. Science 1964, 144, 1581–1583. [Google Scholar] [CrossRef] [PubMed]
- Horisberger, M.A. The action of recombinant bovine interferons on influenza virus replication correlates with the induction of two Mx-related proteins in bovine cells. Virology 1988, 162, 181–186. [Google Scholar] [CrossRef]
- Verhelst, J.; Hulpiau, P.; Saelens, X. Mx proteins: Antiviral gatekeepers that restrain the uninvited. Microbiol. Mol. Biol. Rev. 2013, 77, 551–566. [Google Scholar] [CrossRef] [PubMed]
- Garigliany, M.M.; Cornet, A.; Desmecht, D. Human/bovine chimeric MxA-like GTPases reveal a contribution of N-terminal domains to the magnitude of anti-influenza A activity. J. Interferon Cytokine Res. 2012, 32, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E. Influenza virus inhibitor in human milk and cow’s milk. Part 1. Comparative studies on neuraminic acid content and the level of the inhibitor titer in the hemagglutination test. Z. Kinderheilkd. 1960, 84, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Krizanova, O.; Rathova, V.; Kociskova, D.; Szanto, J. Purification and properties of beta inhibitor from bovine serum. Bratisl. Lek. Listy 1963, 2, 22–30. [Google Scholar] [PubMed]
- Moisa, I. Sensitivity of hemagglutinins of some influenza viruses to non-specific inhibitors from the serum of various species of animals. Stud. Cercet. Virusol. 1972, 23, 283–300. [Google Scholar] [PubMed]
- Shortridge, K.F.; Lansdell, A. Serum inhibitors of A 2 -Hong Kong influenza virus haemagglutination. Microbios 1972, 6, 213–219. [Google Scholar] [PubMed]
- Osipova, Z.A.; Medvedeva, M.N. Effect of cattle serum inhibitors on persistence of influenza virus in MDSK cells. Mikrobiol. Zhurnal 1980, 42, 390–391. [Google Scholar]
- Hartshorn, K.L.; Holmskov, U.; Hansen, S.; Zhang, P.; Meschi, J.; Mogues, T.; White, M.R.; Crouch, E.C. Distinctive anti-influenza properties of recombinant collectin 43. Biochem. J. 2002, 366, 87–96. [Google Scholar] [CrossRef]
- Singh, N.; Pandey, A.; Jayashankar, L.; Mittal, S.K. Bovine adenoviral vector-based H5N1 influenza vaccine overcomes exceptionally high levels of pre-existing immunity against human adenovirus. Mol. Ther. 2008, 16, 965–971. [Google Scholar] [CrossRef] [PubMed]
- Sayedahmed, E.E.; Hassan, A.O.; Kumari, R.; Cao, W.; Gangappa, S.; York, I.; Sambhara, S.; Mittal, S.K. A bovine adenoviral vector-based H5N1 influenza -vaccine provides enhanced immunogenicity and protection at a significantly low dose. Mol. Ther. Methods Clin. Dev. 2018, 10, 210–222. [Google Scholar] [CrossRef] [PubMed]
- Mailybayeva, A.; Yespembetov, B.; Ryskeldinova, S.; Zinina, N.; Sansyzbay, A.; Renukaradhya, G.J.; Petrovsky, N.; Tabynov, K. Improved influenza viral vector based Brucella abortus vaccine induces robust B and T-cell responses and protection against Brucella melitensis infection in pregnant sheep and goats. PLoS ONE 2017, 12, e0186484. [Google Scholar] [CrossRef] [PubMed]
- Tabynov, K. Influenza viral vector based Brucella abortus vaccine: A novel vaccine candidate for veterinary practice. Expert Rev. Vaccines 2016, 15, 1237–1239. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Hika, B.; Liu, R.; Sheng, Z.; Hause, B.M.; Li, F.; Wang, D. The hemagglutinin-esterase fusion glycoprotein Is a primary determinant of the exceptional thermal and acid stability of influenza D virus. mSphere 2017, 2, e00254-17. [Google Scholar] [CrossRef] [PubMed]
| Description | Year | Ref. |
|---|---|---|
| Natural infection | ||
| Influenza outbreak in Japan | 1951 | [76] |
| *Incidence, level of influenza, and adeno virus antibodies in domestic animal species | 1969 | [25] |
| *Antigenic characteristics of influenza viruses from domestic animals and birds (USSR) | 1973 | [89] |
| *Influenza outbreak in cattle | 1973 | [90] |
| Isolation of influenza A strains from cattle | 1973 | [46] |
| *Study of cattle influenza | 1976 | [92] |
| *Influenza in cattle | 1977 | [91] |
| Hong Kong influenza A strains in calves | 1977 | [48] |
| *Pathological anatomical examination of the lung from bovine influenza | 1977 | [94] |
| *Influenza of cattle | 1978 | [93] |
| Unexplained sporadic milk drop in cows | 1997 | [95] |
| Bovine influenza | 1998 | [97] |
| Influenza A in dairy cows with sporadic milk syndrome | 1999 | [98] |
| Evidence of antibodies in sera/nasal samples against human influenza viruses from 17 outbreaks of respiratory disease with milk drop syndrome and diarrhea in cattle in 1998–1999 | 2002 | [99] |
| Wild animals as a reservoir for different bacterial and viral diseases including avian influenza | 2002 | [102] |
| Experimental infection | ||
| *Experimental infection of bovines with human influenza virus | 1954 | [26] |
| *Experimental inoculation of calves with influenza virus A/csf/Udmurtiia/116/73 | 1977 | [27] |
| Experimental infection of influenza in yak and presence of H3N2 antibodies | 1974 | [103] |
| Recombinant vaccinia virus expressing HA in cattle, sheep, and poultry | 1986 | [104] |
| Intranasal inoculation of calves with live swine influenza virus | 1987 | [105] |
| Experimental inoculation of a cat derived highly pathogenic avian influenza virus in calves | 2008 | [106] |
| Experimental inoculation of highly pathogenic avian influenza virus H5N1 in cattle egrets | 2011 | [107] |
| Description | Year | Ref. |
|---|---|---|
| Influenza A and B specific antibodies in domestic and wild animals | 1965 | [118] |
| Serological study in Ottawa based on immunoprecipitation test found influenza A antibodies in sheep and goat among 14 species tested | 1975 | [119] |
| Serosurveillance of swine H1N1 in cattle and swine in Great Britain | 1978 | [120] |
| Serological screening of influenza B and C in cattle, horses and other animals in Japan | 1978 | [56] |
| Serological evidence of influenza A in cattle in Japan | 1978 | [111] |
| Influenza-specific antibodies not detected in indigenous and non-indigenous sheep breeds of Northern Ireland | 1984 | [121] |
| Influenza in ruminants: a review with information regarding viruses isolated from the cattle | 1984 | [47] |
| Single radial hemolysis to measure influenza antibody in cattle serum | 1986 | [123] |
| Swine influenza virus as a component in the respiratory disease complex in calves | 1986 | [122] |
| Presence of influenza A specific antibodies in cattle with respiratory disease and reduced milk yield | 1998 | [96] |
| Vaccination study of foot and mouth disease and influenza in cattle and sheep | 1998 | [125] |
| Influenza A antibodies associated with an acute reduction in milk yield in cattle in Britain | 2008 | [100] |
| Serosurveillance study of avian influenza H5N1 in cattle, buffaloes, sheep, goat, and other animals in Egypt | 2013 | [124] |
| Description | Year | Ref. |
|---|---|---|
| Studies on the cellular enzymes and their role in the cytopathic effect of influenza in cell cultures | 1967 | [181] |
| Replication of WSN influenza virus in high titers in MDBK cell line at high and low multiplicity of infection | 1969 | [144] |
| *Adaptation of the influenza virus in calf kidney cell cultures | 1969 | [182] |
| Study of infective and incomplete influenza virions grown in MDBK and HeLa cells | 1970 | [145] |
| Calf serum suppressed plaque formation of many influenza virus strains in different cell lines | 1970 | [183] |
| Influenza B virus propagation in bovine fetal kidney cell cultures: incomplete virus formation | 1970 | [148] |
| Influenza B virus forms plaque in primary calf kidney cells | 1971 | [156] |
| Interaction of swine influenza and bovine mycoplasma in bovine tracheal cultures | 1971 | [177] |
| *Influenza virus adaptation (A2 (Hong Kong) 68) in calf kidney cell cultures | 1972 | [184] |
| Use of bovine kidney cells for the propagation of ts recombinant viruses | 1972 | [173] |
| Use of primary bovine kidney cells at 25 °C for growing low temperature adapted vaccine virus | 1973 | [174] |
| Influenza virions grown in Madin–Darby Bovine Kidney (MDBK) cells without calf serum have more uncleaved HA especially in the early phase indicating that HA cleavage is both host cell and strain dependent | 1973 | [166] |
| Influenza virus grown in MDBK cells in the presence of medium containing 2% calf serum caused cleavage of HA polypeptide, to HA1 and HA2 unlike serum-free medium due to the plasminogen component in the sera | 1973 | [167] |
| Use of bovine kidney and trachea organ cultures for influenza for studying the virus reactogenic and immunogenic properties | 1975 | [178] |
| Study of the polypeptide composition of incomplete influenza viruses propagated in MDBK cells | 1975 | [185] |
| Use of lung and trachea organ cultures from bovine and other species for the influenza studies | 1976 | [176] |
| Use of bovine embryo kidney cells, roller cultures for high titer influenza virus propagation | 1977 | [157] |
| Description | Year | Ref. |
|---|---|---|
| Inhibition of influenza virus hemagglutination by cow’s milk | 1949 | [189] |
| *Inhibition of influenza virus hemagglutination by bovine amniotic fluid factor | 1954 | [193] |
| Mucoprotein from bovine submaxillary glands with restricted hemagglutination inhibition activity against influenza virus | 1955 | [192] |
| Influenza virus inhibitor in human and cow milk | 1960 | [226] |
| *Beta inhibitors in bovine serum | 1963 | [227] |
| Use of sera from different species of animals to study periodate induced hemagglutination inhibitor | 1968 | [207] |
| Influenza A2/Hong Kong strains were sensitive to periodate-resistant inhibitors in normal bovine serum | 1971 | [206] |
| *Serum inhibitors of hemagglutination | 1972 | [228] |
| Serum inhibitors of hemagglutination of A2/Hong Kong strains | 1972 | [229] |
| *Characterization of non-specific inhibitors of hemagglutination of influenza A virus in the sera of different species of animals and birds | 1977 | [208] |
| *Serum inhibitors of cattle and influenza virus persistence in Madin–Darby Swine Kidney (MDSK) cells | 1980 | [230] |
| Conglutinin as bovine serum beta inhibitor of influenza virus hemagglutination and infectivity of H1 and H3 subtypes | 1992 | [212] |
| Isolation of influenza virus inhibitor, conglutinin | 1992 | [214] |
| Opsonizing activity of conglutinin against influenza A virus | 1993 | [213] |
| Recombinant bovine conglutinin deficient in N-terminal and collagenous domains and its activity against influenza virus | 1996 | [211] |
| Bovine collectins and its antiviral activity against rota virus | 1998 | [216] |
| CL-43, bovine serum collectin, and its antiviral activity by inhibiting the hemagglutination activity of influenza A virus | 2002 | [231] |
| Bovine collectins_Conglutinin, CL-43, and CL-46 | 2006 | [218] |
| Recombinant trimeric neck and carbohydrate recognition domains (NCRD) of bovine conglutinin and CL-46 demonstrated a higher level of intrinsic antiviral activity against influenza A virus | 2010 | [217] |
| Aprotinin, a natural polypeptide of bovine lung origin can inhibit the HA cleavage of pdmH1N1 and its replication in different host systems | 2011 | [219] |
| Bovine colostrum can enhance natural killer cell activity and thus boosts the immune response against influenza in a mouse model | 2014 | [196] |
| Peptide inhibitors derived from lactoferrin C-lobe possess broad anti-influenza activity and prevented influenza hemagglutination and infection | 2012, 2017 | [199,203] |
| Bovine lactoferrin interferes with the fusion of HA glycoprotein and thus inhibits influenza A infection | 2019 | [204] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sreenivasan, C.C.; Thomas, M.; Kaushik, R.S.; Wang, D.; Li, F. Influenza A in Bovine Species: A Narrative Literature Review. Viruses 2019, 11, 561. https://doi.org/10.3390/v11060561
Sreenivasan CC, Thomas M, Kaushik RS, Wang D, Li F. Influenza A in Bovine Species: A Narrative Literature Review. Viruses. 2019; 11(6):561. https://doi.org/10.3390/v11060561
Chicago/Turabian StyleSreenivasan, Chithra C., Milton Thomas, Radhey S. Kaushik, Dan Wang, and Feng Li. 2019. "Influenza A in Bovine Species: A Narrative Literature Review" Viruses 11, no. 6: 561. https://doi.org/10.3390/v11060561
APA StyleSreenivasan, C. C., Thomas, M., Kaushik, R. S., Wang, D., & Li, F. (2019). Influenza A in Bovine Species: A Narrative Literature Review. Viruses, 11(6), 561. https://doi.org/10.3390/v11060561






