Transcriptome Response of Female Culicoides sonorensis Biting Midges (Diptera: Ceratopogonidae) to Early Infection with Epizootic Hemorrhagic Disease Virus (EHDV-2)
Abstract
1. Introduction
2. Materials and Methods
2.1. Virus Culture
2.2. Culicoides sonorensis Rearing and Treatment
2.3. Culicoides sonorensis Processing for Transcriptome Analysis
2.4. Transcriptome Pre-Processing, Construction and Sequencing of Libraries
2.5. De novo Assembly and Annotation
2.6. Analysis of Differential Unigene Expression and Gene Ontology Enrichment
2.7. KEGG Pathway Analysis
2.8. Analysis of Sensory, Memory and Behavior Genes
2.9. qRT-PCR Validation of Expression of Selected Unigenes
3. Results
3.1. Virus Titer
3.2. Sequencing and Updating of the Culicoides sonorensis Reference Transcriptome
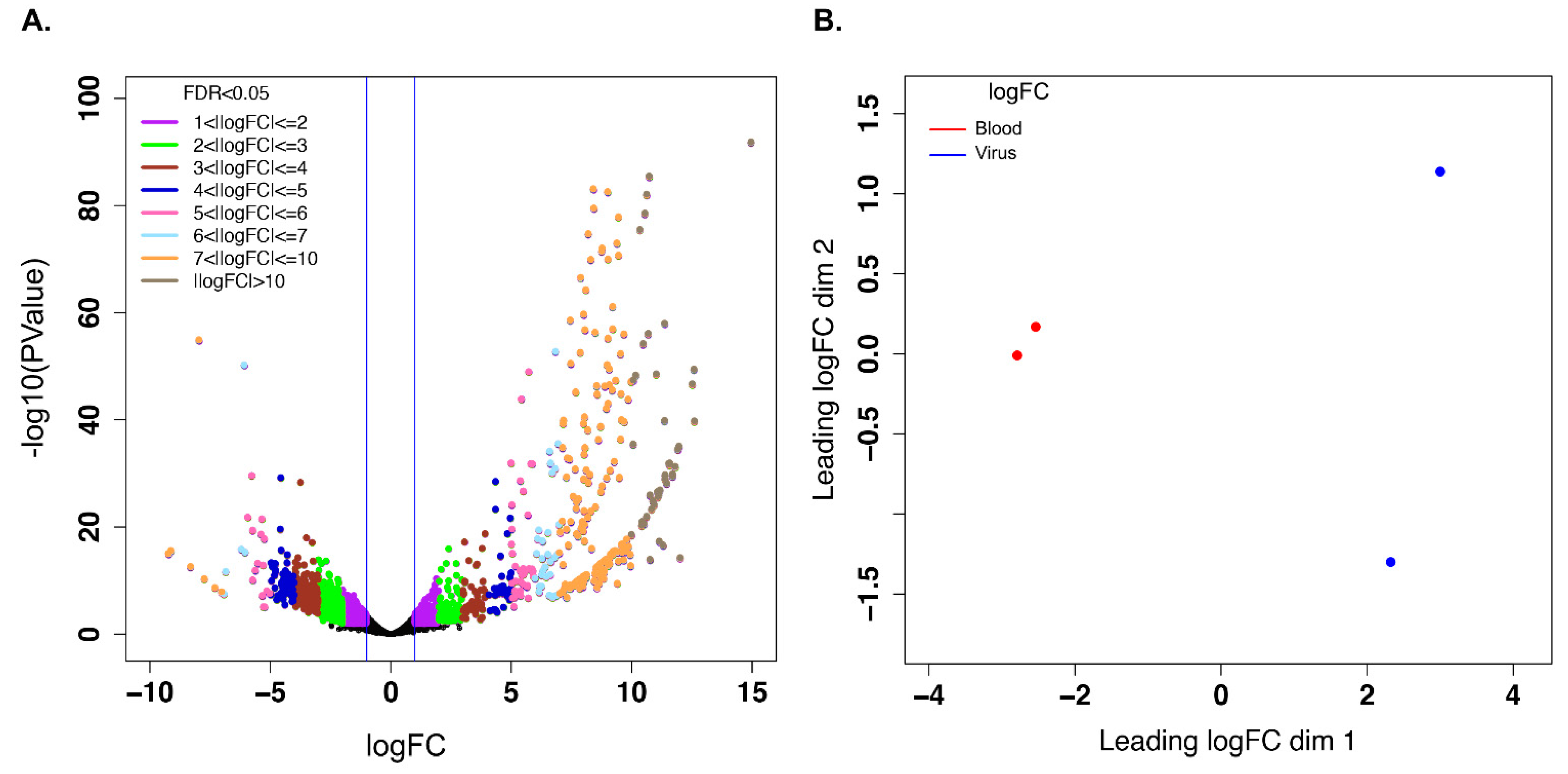
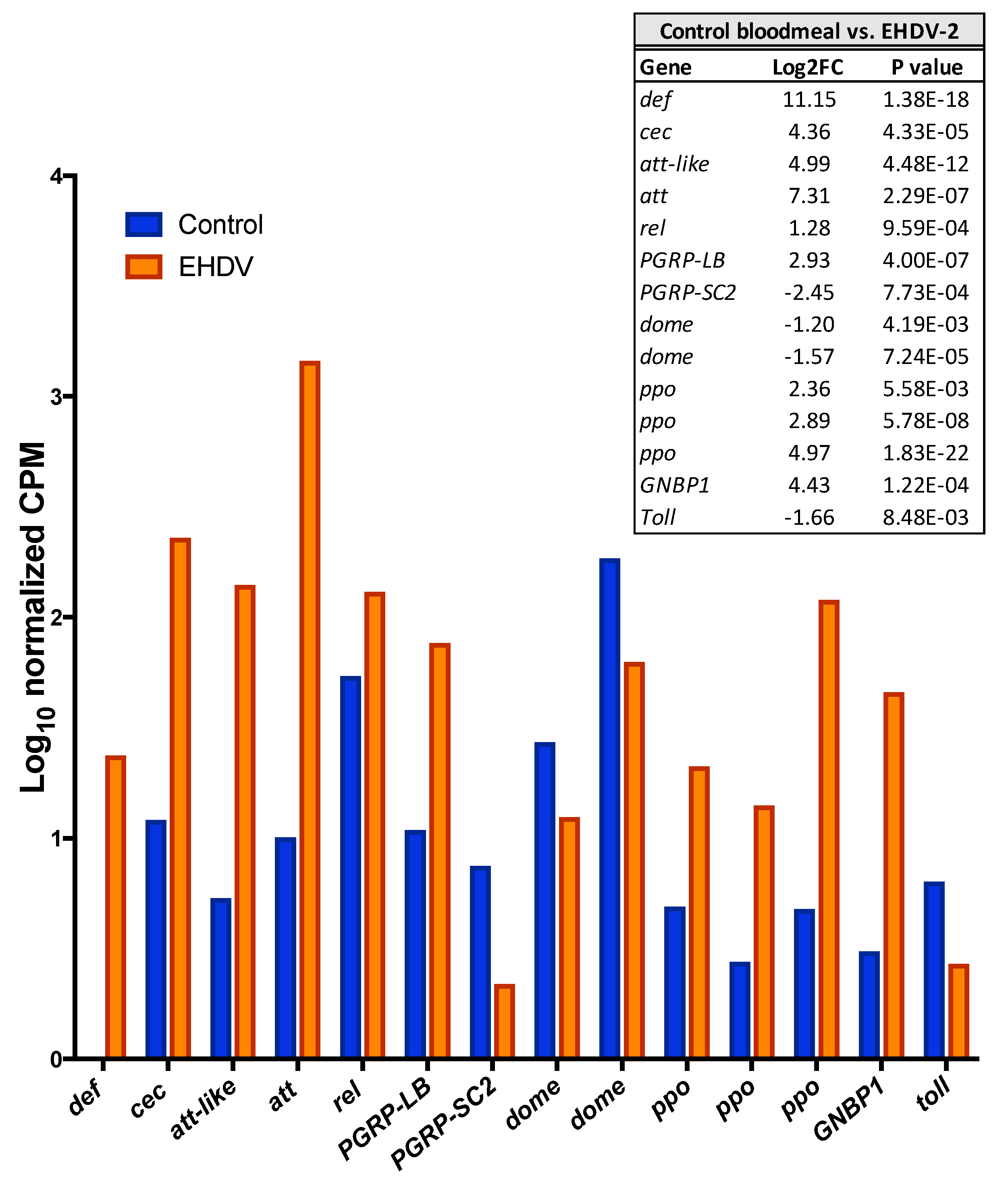
3.3. Unigenes That Were Differentially Expressed during Early EHDV Infection
3.4. Gene Ontology Term Enrichment
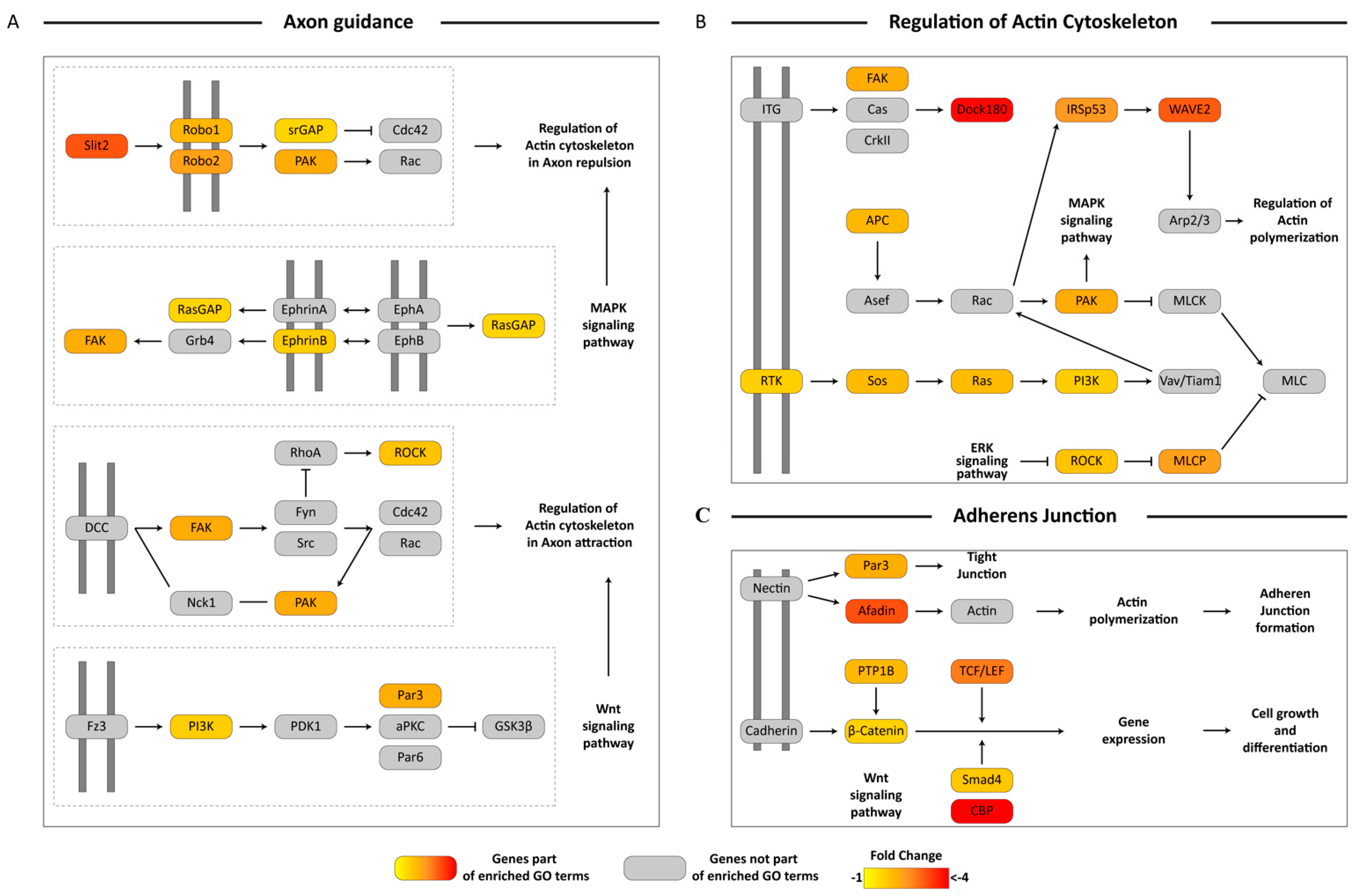
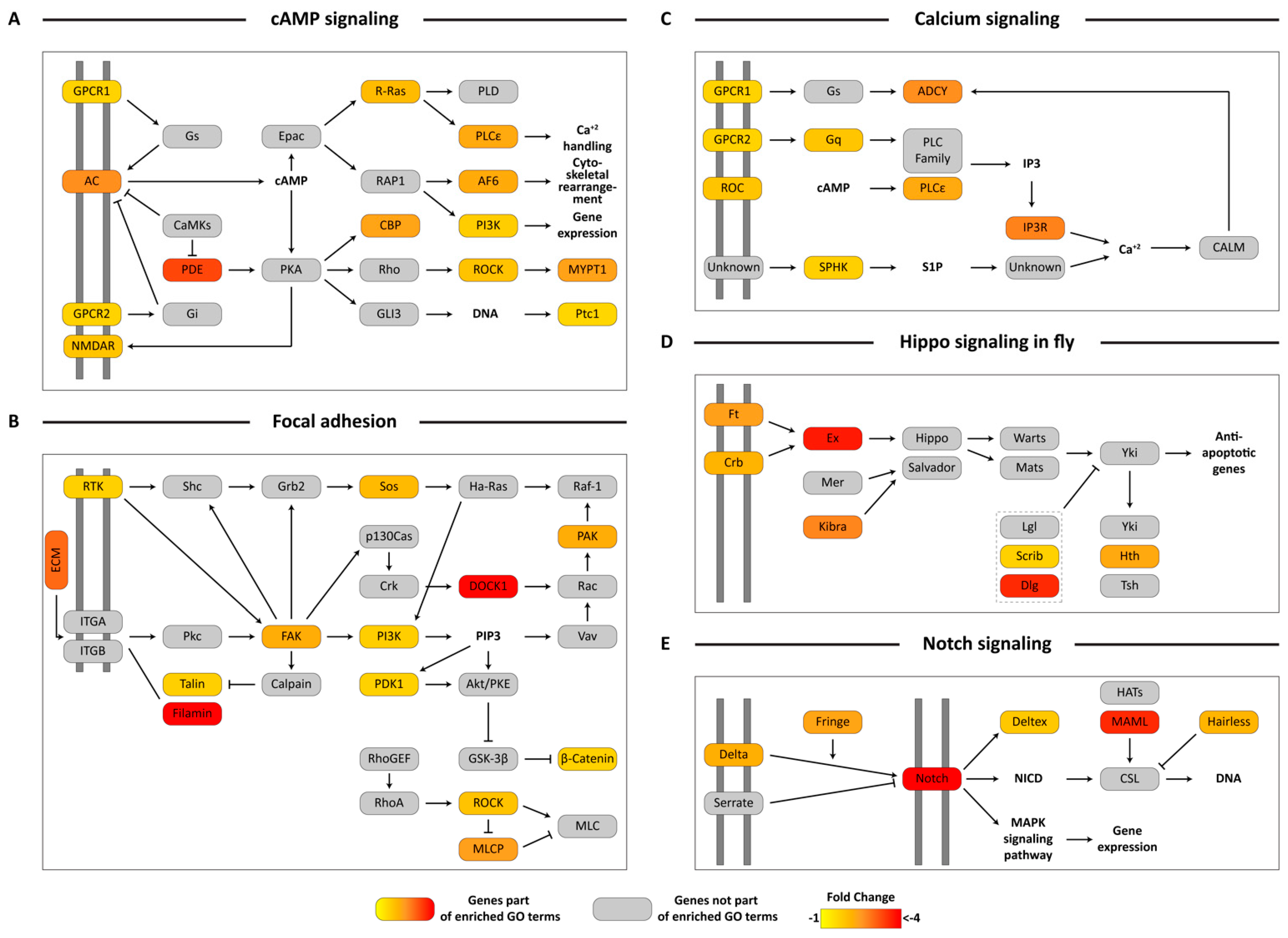
3.5. KEGG Pathway Mapping of Unigenes Associated with Biologically-Relevant GO Terms
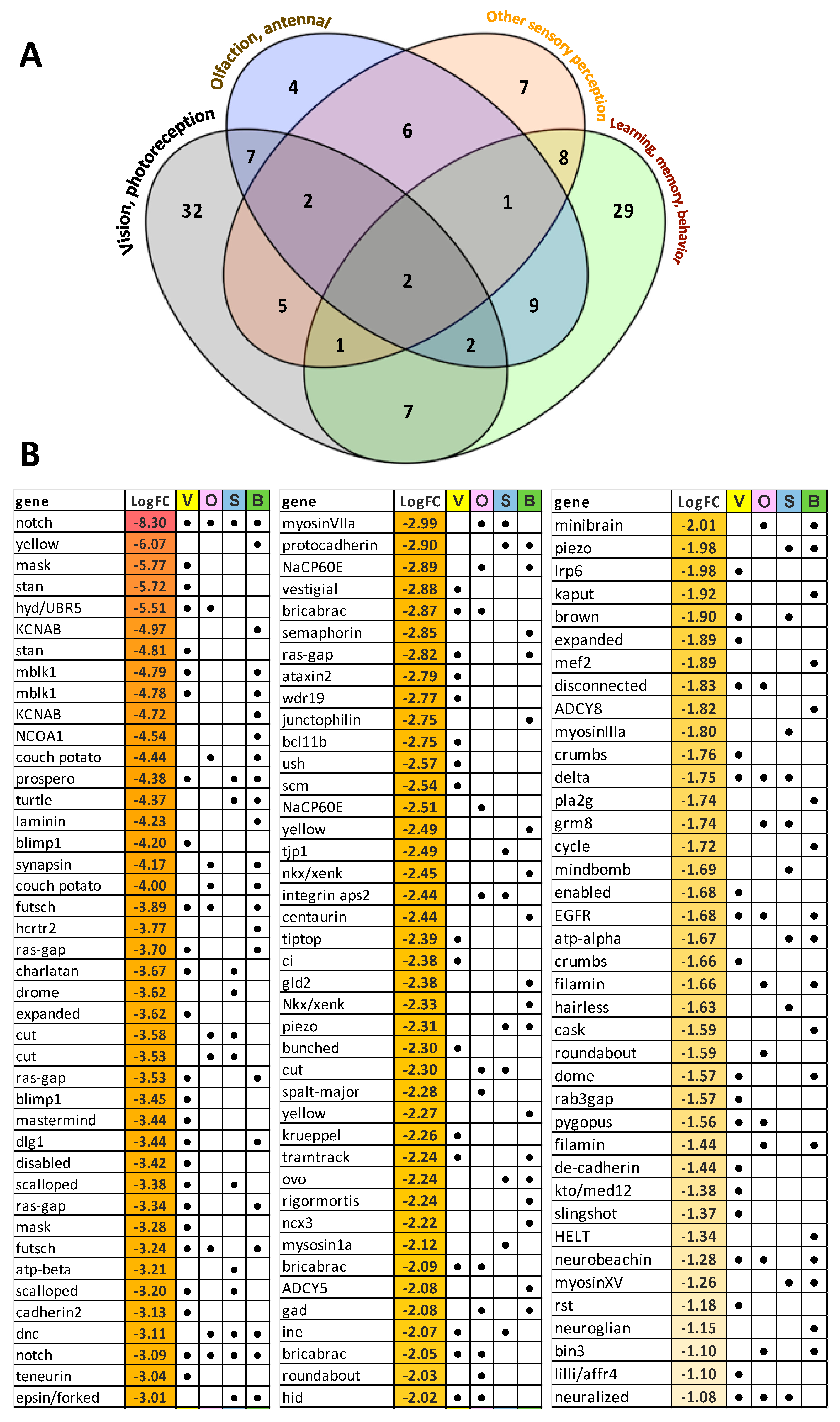
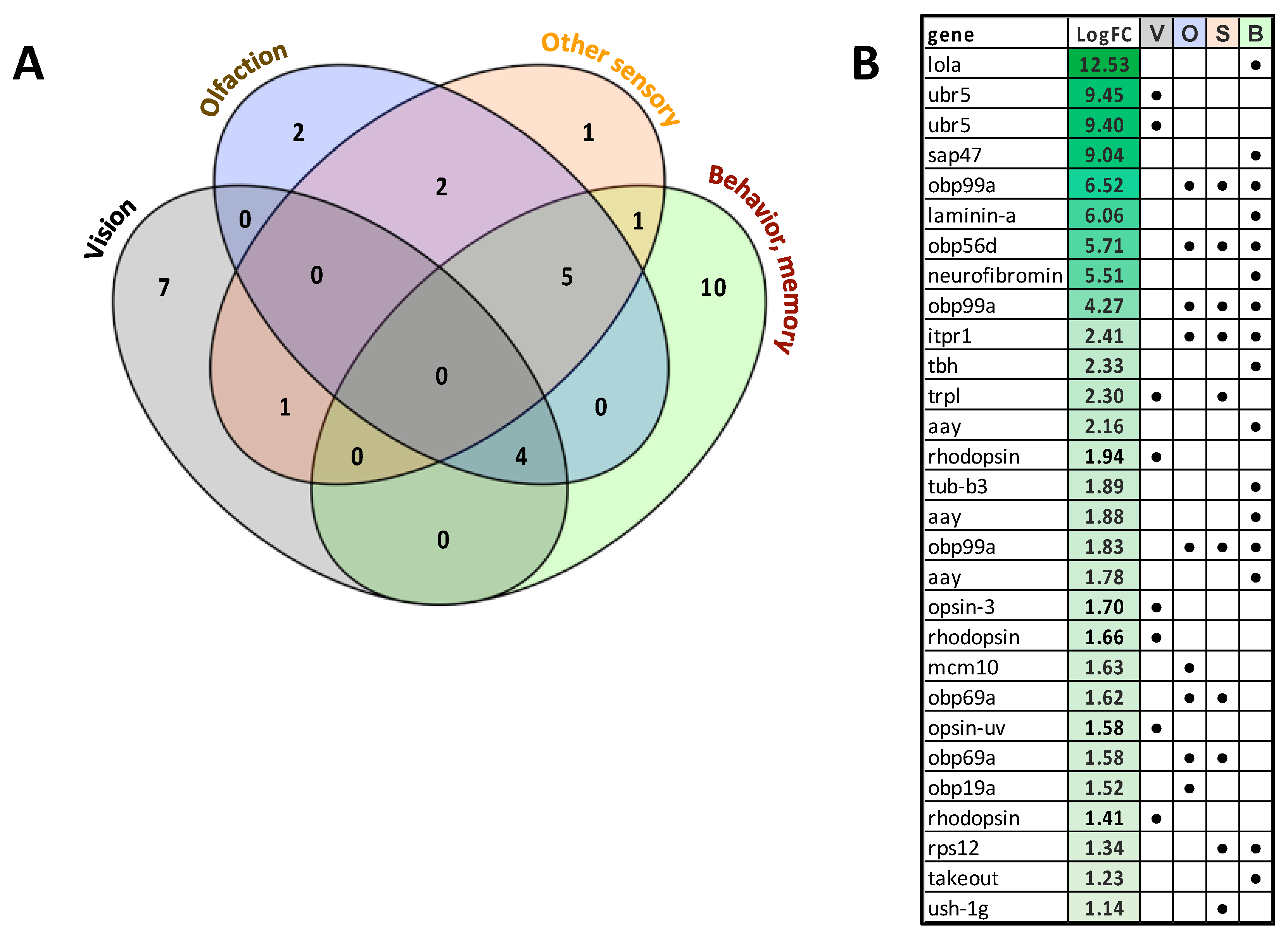
3.6. Analysis of Unigenes Associated with Sensory Processes, Behavior, Learning and Memory
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ruder, M.G.; Lysyk, T.J.; Stallknecht, D.E.; Foil, L.D.; Johnson, D.J.; Chase, C.C.; Dargatz, D.A.; Gibbs, E.P.J. Transmission and epidemiology of bluetongue and epizootic hemorrhagic disease in North America: Current perspectives, research gaps, and future directions. Vector-Borne Zoonotic Dis. 2015, 15, 348–363. [Google Scholar] [CrossRef] [PubMed]
- Ruder, M.G.; Johnson, D.; Ostlund, E.; Allison, A.B.; Kienzle, C.; Phillips, J.E.; Poulson, R.L.; Stallknecht, D.E. The first 10 years (2006–15) of epizootic hemorrhagic disease virus serotype 6 in the USA. J. Wildl. Dis. 2017, 53, 901–905. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Beringer, J.; Hansen, L.P.; Stallknecht, D.E. An epizootic of hemorrhagic disease in white-tailed deer in Missouri. J. Wildl. Dis. 2000, 36, 588–591. [Google Scholar] [CrossRef] [PubMed]
- Stevens, G.; McCluskey, B.; King, A.; O’Hearn, E.; Mayr, G. Review of the 2012 epizootic hemorrhagic disease outbreak in domestic ruminants in the United States. PLoS ONE 2015, 10, e0133359. [Google Scholar] [CrossRef] [PubMed]
- Gaydos, J.K.; Crum, J.M.; Davidson, W.R.; Cross, S.S.; Owen, S.F.; Stallknecht, D.E. Epizootiology of an epizootic hemorrhagic disease outbreak in West Virginia. J. Wildl. Dis. 2004, 40, 383–393. [Google Scholar] [CrossRef] [PubMed][Green Version]
- McGregor, B.L.; Erram, D.; Acevedo, C.; Alto, B.W.; Burkett-Cadena, N.D. Vector competence of Culicoides sonorensis (Diptera: Ceratopogonidae) for epizootic hemorrhagic disease virus serotype 2 strains from Canada and Florida. Viruses 2019, 11, 367. [Google Scholar] [CrossRef]
- Mendiola, S.Y.; Mills, M.K.; Maki, E.; Drolet, B.S.; Wilson, W.C.; Berghaus, R.; Stallknecht, D.E.; Breitenbach, J.; McVey, D.S.; Ruder, M.G. EHDV-2 infection prevalence varies in Culicoides sonorensis after feeding on infected white-tailed deer over the course of viremia. Viruses 2019, 11, 371. [Google Scholar] [CrossRef] [PubMed]
- Ruder, M.G.; Howerth, E.W.; Stallknecht, D.E.; Allison, A.B.; Carter, D.L.; Drolet, B.S.; Klement, E.; Mead, D.G. Vector competence of Culicoides sonorensis (Diptera: Ceratopogonidae) to epizootic hemorrhagic disease virus serotype 7. Parasit. Vectors 2012, 5, 236. [Google Scholar] [CrossRef]
- Mills, M.K.; Ruder, M.G.; Nayduch, D.; Michel, K.; Drolet, B.S. Dynamics of epizootic hemorrhagic disease virus infection within the vector, Culicoides sonorensis (Diptera: Ceratopogonidae). PLoS ONE 2017, 12, e0188865. [Google Scholar] [CrossRef] [PubMed]
- Mills, M.K.; Michel, K.; Pfannenstiel, R.S.; Ruder, M.G.; Veronesi, E.; Nayduch, D. Culicoides–virus interactions: Infection barriers and possible factors underlying vector competence. Curr. Opin. Insect Sci. 2017, 22, 7–15. [Google Scholar] [CrossRef]
- Nayduch, D.; Lee, M.B.; Saski, C.A. The reference transcriptome of the adult female biting midge (Culicoides sonorensis) and differential gene expression profiling during teneral, blood, and sucrose feeding conditions. PLoS ONE 2014, 9, e98123. [Google Scholar] [CrossRef]
- Nayduch, D.; Lee, M.B.; Saski, C.A. Gene discovery and differential expression analysis of humoral immune response elements in female Culicoides sonorensis (Diptera: Ceratopogonidae). Parasit Vectors 2014, 7, 388. [Google Scholar] [CrossRef]
- Cheng, G.; Liu, Y.; Wang, P.; Xiao, X. Mosquito defense strategies against viral infection. Trends Parasitol. 2016, 32, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Lu, Z.; Strand, M.R.; Jiang, H. Antiviral, anti-parasitic, and cytotoxic effects of 5, 6-dihydroxyindole (DHI), a reactive compound generated by phenoloxidase during insect immune response. Insect Biochm. Mol. Biol. 2011, 41, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Andres, J.; Rani, S.; Varjak, M.; Chase-Topping, M.E.; Beck, M.H.; Ferguson, M.C.; Schnettler, E.; Fragkoudis, R.; Barry, G.; Merits, A. Phenoloxidase activity acts as a mosquito innate immune response against infection with Semliki Forest virus. Plos Pathog. 2012, 8, e1002977. [Google Scholar] [CrossRef] [PubMed]
- Kingsolver, M.B.; Huang, Z.; Hardy, R.W. Insect antiviral innate immunity: Pathways, effectors, and connections. J. Mol. Biol. 2013, 425, 4921–4936. [Google Scholar] [CrossRef] [PubMed]
- Nunamaker, R.A.; De León, A.A.P.; Campbell, C.L.; Lonning, S.M. Oral infection of Culicoides sonorensis (Diptera: Ceratopogonidae) by vesicular stomatitis virus. J. Med. Entomol. 2000, 37, 784–786. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A. The Pfam protein families database in 2019. Nucleic Acids Res. 2018, 47, D427–D432. [Google Scholar] [CrossRef]
- Consortium, T.U. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2018, 47, D506–D515. [Google Scholar] [CrossRef]
- Wu, T.D.; Watanabe, C.K. GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 2005, 21, 1859–1875. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Statist. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef] [PubMed]
- Volders, K.; Scholz, S.; Slabbaert, J.R.; Nagel, A.C.; Verstreken, P.; Creemers, J.W.; Callaerts, P.; Schwärzel, M. Drosophila rugose is a functional homolog of mammalian Neurobeachin and affects synaptic architecture, brain morphology, and associative learning. J. Neurosci. 2012, 32, 15193–15204. [Google Scholar] [CrossRef] [PubMed]
- Giniger, E.; Tietje, K.; Jan, L.Y.; Jan, Y.N. lola encodes a putative transcription factor required for axon growth and guidance in Drosophila. Development 1994, 120, 1385–1398. [Google Scholar]
- Prokop, A.; Beaven, R.; Qu, Y.; Sánchez-Soriano, N. Using fly genetics to dissect the cytoskeletal machinery of neurons during axonal growth and maintenance. J. Cell Sci. 2013, 126, 2331–2341. [Google Scholar] [CrossRef]
- Fu, H.; Leake, C.J.; Mertens, P.P.; Mellor, P.S. The barriers to bluetongue virus infection, dissemination and transmission in the vector, Culicoides variipennis (Diptera: Ceratopogonidae). Arch. Virol. 1999, 144, 747–761. [Google Scholar] [CrossRef]
- Drolet, B.S.; Campbell, C.L.; Stuart, M.A.; Wilson, W.C. Vector competence of Culicoides sonorensis (Diptera: Ceratopogonidae) for vesicular stomatitis virus. J. Med. Entomol. 2005, 42, 409–418. [Google Scholar] [CrossRef]
- Ammar, E.-D.; Hogenhout, S.A. A neurotropic route for Maize mosaic virus (Rhabdoviridae) in its planthopper vector Peregrinus maidis. Virus Res. 2008, 131, 77–85. [Google Scholar] [CrossRef]
- McDermott, E.G.; Mayo, C.E.; Gerry, A.C.; Laudier, D.; MacLachlan, N.J.; Mullens, B.A. Bluetongue virus infection creates light averse Culicoides vectors and serious errors in transmission risk estimates. Parasit. Vectors 2015, 8, 460. [Google Scholar] [CrossRef]
- Grimstad, P.R.; Ross, Q.E.; Craig, G.B., Jr. Aedes triseriatus (Diptera: Culicidae) and La Crosse Virus: II. Modification of mosquito feeding behavior by virus infection. J. Med. Entomol. 1980, 17, 1–7. [Google Scholar] [CrossRef]
- Platt, K.B.; Linthicum, K.J.; Myint, K.S.; Innis, B.L.; Lerdthusnee, K.; Vaughn, D.W. Impact of dengue virus infection on feeding behavior of Aedes aegypti. Am. J. Trop. Med. Hyg. 1997, 57, 119–125. [Google Scholar] [CrossRef]
- Lefcort, H.; Durden, L.A. The effect of infection with Lyme disease spirochetes (Borrelia burgdorferi) on the phototaxis, activity, and questing height of the tick vector Ixodes scapularis. Parasitology 1996, 113, 97–103. [Google Scholar] [CrossRef]
- Beach, R.; Kiilu, G.; Leeuwenburg, J. Modification of sand fly biting behavior by Leishmania leads to increased parasite transmission. Am. J. Trop. Med. Hyg. 1985, 34, 278–282. [Google Scholar] [CrossRef]
- Jackson, B.T.; Brewster, C.C.; Paulson, S.L. La Crosse virus infection alters blood feeding behavior in Aedes triseriatus and Aedes albopictus (Diptera: Culicidae). J. Med. Entomol. 2014, 49, 1424–1429. [Google Scholar] [CrossRef] [PubMed]
- Campbell, C.L.; Wilson, W.C. Differentially expressed midgut transcripts in Culicoides sonorensis (Diptera: Ceratopogonidae) following Orbivirus (Reoviridae) oral feeding. Insect Mol. Biol. 2002, 11, 595–604. [Google Scholar] [CrossRef] [PubMed]
| Time (h) | % CPE Positive * | TCID50 per midge (n) ** |
|---|---|---|
| 0 | 100 (10/10) | 2.97 (4), 3.1 (3), 3.3 (3) |
| 36 | 100 (10/10) | <2.3 (6), 2.3 (2), 2.5, 2.8 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nayduch, D.; Shankar, V.; Mills, M.K.; Robl, T.; Drolet, B.S.; Ruder, M.G.; Scully, E.D.; Saski, C.A. Transcriptome Response of Female Culicoides sonorensis Biting Midges (Diptera: Ceratopogonidae) to Early Infection with Epizootic Hemorrhagic Disease Virus (EHDV-2). Viruses 2019, 11, 473. https://doi.org/10.3390/v11050473
Nayduch D, Shankar V, Mills MK, Robl T, Drolet BS, Ruder MG, Scully ED, Saski CA. Transcriptome Response of Female Culicoides sonorensis Biting Midges (Diptera: Ceratopogonidae) to Early Infection with Epizootic Hemorrhagic Disease Virus (EHDV-2). Viruses. 2019; 11(5):473. https://doi.org/10.3390/v11050473
Chicago/Turabian StyleNayduch, Dana, Vijay Shankar, Mary K. Mills, Tanner Robl, Barbara S. Drolet, Mark G. Ruder, Erin D. Scully, and Christopher A. Saski. 2019. "Transcriptome Response of Female Culicoides sonorensis Biting Midges (Diptera: Ceratopogonidae) to Early Infection with Epizootic Hemorrhagic Disease Virus (EHDV-2)" Viruses 11, no. 5: 473. https://doi.org/10.3390/v11050473
APA StyleNayduch, D., Shankar, V., Mills, M. K., Robl, T., Drolet, B. S., Ruder, M. G., Scully, E. D., & Saski, C. A. (2019). Transcriptome Response of Female Culicoides sonorensis Biting Midges (Diptera: Ceratopogonidae) to Early Infection with Epizootic Hemorrhagic Disease Virus (EHDV-2). Viruses, 11(5), 473. https://doi.org/10.3390/v11050473





