Isolation and Characterization of Lactobacillus brevis Phages
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Cultivation Media
2.2. Phage Isolation and Enrichment
2.3. Phage Detection, Purification and Host Range Analysis
2.4. Phage Concentration and Purification
2.5. Transmission Electron Microscopy
2.6. Phage DNA Extraction and Sequencing
2.7. Phage Structural Proteome and Mass-Spectrometry
2.8. Proteomic Tree
2.9. Phage Activity against Lb. brevis Beer-Spoiling Strains
2.10. Nucleotide Sequence Accession Numbers
3. Results
3.1. Phage Isolation and Host Range Profile
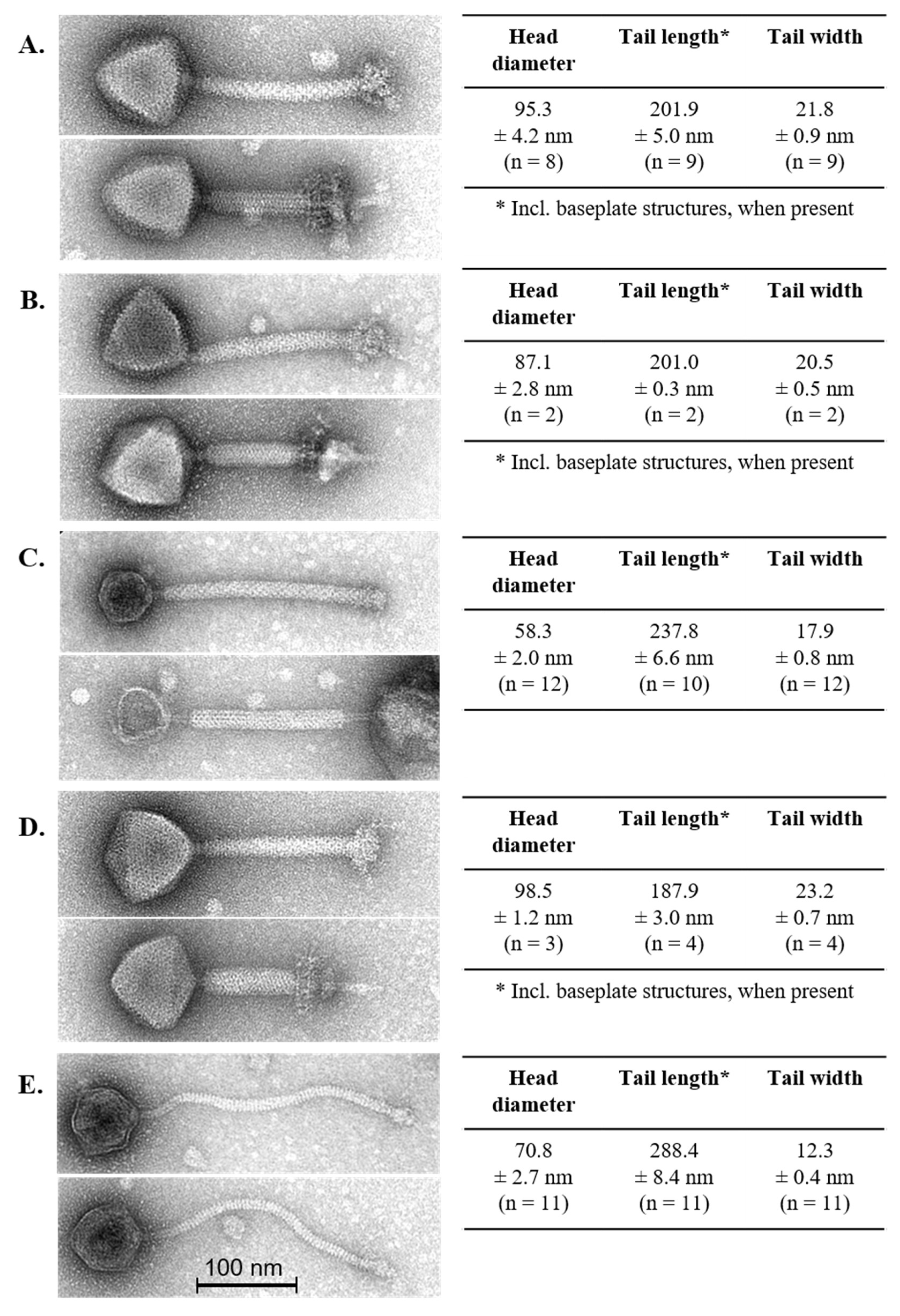
3.2. Phage Morphology
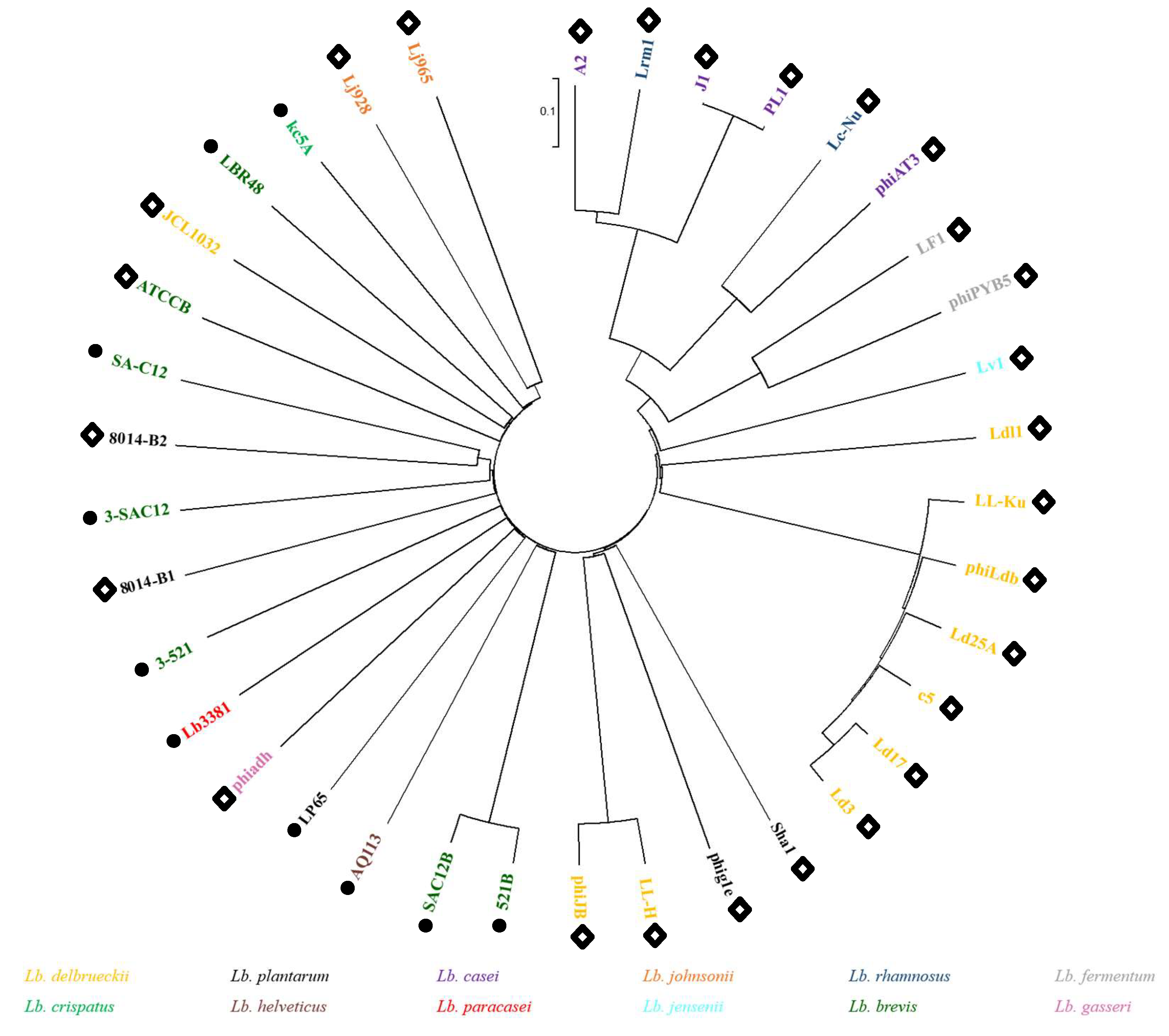
3.3. Lb. brevis Phages Comparative Analysis and Grouping
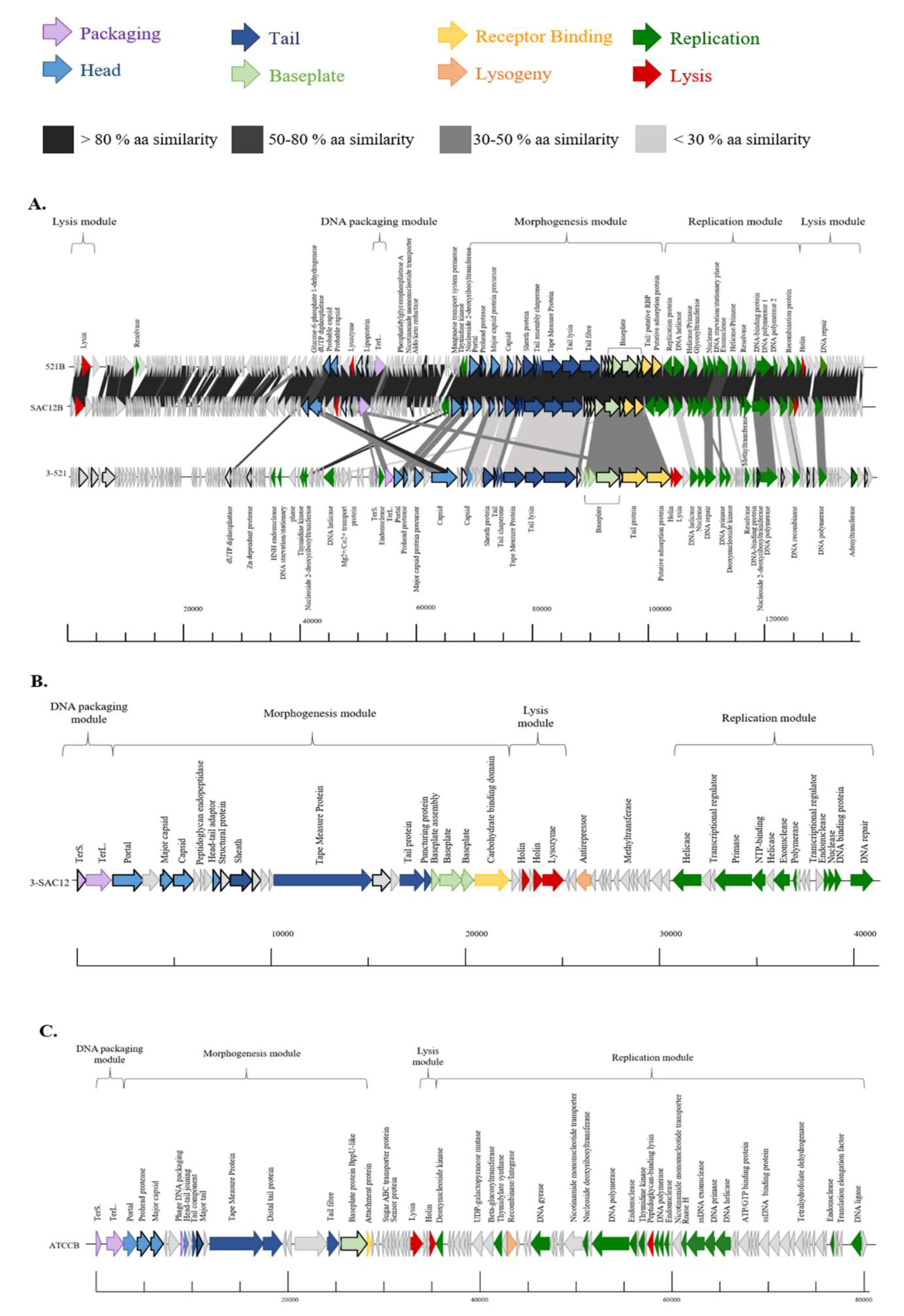
3.4. Genome Analysis
3.5. Morphogenesis Module
3.6. Structural Proteome
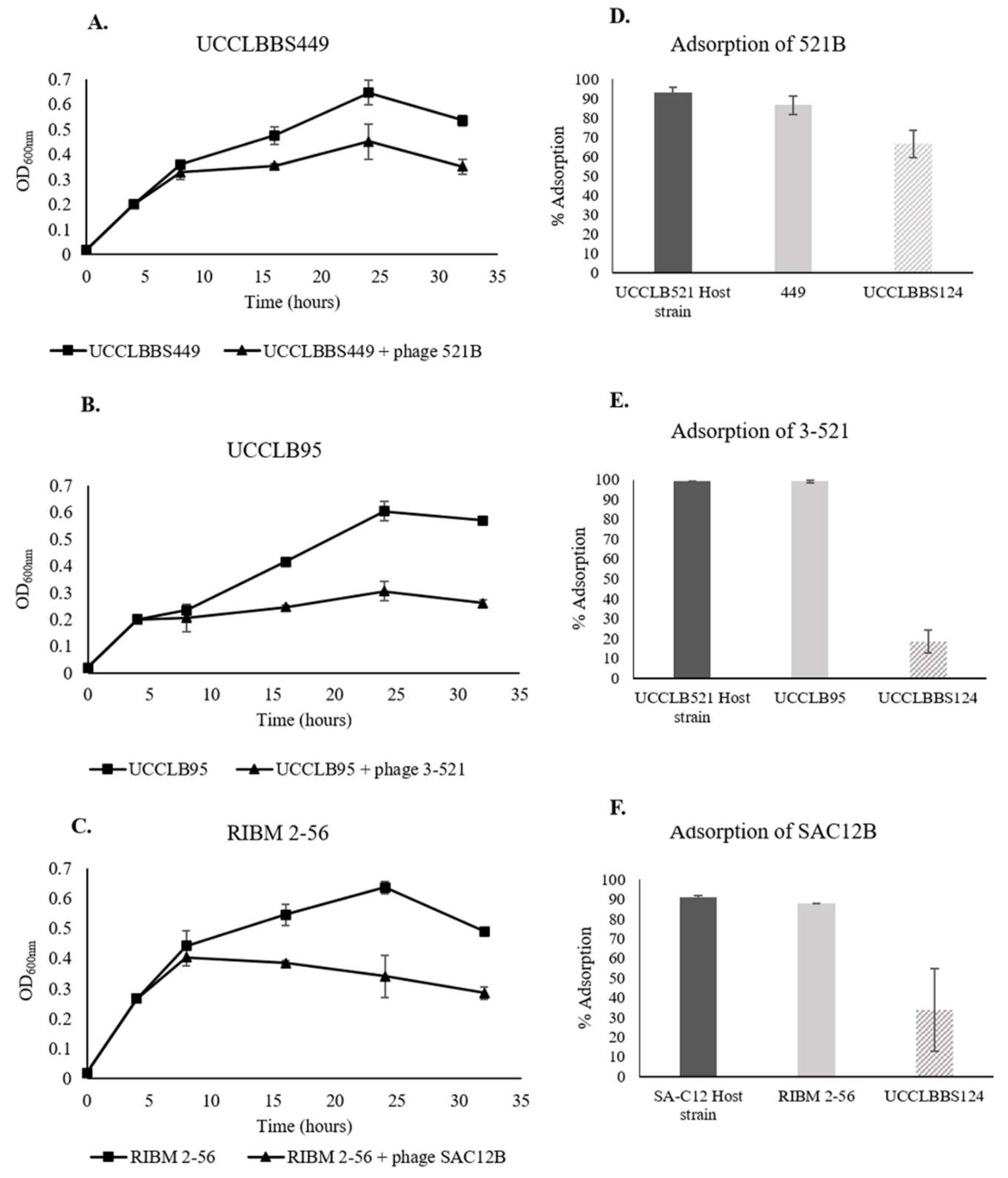
3.7. Phage Activity against Lb. brevis Beer-Spoiling Strains
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vos, P.; Garrity, G.; Jones, D.; Krieg, N.R.; Ludwig, W.; Rainey, F.A.; Schleifer, K.-H.; Whitman, W. Bergey’s Manual of Systematic Bacteriology Volume 3: The Firmicutes; Springer Science and Business Media: Athens, GA, USA, 2011; pp. 464–511. [Google Scholar]
- Salvetti, E.; Torriani, S.; Felis, G.E. The genus Lactobacillus: A taxonomic update. Probiotics Antimicrob. Proteins 2012, 4, 217–226. [Google Scholar] [CrossRef]
- Felis, G.E.; Dellaglio, F. Taxonomy of Lactobacilli and Bifidobacteria. Curr. Issues Intest. Microbiol. 2007, 8, 44–61. [Google Scholar]
- Bergsveinson, J.; Pittet, V.; Ewen, E.; Baecker, N.; Ziola, B. Genome sequence of rapid beer-spoiling isolate Lactobacillus brevis BSO 464. Genome Announce. 2015, 3, e01411–e01415. [Google Scholar] [CrossRef]
- Sakamoto, K.; Konings, W.N. Beer spoilage bacteria and hop resistance. Int. J. Food Microbiol. 2003, 89, 105–124. [Google Scholar] [CrossRef]
- Suzuki, K.; Iijima, K.; Sakamoto, K.; Sami, M.; Yamashita, H. A review of hop resistance in beer spoilage lactic acid bacteria. J. Institute Brewing 2006, 112, 173–191. [Google Scholar] [CrossRef]
- Vaughan, A.; O’Sullivan, T.; van Sinderen, D. Enhancing the microbiological stability of malt and beer—A review. J. Institute Brewing 2005, 111, 355–371. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Bamforth, C.W. The Microbiology of malting and brewing. Microbiol. Mol. Biol. Rev. 2013, 77, 157–172. [Google Scholar] [CrossRef]
- Back, W.; Leibhard, M.; Bohak, I. Flash pasteurisation-membrane filtration. Biological security in comparison. Brauwelt Int. 1990, 12, 42–49. [Google Scholar]
- Davidson, P.M.; Harrison, M.A. Resistance and adaptation to food antimicrobials, sanitizers, and other process controls. Food Technol. 2002, 56, 69–78. [Google Scholar]
- Basanta, A.; Sánchez, J.; Gómez-Sala, B.; Herranz, C.; Hernández, P.E.; Cintas, L.M. Antimicrobial activity of Enterococcus faecium L50, a strain producing enterocins L50 (L50A and L50B), P and Q, against beer-spoilage lactic acid bacteria in broth, wort (hopped and unhopped), and alcoholic and non-alcoholic lager beers. Int. J. Food Microbiol. 2008, 125, 293–307. [Google Scholar] [CrossRef]
- Gil, G.; del Monaco, S.; Cerrutti, P.; Galvagno, M. Selective antimicrobial activity of chitosan on beer spoilage bacteria and brewing yeasts. Biotechnol. Lett. 2004, 26, 569–574. [Google Scholar] [CrossRef]
- Vaughan, A.; Rouse, S.; van Sinderen, D. Investigating the antimicrobial efficacy of a Lactococcal bacteriocin for the development of microbiologically stable beer. J. Institute Brewing 2004, 110, 181–188. [Google Scholar] [CrossRef]
- Mahony, J.; McAuliffe, O.; Ross, R.P.; van Sinderen, D. Bacteriophages as biocontrol agents of food pathogens. Curr. Opin. Biotechnol. 2011, 22, 157–163. [Google Scholar] [CrossRef]
- Coffey, B.; Mills, S.; Coffey, A.; McAuliffe, O.; Ross, R.P. Phage and their lysins as biocontrol agents for food safety applications. Annual Rev. Food Sci. Technol. 2010, 1, 449–468. [Google Scholar] [CrossRef]
- Endersen, L.; O’Mahony, J.; Hill, C.; Ross, R.P.; McAuliffe, O.; Coffey, A. phage therapy in the food industry. Annual Rev. Food Sci. Technol. 2014, 5, 327–349. [Google Scholar] [CrossRef]
- Deasy, T.; Mahony, J.; Neve, H.; Heller, K.J.; van Sinderen, D. Isolation of a virulent Lactobacillus brevis phage and its application in the control of beer spoilage. J. Food Protect. 2011, 74, 2157–2161. [Google Scholar] [CrossRef]
- Brüssow, H.; Desiere, F. Comparative phage genomics and the evolution of Siphoviridae: Insights from dairy phages. Mol. Microbiol. 2001, 39, 213–222. [Google Scholar] [CrossRef]
- Jang, S.H.; Yoon, B.H.; Chang, H.I. Complete nucleotide sequence of the temperate bacteriophage LBR48, a new member of the family Myoviridae. Archi. Virol. 2011, 156, 319–322. [Google Scholar] [CrossRef]
- Svensson, U.; Christiansson, A. Methods for phage monitoring. Bulletin Inter. Dairy Federation 1991, 263, 29–39. [Google Scholar]
- Makarova, K.; Slesarev, A.; Wolf, Y.; Sorokin, A.; Mirkin, B.; Koonin, E.; Pavlov, A.; Pavlova, N.; Karamychev, V.; Polouchine, N.; et al. Comparative genomics of the lactic acid bacteria. Proc. Nat. Acad. Sci. USA 2006, 103, 15611–15616. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Purification of bacteriophage λ particles by isopycnic centrifugation through CsCl gradients. Cold Spring Harbor Protocols 2006, 1, pdb-prot3968. [Google Scholar] [CrossRef]
- Casey, E.; Mahony, J.; Neve, H.; Noben, J.-P.; Dal Bello, F.; van Sinderen, D. Novel phage group infecting Lactobacillus delbrueckii subsp. lactis, as revealed by genomic and proteomic analysis of bacteriophage Ldl1. Appl. Environ. Microbiol. 2015, 81, 1319–1326. [Google Scholar] [CrossRef]
- Lugli, G.A.; Milani, C.; Mancabelli, L.; van Sinderen, D.; Ventura, M. MEGAnnotator: A user-friendly pipeline for microbial genomes assembly and annotation. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformat. 2010, 11, 119. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Soding, J.; Biegert, A.; Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005, 33, W244–W248. [Google Scholar] [CrossRef]
- Rutherford, K.; Parkhill, J.; Crook, J.; Horsnell, T.; Rice, P.; Rajandream, M.A.; Barrell, B. Artemis: sequence visualization and annotation. Bioinformatics 2000, 16, 944–945. [Google Scholar] [CrossRef]
- Holtappels, M.; Vrancken, K.; Schoofs, H.; Deckers, T.; Remans, T.; Noben, J.-P.; Valcke, R. A comparative proteome analysis reveals flagellin, chemotaxis regulated proteins and amylovoran to be involved in virulence differences between Erwinia amylovora strains. J. Proteomics 2015, 123, 54–69. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protocols Bioinformat. 2003, 1, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings Bioinformat. 2008, 9, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Collins, B.; Bebeacua, C.; Mahony, J.; Blangy, S.; Douillard, F.P.; Veesler, D.; Cambillau, C.; van Sinderen, D. Structure and functional analysis of the host recognition device of lactococcal phage Tuc2009. J. Virol. 2013, 87, 8429–8440. [Google Scholar] [CrossRef] [PubMed]
- Matthews, R.E.F.; Maurin, J. Classification and nomenclature of viruses. Ann. Microbiol. 1979, 1, 133–136. [Google Scholar] [CrossRef]
- Zago, M.; Scaltriti, E.; Rossetti, L.; Guffanti, A.; Armiento, A.; Fornasari, M.E.; Grolli, S.; Carminati, D.; Brini, E.; Pavan, P.; et al. Characterization of the genome of the dairy Lactobacillus helveticus bacteriophage ΦAQ113. Appl. Environment. Microbiol. 2013, 79, 4712–4718. [Google Scholar] [CrossRef]
- Tuohimaa, A.; Riipinen, K.-A.; Brandt, K.; Alatossava, T. The genome of the virulent phage Lc-Nu of probiotic Lactobacillus rhamnosus, and comparative genomics with Lactobacillus casei phages. Archi. Virol. 2006, 151, 947. [Google Scholar] [CrossRef] [PubMed]
- Alemayehu, D.; Ross, R.P.; O’Sullivan, O.; Coffey, A.; Stanton, C.; Fitzgerald, G.F.; McAuliffe, O. Genome of a virulent bacteriophage Lb338–1 that lyses the probiotic Lactobacillus paracasei cheese strain. Gene 2009, 448, 29–39. [Google Scholar] [CrossRef]
- Chibani-Chennoufi, S.; Dillmann, M.-L.; Marvin-Guy, L.; Rami-Shojaei, S.; Brüssow, H. Lactobacillus plantarum bacteriophage LP65: A new member of the SPO1-like genus of the family Myoviridae. J. Bacteriol. 2004, 186, 7069–7083. [Google Scholar] [CrossRef]
- Rossmann, M.G.; Mesyanzhinov, V.V.; Arisaka, F.; Leiman, P.G. The bacteriophage T4 DNA injection machine. Curr. Opin. Struct. Biol. 2004, 14, 171–180. [Google Scholar] [CrossRef]
- Abedon, S.T. Lysis from without. Bacteriophage 2011, 1, 46–49. [Google Scholar] [CrossRef]
| Lactobacillus brevis Strains | Isolation Source |
|---|---|
| ATCC367 [21] | Silage |
| UCCLBBS124 | Beer |
| UCCLB521 | Brewery |
| UCCLB556 | Brewery |
| SA-C12 | Silage |
| UCCLBBS449 | Beer |
| UCCLB94 | Beer |
| UCCLB95 | Beer |
| RIBM 2-56 | Beer |
| Lb. brevis Non-Beer Spoiling Strains | Lb. brevis Beer-Spoiling Strains | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ATCC367 | UCCLB521 | UCCLB556 | SA-C12 | UCCLBBS124 | UCCLBBS449 | UCCLB94 | UCCLB95 | RIBM 2-56 | ||
| Phage | 3-521 | - | + * | - | - | - | - | - | ~ | - |
| 521B | - | + * | - | - | - | ~ | - | - | - | |
| 3-SAC12 | - | - | - | + * | - | - | - | - | - | |
| SAC12B | - | + | - | + * | - | - | - | - | ~ | |
| ATCCB | + * | - | - | - | - | - | - | - | - | |
| Phage | Sample (Date) | Isolation Source | Genome Size (bp) | ORFs | GC Content (%) | % nt Identity (% coverage) | |
|---|---|---|---|---|---|---|---|
| Myoviridae | 3-521 | S1 (2017) | Wastewater (Ireland) | 140,816 | 155 | 36.93 | |
| 521B | S2 (2018) | Wastewater (Ireland) | 136,442 | 188 | 32.27 | 97 (88) with SAC12B | |
| SAC12B | S2 (2018) | Wastewater (Ireland) | 136,608 | 191 | 32.41 | 97 (88) with 521B | |
| 3-SAC12 | S1 (2017) | Wastewater (Ireland) | 41,292 | 61 | 40.01 | ||
| Siphoviridae | ATCCB | S2 (2018) | Wastewater (Ireland) | 80,538 | 96 | 30.80 |
| Phage | ORF | Putative Function | No. of Peptides | Sequence Coverage (%) |
|---|---|---|---|---|
| 521B | 80 | Probable capsid protein | 8 | 29.4 |
| 81 | Probable capsid protein | 9 | 28.4 | |
| 86 | Structural protein | 3 | 16.1 | |
| 88 | Lipoprotein | 5 | 50.8 | |
| 106 | Structural protein | 4 | 37.9 | |
| 121 | Portal protein | 12 | 28.3 | |
| 122 | Structural protein | 2 | 17.4 | |
| 123 | Caudovirus prohead protease | 4 | 20.8 | |
| 125 | Major capsid protein precursor | 19 | 59.7 | |
| 128 | Capsid protein | 3 | 16.8 | |
| 130 | Gp91 | 8 | 35 | |
| 132 | Major tail sheath protein | 16 | 40.4 | |
| 133 | Tail protein | 5 | 59.9 | |
| 136 | Tape measure protein | 28 | 31.2 | |
| 137 | Tail lysin | 12 | 15.1 | |
| 138 | Structural component of the tail fibre | 8 | 10 | |
| 140 | Structural protein | 2 | 15.1 | |
| 141 | Structural protein | 5 | 42.2 | |
| 142 | Baseplate protein | 3 | 23.1 | |
| 143 | Baseplate J-like protein | 6 | 15.1 | |
| 144 | Baseplate protein | 7 | 9.4 | |
| 146 | Tail protein | 15 | 34.1 | |
| 147 | Putative adsorption protein | 9 | 22.2 | |
| 156 | DNA starvation/stationary phase protein | 6 | 48 | |
| 185 | Structural protein | 3 | 38.5 | |
| 3-521 | 10 | dUTP diphosphatase | 2 | 9.9 |
| 19 | Zn-dependent protease | 5 | 23.7 | |
| 52 | Portal protein | 3 | 7.9 | |
| 53 | Prohead protease | 1 | 8.3 | |
| 55 | Major capsid protein | 19 | 51.1 | |
| 57 | Phage capsid and scaffold | 19 | 18.6 | |
| 60 | Structural protein | 4 | 20.1 | |
| 65 | Tail sheath protein | 14 | 25 | |
| 66 | Putative tail protein | 6 | 53.3 | |
| 69 | Tape measure protein | 14 | 17.4 | |
| 70 | Tail lysin protein | 16 | 21.1 | |
| 71 | gp673 | 2 | 1.7 | |
| 72 | Structural protein | 3 | 15.3 | |
| 76 | Baseplate protein | 4 | 3 | |
| 77 | Structural protein | 2 | 18 | |
| 78 | Tail protein | 22 | 24.5 | |
| 79 | Tail associated protein | 23 | 20.1 | |
| 98 | Nucleoside 2-deoxyribosyltransferase | 2 | 12.4 | |
| 101 | Structural protein | 1 | 6.1 | |
| 106 | Structural protein | 7 | 62 | |
| 108 | Tail protein | 1 | 18 | |
| 117 | Adenyltransferase | 6 | 16.9 | |
| 119 | ADP-ribose pyrophosphatase | 1 | 5.3 | |
| 124 | Structural protein | 6 | 15.7 | |
| 126 | AAA superfamily ATPase | 9 | 26.2 | |
| 128 | Phosphatase | 4 | 5.4 | |
| 3-SAC12 | 1 | Terminase small subunit | 1 | 6.6 |
| 3 | Portal protein | 13 | 31.4 | |
| 5 | Major capsid protein | 4 | 26.3 | |
| 6 | Capsid protein | 9 | 26.7 | |
| 10 | Putative head-tail adaptor | 1 | 9.8 | |
| 11 | Structural protein | 1 | 9 | |
| 12 | Sheath protein | 3 | 11.8 | |
| 13 | Structural protein | 2 | 22.4 | |
| 17 | Structural protein | 4 | 13.6 | |
| ATCCB | 70 | Baseplate protein | 6 | 9.2 |
| 79 | Major tail protein | 6 | 50.2 | |
| 86 | Major capsid protein | 8 | 25.9 | |
| 87 | Prohead protease | 2 | 7.5 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feyereisen, M.; Mahony, J.; Lugli, G.A.; Ventura, M.; Neve, H.; Franz, C.M.A.P.; Noben, J.-P.; O’Sullivan, T.; van Sinderen, D. Isolation and Characterization of Lactobacillus brevis Phages. Viruses 2019, 11, 393. https://doi.org/10.3390/v11050393
Feyereisen M, Mahony J, Lugli GA, Ventura M, Neve H, Franz CMAP, Noben J-P, O’Sullivan T, van Sinderen D. Isolation and Characterization of Lactobacillus brevis Phages. Viruses. 2019; 11(5):393. https://doi.org/10.3390/v11050393
Chicago/Turabian StyleFeyereisen, Marine, Jennifer Mahony, Gabriele A. Lugli, Marco Ventura, Horst Neve, Charles M. A. P. Franz, Jean-Paul Noben, Tadhg O’Sullivan, and Douwe van Sinderen. 2019. "Isolation and Characterization of Lactobacillus brevis Phages" Viruses 11, no. 5: 393. https://doi.org/10.3390/v11050393
APA StyleFeyereisen, M., Mahony, J., Lugli, G. A., Ventura, M., Neve, H., Franz, C. M. A. P., Noben, J.-P., O’Sullivan, T., & van Sinderen, D. (2019). Isolation and Characterization of Lactobacillus brevis Phages. Viruses, 11(5), 393. https://doi.org/10.3390/v11050393







