Mosquito Small RNA Responses to West Nile and Insect-Specific Virus Infections in Aedes and Culex Mosquito Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Virus Infections
2.2. Eradication of Cell Fusing agent Virus from Aag2 Cells
2.3. RNA Isolation and Deep Sequencing of Small RNAs
2.4. Small RNA Analysis
2.5. De Novo Contig Assembly from Small RNA Reads
3. Results
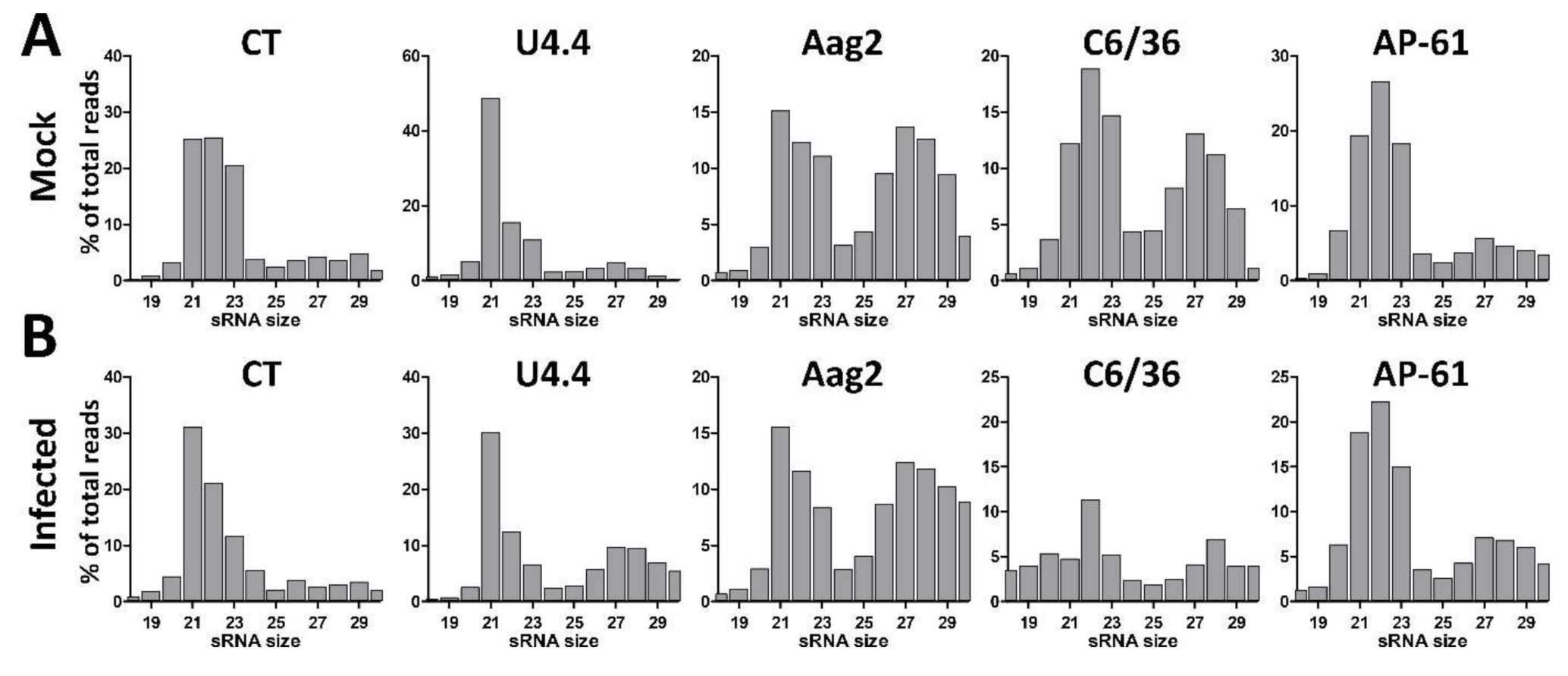
3.1. Small RNA Profiles in Uninfected and WNV-Infected Aedes and Culex spp. Mosquito Cells
3.2. Discovery of Insect-Specific Viruses by De Novo Assembly of Small RNA Reads
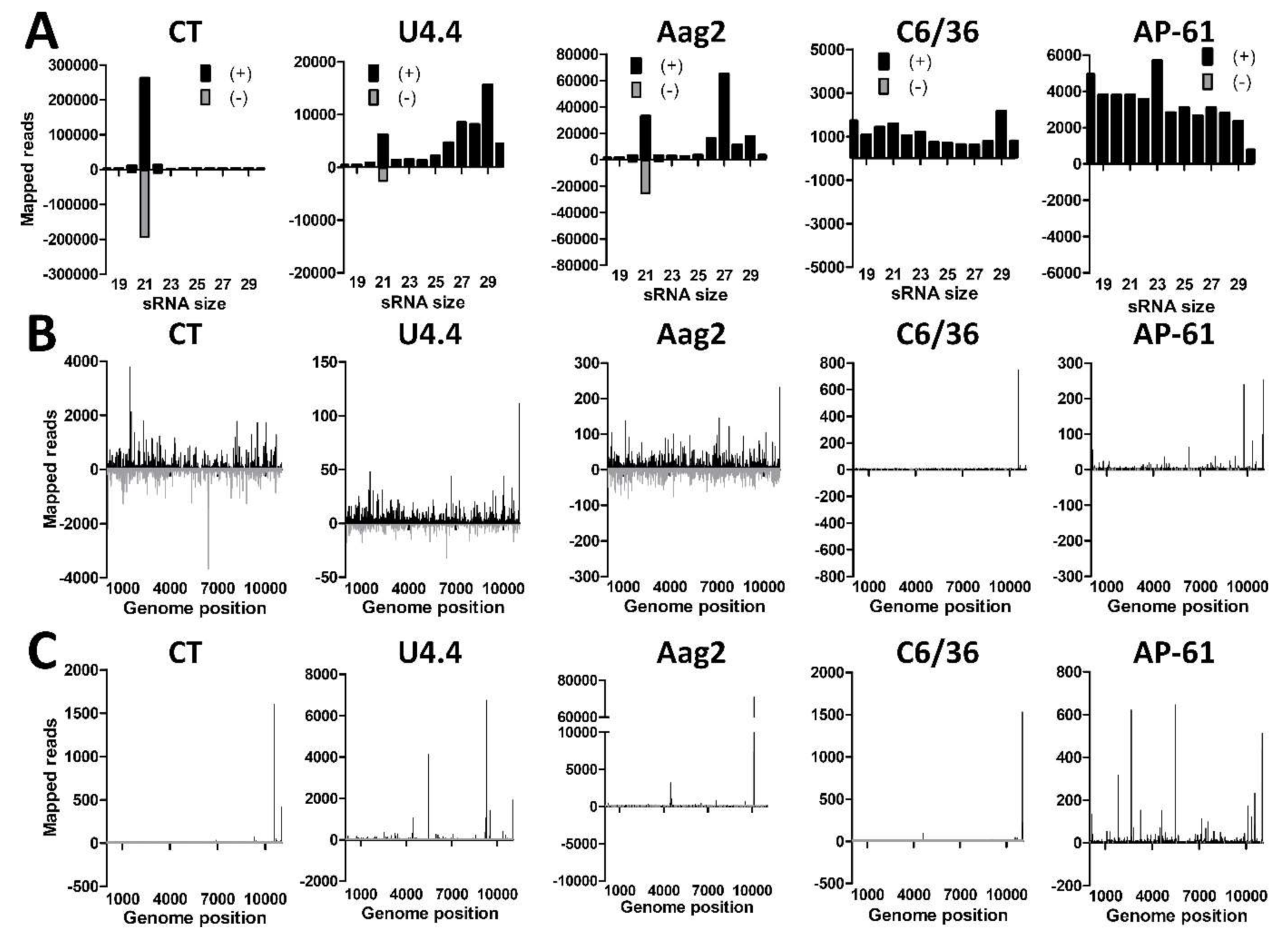
3.3. Small RNA Profiles of Insect-Specific Viruses in Culex Tarsalis cells Reveal the Production of Viral siRNAs and piRNAs
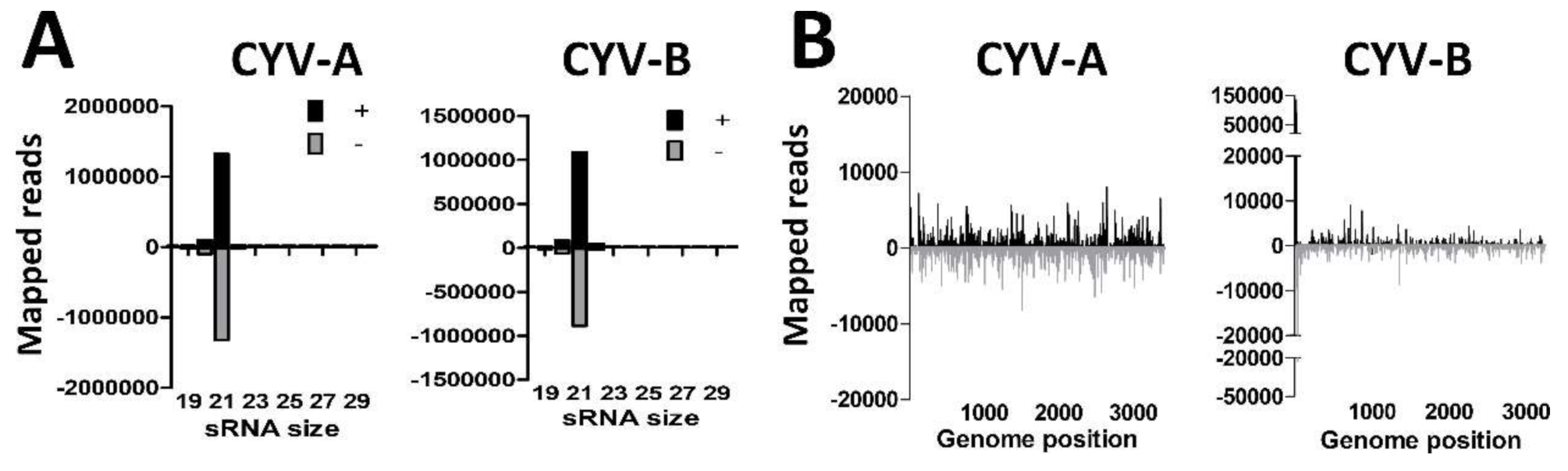
3.4. U4.4 Cells are Persistently Infected with Culex Y Virus and Demonstrate a Potent siRNA Response
3.5. Aag2 Cells Produce siRNAs and piRNAs to Insect-Specific Virus Infection
3.6. Differential Small RNA Responses to WNV-Infection in Culex and Aedes Mosquito Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Greninger, A.L. A decade of RNA virus metagenomics is (not) enough. Virus Res. 2018, 244, 218–229. [Google Scholar] [CrossRef] [PubMed]
- Weger-Lucarelli, J.; Rückert, C.; Grubaugh, N.D.; Misencik, M.J.; Armstrong, P.M.; Stenglein, M.D.; Ebel, G.D.; Brackney, D.E. Adventitious viruses persistently infect three commonly used mosquito cell lines. Virology 2018, 521, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Webster, C.L.; Longdon, B.; Lewis, S.H.; Obbard, D. Twenty five new viruses associated with the Drosophilidae (Diptera). bioRxiv 2016, 12, 041665. [Google Scholar] [CrossRef] [PubMed]
- Webster, C.L.; Waldron, F.M.; Robertson, S.; Crowson, D.; Ferrari, G.; Quintana, J.F.; Brouqui, J.M.; Bayne, E.H.; Longdon, B.; Buck, A.H.; et al. The discovery, distribution, and evolution of viruses associated with Drosophila melanogaster. PLOS Biol. 2015, 13, e1002210. [Google Scholar] [CrossRef] [PubMed]
- Goenaga, S.; Kenney, J.L.; Duggal, N.K.; Delorey, M.; Ebel, G.D.; Zhang, B.; Levis, S.; Enria, D.; Brault, A. Potential for co-infection of a mosquito-specific flavivirus, nhumirim virus, to block west nile virus transmission in mosquitoes. Viruses 2015, 7, 5801–5812. [Google Scholar] [CrossRef] [PubMed]
- Hobson-Peters, J.; Yam, A.W.Y.; Lu, J.W.F.; Setoh, Y.X.; May, F.J.; Kurucz, N.; Walsh, S.; Prow, N.A.; Davis, S.S.; Weir, R.; et al. A new insect-specific Flavivirus from northern Australia suppresses replication of West Nile virus and Murray Valley encephalitis virus in co-infected mosquito cells. PLoS ONE 2013, 8, e56534. [Google Scholar] [CrossRef] [PubMed]
- Burivong, P.; Pattanakitsakul, S.N.; Thongrungkiat, S.; Malasit, P.; Flegel, T.W. Markedly reduced severity of Dengue virus infection in mosquito cell cultures persistently infected with Aedes albopictus densovirus (AalDNV). Virology 2004, 329, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Bolling, B.G.; Olea-Popelka, F.J.; Eisen, L.; Moore, C.G.; Blair, C.D. Transmission dynamics of an insect-specific flavivirus in a naturally infected Culex pipiens laboratory colony and effects of co-infection on vector competence for West Nile virus. Virology 2012, 427, 90–97. [Google Scholar] [CrossRef]
- Kenney, J.L.; Solberg, O.D.; Langevin, S.A.; Brault, A.C.; Collins, F.; Labs, N. Characterization of a novel insect-specific flavivirus from Brazil: Potential for inhibition of infection of arthropod cells with medically important flaviviruses. J. Gen. Virol. 2015, 95, 2796–2808. [Google Scholar] [CrossRef] [PubMed]
- Romo, H.; Kenney, J.L.; Blitvich, B.J.; Brault, A.C. Restriction of Zika virus infection and transmission in Aedes aegypti mediated by an insect-specific flavivirus. Emerg. Microbes Infect. 2018, 7, 181. [Google Scholar] [CrossRef]
- Hall-Mendelin, S.; McLean, B.J.; Bielefeldt-Ohmann, H.; Hobson-Peters, J.; Hall, R.A.; Van Den Hurk, A.F. The insect-specific Palm Creek virus modulates West Nile virus infection in and transmission by Australian mosquitoes. Parasites Vectors 2016, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Blair, C.D. Mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission. Future Microbiol. 2011, 6, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Blair, C.; Olson, K. The role of RNA interference (RNAi) in arbovirus-vector interactions. Viruses 2015, 7, 820–843. [Google Scholar] [CrossRef]
- Elbashir, S.M.; Martinez, J.; Patkaniowska, A.; Lendeckel, W.; Tuschl, T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001, 20, 6877–6888. [Google Scholar] [CrossRef]
- Bronkhorst, A.W.; van Rij, R.P. The long and short of antiviral defense: Small RNA-based immunity in insects. Curr Opin Virol. 2014, 7, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Kawaoka, S.; Izumi, N.; Katsuma, S.; Tomari, Y. 3′ end formation of PIWI-interacting RNAs in vitro. Mol. Cell. 2011, 43, 1015–1022. [Google Scholar] [CrossRef] [PubMed]
- Sienski, G.; Dönertas, D.; Brennecke, J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 2012, 151, 964–980. [Google Scholar] [CrossRef]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef] [PubMed]
- Gunawardane, L.S.; Saito, K.; Nishida, K.M.; Miyoshi, K.; Kawamura, Y.; Nagami, T.; Siomi, H.; Siomi, M.C. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 2007, 315, 1587–1590. [Google Scholar] [CrossRef] [PubMed]
- Lewis, S.H.; Salmela, H.; Obbard, D.J. Duplication and diversification of dipteran argonaute genes, and the evolutionary divergence of Piwi and Aubergine. Genome Biol. Evol. 2016, 8, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Campbell, C.L.; Black, W.C.; Hess, A.M.; Foy, B.D. Comparative genomics of small RNA regulatory pathway components in vector mosquitoes. BMC Genom. 2008, 9, 425. [Google Scholar] [CrossRef] [PubMed]
- Miesen, P.; Ivens, A.; Buck, A.H.; van Rij, R.P. Small RNA profiling in dengue virus 2-infected Aedes mosquito cells reveals viral piRNAs and novel host miRNAs. PLoS Negl. Trop. Dis. 2016, 10, e0004452. [Google Scholar] [CrossRef] [PubMed]
- Miesen, P.; Girardi, E.; van Rij, R.P. Distinct sets of PIWI proteins produce arbovirus and transposon-derived piRNAs in Aedes aegypti mosquito cells. Nucleic Acids Res. 2015, 43, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Vodovar, N.; Bronkhorst, A.W.; van Cleef, K.W.R.; Miesen, P.; Blanc, H.; van Rij, R.P.; Saleh, M.C. Arbovirus-derived piRNAs exhibit a ping-pong signature in mosquito cells. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Varjak, M.; Donald, C.L.; Mottram, T.J.; Sreenu, V.B.; Merits, A.; Maringer, K.; Schnettler, E.; Kohl, A. Characterization of the Zika virus induced small RNA response in Aedes aegypti cells. PLoS Negl. Trop. Dis. 2017, 11, e0006010. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, I.; Jansen, S.; Fall, G.; Lorenzen, S.; Rudolf, M.; Huber, K.; Heitmann, A.; Schicht, S.; Watson, M.; Castelli, I.; et al. RNA interference restricts Rift Valley Fever virus in multiple insect systems. mSphere 2017, 2, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, I.; Shi, X.; McFarlane, M.; Watson, M.; Blomström, A.-L.; Skelton, J.K.; Kohl, A.; Elliott, R.M.; Schnettler, E. The antiviral RNAi response in vector and non-vector cells against Orthobunyaviruses. PLoS Negl. Trop. Dis. 2017, 11, e0005272. [Google Scholar] [CrossRef] [PubMed]
- Schnettler, E.; Donald, C.L.; Human, S.; Watson, M.; Siu, R.W.C.; McFarlane, M.; Fazakerley, J.K.; Kohl, A.; Fragkoudis, R. Knockdown of piRNA pathway proteins results in enhanced Semliki Forest virus production in mosquito cells. J. Gen. Virol. 2013, 94, 1680–1689. [Google Scholar] [CrossRef] [PubMed]
- Schnettler, E.; Ratinier, M.; Watson, M.; Shaw, A.E.; McFarlane, M.; Varela, M.; Elliott, R.M.; Palmarini, M.; Kohl, A. RNA interference targets arbovirus replication in Culicoides cells. J. Virol. 2013, 87, 2441–2454. [Google Scholar] [CrossRef] [PubMed]
- Miesen, P.; Joosten, J.; van Rij, R.P. PIWIs go viral: Arbovirus-derived piRNAs in vector mosquitoes. PLoS Pathog. 2016, 12, 1006017. [Google Scholar] [CrossRef] [PubMed]
- Joosten, J.; Miesen, P.; Taşköprü, E.; Pennings, B.; Jansen, P.W.; Huynen, M.A.; Vermeulen, M.; Van Rij, R.P. The Tudor protein Veneno assembles the ping-pong amplification complex that produces viral piRNAs in Aedes mosquitoes. Nucleic Acids Res. 2018, 1, 1–14. [Google Scholar] [CrossRef]
- Fros, J.J.; Miesen, P.; Vogels, C.B.; Gaibani, P.; Sambri, V.; Martina, B.E.; Koenraadt, C.J.; van Rij, R.P.; Vlak, J.M.; Takken, W.; et al. Comparative Usutu and West Nile virus transmission potential by local Culex pipiens mosquitoes in north-western Europe. One Health 2015, 1, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, F.; Chiles, R.E.; Fang, Y.; Green, E.N.; Reisen, W.K. Effects of time after infection, mosquito genotype, and infectious viral dose on the dynamics of Culex tarsalis vector competence for western equine encephalomyelitis virus. J. Am. Mosq. Control Assoc. 2006, 22, 272–281. [Google Scholar] [CrossRef]
- Goddard, L.B.; Roth, A.E.; Reisen, W.K.; Scott, T.W. Vector competence of California mosquitoes for West Nile virus. Emerg. Infect. Dis. 2002, 8, 1385–1391. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, C.J.; Gubler, D.J. Vector competence of geographic strains of Aedes albopictus and Aedes polynesiensis and certain other Aedes (Stegomyia) mosquitoes for Ross River virus. J. Am. Mosq. Control Assoc. 1987, 3, 142–147. [Google Scholar]
- Vanlandingham, D.L.; Higgs, S.; Huang, Y.-J.S. Aedes albopictus (Diptera: Culicidae) and mosquito-borne viruses in the USA. J. Med. Entomol. 2016, 53, 1024–1028. [Google Scholar] [CrossRef]
- Souza-Neto, J.A.; Powell, J.R.; Bonizzoni, M. Aedes aegypti vector competence studies: A review. Infect. Genet. Evol. 2018, 67, 191–209. [Google Scholar] [CrossRef] [PubMed]
- Singh, K. Cell cultures derived from larvae of Aedes albopictus (Skuse) and Aedes aegypti (L.). Curr. Sci. 1967, 36, 506–508. [Google Scholar]
- Lan, Q.; Fallon, A.M. Small heat shock proteins distinguish between two mosquito species and confirm identity of their cell lines. Am. J. Trop. Med. Hyg. 1990, 43, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Varma, M.G.; Pudney, M.; Leake, C.J.; Peralta, P.H. Isolations in a mosquito (Aedes pseudoscutellaris) cell line (Mos. 61) of yellow fever virus strains from original field material. Intervirology 1975, 6, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.; Ball, G.H. Comparison of amino acid utilization by cell lines of Culex tarsalis and of Culex pipiens. In Int. Conf. Invertebrate Tissue Culture Applications in Medicine, Biology, and Agriculture; Academic Press: Waltham, MA, USA, 1976; pp. 263–266. [Google Scholar]
- Göertz, G.P.; Fros, J.J.; Miesen, P.; Vogels, C.B.F.; van der Bent, M.L.; Geertsema, C.; Koenraadt, C.J.M.; van Rij, R.P.; van Oers, M.M.; Pijlman, G.P. Noncoding subgenomic Flavivirus RNA is processed by the mosquito RNA interference machinery and determines West Nile virus transmission by Culex pipiens mosquitoes. J. Virol. 2016, 90, 10145–10159. [Google Scholar] [CrossRef] [PubMed]
- Goecks, J.; Nekrutenko, A.; Taylor, J. Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010, 11, R86. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Whitfield, Z.J.; Dolan, P.T.; Kunitomi, M.; Tassetto, M.; Seetin, M.G.; Oh, S.; Heiner, C.; Paxinos, E.; Andino, R. The diversity, structure, and function of heritable adaptive immunity sequences in the Aedes aegypti genome. Curr. Biol. 2017, 27, 3511–3519. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.R.; Koren, S.; Dilley, K.A.; Puri, V.; Brown, D.M.; Harkins, D.M.; Thibaud-Nissen, F.; Rosen, B.; Chen, X.G.; Tu, Z.; et al. Analysis of the Aedes albopictus C6/36 genome provides insight into cell line utility for viral propagation. Gigascience 2018, 7, 1–16. [Google Scholar] [CrossRef]
- Antoniewski, C. Computing siRNA and piRNA overlap signatures. Methods Mol. Biol. 2014, 135–146. [Google Scholar] [CrossRef]
- Carissimo, G.; Eiglmeier, K.; Reveillaud, J.; Holm, I.; Diallo, M.; Diallo, D.; Vantaux, A.; Kim, S.; Ménard, D.; Siv, S.; et al. Identification and characterization of two novel RNA viruses from Anopheles gambiae species complex mosquitoes. PLoS ONE 2016, 11, e0153881. [Google Scholar] [CrossRef]
- Carissimo, G.; Van Den Beek, M.; Vernick, K.D.; Antoniewski, C. Metavisitor, a suite of galaxy tools for simple and rapid detection and discovery of viruses in deep sequence data. PLoS ONE 2017, 12, e0168397. [Google Scholar] [CrossRef]
- Chandler, J.A.; Liu, R.M.; Bennett, S.N. RNA shotgun metagenomic sequencing of northern California (USA) mosquitoes uncovers viruses, bacteria, and fungi. Front Microbiol. 2015, 6, 1–16. [Google Scholar] [CrossRef]
- Stollar, V.; Thomas, V.L. An agent in the Aedes aegypti cell line (Peleg) which causes fusion of Aedes albopictus cells. Virology 1975, 64, 367–377. [Google Scholar] [CrossRef]
- Franzke, K.; Leggewie, M.; Sreenu, V.B.; Jansen, S.; Heitmann, A.; Welch, S.R.; Brennan, B.; Elliott, R.M.; Tannich, E.; Becker, S.C.; et al. Detection, infection dynamics and small RNA response against Culex Y virus in mosquito-derived cells. J. Gen. Virol. 2018, 1–7. [Google Scholar] [CrossRef]
- Palatini, U.; Miesen, P.; Carballar-Lejarazu, R.; Ometto, L.; Rizzo, E.; Tu, Z.; Rij, R.P.; Bonizzoni, M. Comparative genomics shows that viral integrations are abundant and express piRNAs in the arboviral vectors Aedes aegypti and Aedes albopictus. BMC Genom. 2017, 18, 512. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Frangeul, L.; Dickson, L.B.; Blanc, H.; Verdier, Y.; Vinh, J.; Lambrechts, L.; Saleh, M.C. Uncovering the repertoire of endogenous flaviviral elements in Aedes mosquito genomes. J. Virol. 2017, 91, e00571-17. [Google Scholar] [CrossRef] [PubMed]
- Arensburger, P.; Hice, R.H.; Wright, J.A.; Craig, N.L.; Atkinson, P.W. The mosquito Aedes aegypti has a large genome size and high transposable element load but contains a low proportion of transposon-specific piRNAs. BMC Genom. 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Luo, Y.; Lu, R.; Lau, N.; Lai, E.C.; Li, W.-X.; Ding, S.W. Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc Natl Acad. Sci. USA 2010, 107, 1606–1611. [Google Scholar] [CrossRef]
- Van Rij, R.P.; Berezikov, E. Small RNAs and the control of transposons and viruses in Drosophila. Trends Microbiol. 2009, 17, 163–171. [Google Scholar] [CrossRef]
- Poirier, E.Z.; Goic, B.; Tomé-Poderti, L.; Frangeul, L.; Boussier, J.; Gausson, V.; Blanc, H.; Vallet, T.; Loyd, H.; Levi, L.I.; et al. Dicer-2-dependent generation of viral DNA from defective genomes of RNA viruses modulates antiviral immunity in insects. Cell Host Microbe 2018, 23, 1–13. [Google Scholar] [CrossRef]
- Flynt, A.; Liu, N.; Martin, R.; Lai, E.C. Dicing of viral replication intermediates during silencing of latent Drosophila viruses. Proc. Natl. Acad. Sci. USA 2009, 106, 5270–5275. [Google Scholar] [CrossRef]
- Vodovar, N.; Goic, B.; Blanc, H.; Saleh, M.-C. In silico reconstruction of viral genomes from small RNAs improves virus-derived small interfering RNA profiling. J. Virol. 2011, 85, 11016–11021. [Google Scholar] [CrossRef]
- Brackney, D.E.; Scott, J.C.; Sagawa, F.; Woodward, J.E.; Miller, N.A.; Schilkey, F.D.; Mudge, J.; Wilusz, J.; Olson, K.E.; Blair, C.D.; et al. C6/36 Aedes albopictus cells have a dysfunctional antiviral RNA interference response. PLoS Negl. Trop. Dis. 2010, 4, e856. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, A. Isolation of a Singh’s Aedes albopictus cell clone sensitive to dengue and chikungunya viruses. J. Gen. Virol. 1978, 40, 531–544. [Google Scholar] [CrossRef]
- Varma, M.G.R.; Pudney, M.; Leake, C.J. Cell lines from larvae of Aedes (Stegomyia) Malayensis colless and Aedes (S) pseudoscutellaris (Theobald) and their infection with some arboviruses. Trans. R. Soc. Trop. Med. Hyg. 1974, 68, 374–382. [Google Scholar] [CrossRef]
- Van Cleef, K.W.R.; van Mierlo, J.T.; Miesen, P.; Overheul, G.J.; Fros, J.J.; Schuster, S.; Marklewitz, M.; Pijlman, G.P.; Junglen, S.; van Rij, R.P. Mosquito and Drosophila entomobirnaviruses suppress dsRNA- and siRNA-induced RNAi. Nucleic Acids Res. 2014, 42, 8732–8744. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, J. Wrapping things up about virus RNA replication. Traffic 2005, 6, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Harak, C.; Lohmann, V. Ultrastructure of the replication sites of positive-strand RNA viruses. Virology 2015, 479–480, 418–433. [Google Scholar] [CrossRef] [PubMed]
- Siu, R.W.C.; Fragkoudis, R.; Simmonds, P.; Donald, C.L.; Chase-Topping, M.E.; Barry, G.; Attarzadeh-Yazdi, G.; Rodriguez-Andres, J.; Nash, A.A.; Merits, A.; et al. Antiviral RNA interference responses induced by Semliki Forest virus infection of mosquito cells: Characterization, origin, and frequency-dependent functions of virus-derived small interfering RNAs. J. Virol. 2011, 85, 2907–2917. [Google Scholar] [CrossRef]
- Asgari, S. Role of microRNAs in arbovirus/vector interactions. Viruses 2014, 6, 3514–3534. [Google Scholar] [CrossRef]
- Schultz, M.J.; Frydman, H.M.; Connor, J.H. Dual insect specific virus infection limits arbovirus replication in Aedes mosquito cells. Virology 2018, 518, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Goic, B.; Stapleford, K.A.; Frangeul, L.; Doucet, A.J.; Gausson, V.; Blanc, H.; Schemmel-Jofre, N.; Cristofari, G.; Lambrechts, L.; Vignuzzi, M.; et al. Virus-derived DNA drives mosquito vector tolerance to arboviral infection. Nat. Commun. 2016, 7, 12410. [Google Scholar] [CrossRef]
- Elliott, R.M.; Wilkie, M.L. Persistent infection of Aedes albopictus C6/36 cells by Bunyamwera virus. Virology 1986, 150, 21–32. [Google Scholar] [CrossRef]
- Juárez-Martínez, A.B.; Vega-Almeida, T.O.; Salas-Benito, M.; García-Espitia, M.; de Nova-Ocampo, M.; del Ángel, R.M.; Salas-Benito, J.S. Detection and sequencing of defective viral genomes in C6/36 cells persistently infected with dengue virus. Arch. Virol. 2013, 158, 583–599. [Google Scholar] [CrossRef] [PubMed]
- Karpf, A.R.; Lenches, E.; Strauss, E.G.; Strauss, J.H.; Brown, D.T. Superinfection exclusion of alphaviruses in three mosquito cell lines persistently infected with Sindbis virus. J. Virol. 1997, 71, 7119–7123. [Google Scholar] [PubMed]
- Nicoletti, L.; Verani, P. Growth of the Phlebovirus Toscana in a mosquito (Aedes pseudoscutellaris) cell line (AP-61): Establishment of a persistent infection. Arch. Virol. Austria 1985, 85, 35–45. [Google Scholar] [CrossRef]
- Hillman, B.I.; Cai, G. The family Narnaviridae: Simplest of RNA viruses. In Advances in Virus Research, 1st ed.; Academic Press: Cambridge, MA, USA, 2013. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.D.; Tian, J.H.; Chen, L.J.; Chen, X.; Li, C.X.; Qin, X.C.; Li, J.; Cao, J.P.; Eden, J.S.; et al. Redefining the invertebrate RNA virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef] [PubMed]


| Cell Line | Small RNAs | Contigs | Contigs >200 nts | Virus Blastx Hits | Assemblies |
|---|---|---|---|---|---|
| CT | 37,450,526 | 2,126 | 1,201 | 198 | 32 |
| U4.4 | 37,470,471 | 3,423 | 1,655 | 296 | 46 |
| Aag2 | 37,075,343 | 5,969 | 3,357 | 736 | 153 |
| C6/36 | 37,769,699 | 4,393 | 2,267 | 322 | 72 |
| AP-61 | 29,104,543 | 1,435 | 615 | 83 | 19 |
| Virus | Virus Family | Genome | Segment | Genome Size (nts) | Cell Line | Coverage (nts) | Coverage (%) | Mismatches (n) | Identity (%) |
|---|---|---|---|---|---|---|---|---|---|
| CYV | Birnaviridae | dsRNA | A | 3,429 | U4.4 | 3,429 | 100 | 38 | 99 |
| B | 3,254 | U4.4 | 3,254 | 100 | 30 | 99 | |||
| PCLV | Phenuiviridae | -ssRNA | L | 6,769 | CT | 6,671 | 99 | 114 | 98 |
| Aag2 | 0 | 0 | N/A | N/A | |||||
| M | 3,849 | CT | 2,628 | 68 | 54 | 98 | |||
| Aag2 | 3,213 | 83 | 2 | 100 | |||||
| S | 1,362 | CT | 1,362 | 100 | 62 | 95 | |||
| Aag2 | 345 | 25 | 0 | 100 | |||||
| CLBOV | Flaviviridae | +ssRNA | N/A | 10,735 | CT | 7,674 | 71 | 21 | 100 |
| FHV | Nodaviridae | +ssRNA | 1 | 3,107 | CT | 359 | 12 | 17 | 95 |
| 2 | 1,365 | CT | 333 | 24 | 23 | 93 | |||
| CxNV1 | Narna-like | +ssRNA* | N/A | 3,105 | CT | 3,105 | 100 | 81 | 97 |
| AeDNV | Parvoviridae | ssDNA | N/A | 3,937 | Aag2 | 913 | 23 | 41 | 96 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Göertz, G.P.; Miesen, P.; Overheul, G.J.; van Rij, R.P.; van Oers, M.M.; Pijlman, G.P. Mosquito Small RNA Responses to West Nile and Insect-Specific Virus Infections in Aedes and Culex Mosquito Cells. Viruses 2019, 11, 271. https://doi.org/10.3390/v11030271
Göertz GP, Miesen P, Overheul GJ, van Rij RP, van Oers MM, Pijlman GP. Mosquito Small RNA Responses to West Nile and Insect-Specific Virus Infections in Aedes and Culex Mosquito Cells. Viruses. 2019; 11(3):271. https://doi.org/10.3390/v11030271
Chicago/Turabian StyleGöertz, Giel P., Pascal Miesen, Gijs J. Overheul, Ronald P. van Rij, Monique M. van Oers, and Gorben P. Pijlman. 2019. "Mosquito Small RNA Responses to West Nile and Insect-Specific Virus Infections in Aedes and Culex Mosquito Cells" Viruses 11, no. 3: 271. https://doi.org/10.3390/v11030271
APA StyleGöertz, G. P., Miesen, P., Overheul, G. J., van Rij, R. P., van Oers, M. M., & Pijlman, G. P. (2019). Mosquito Small RNA Responses to West Nile and Insect-Specific Virus Infections in Aedes and Culex Mosquito Cells. Viruses, 11(3), 271. https://doi.org/10.3390/v11030271







