Packaging of Genomic RNA in Positive-Sense Single-Stranded RNA Viruses: A Complex Story
Abstract
1. Introduction
2. General Features of (+)ssRNA Virus Nucleocapsid Assembly
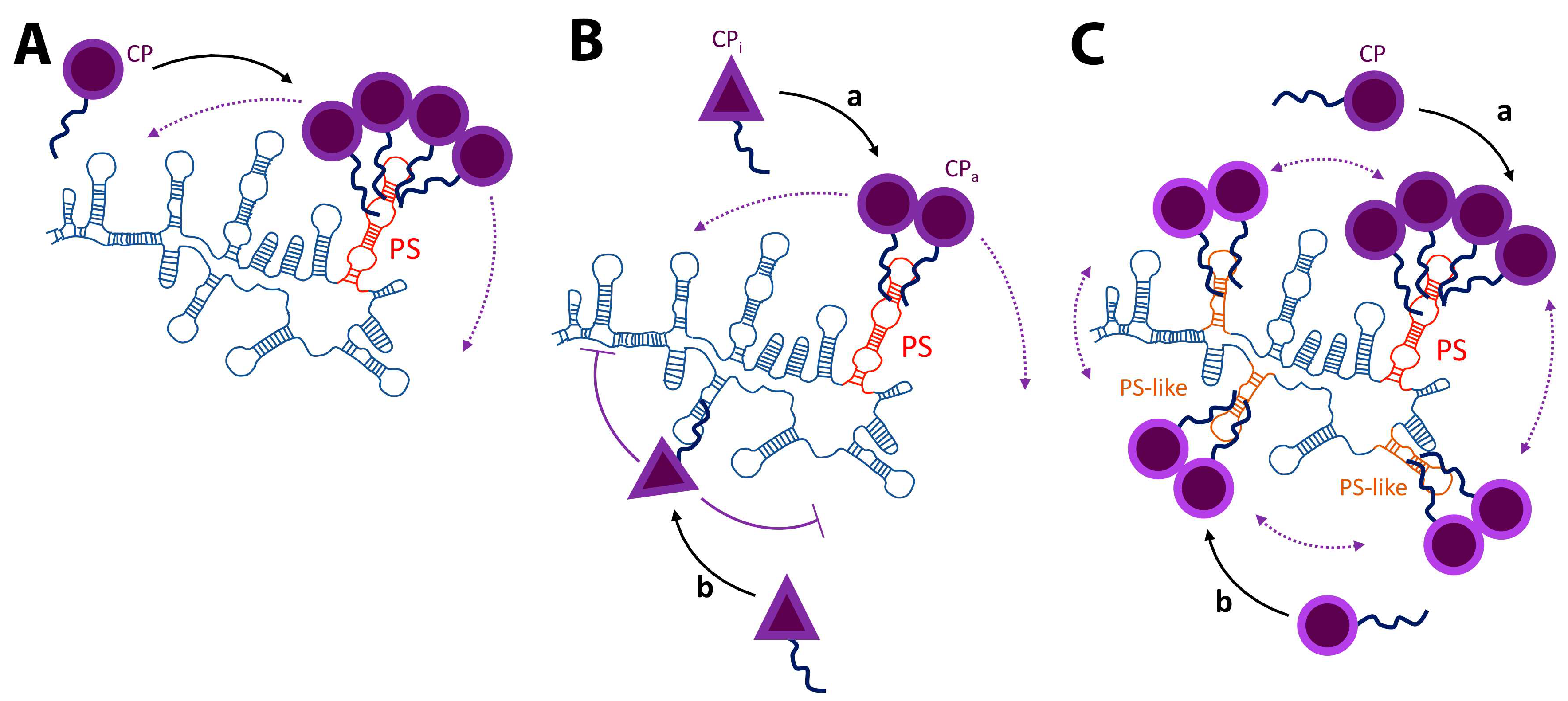
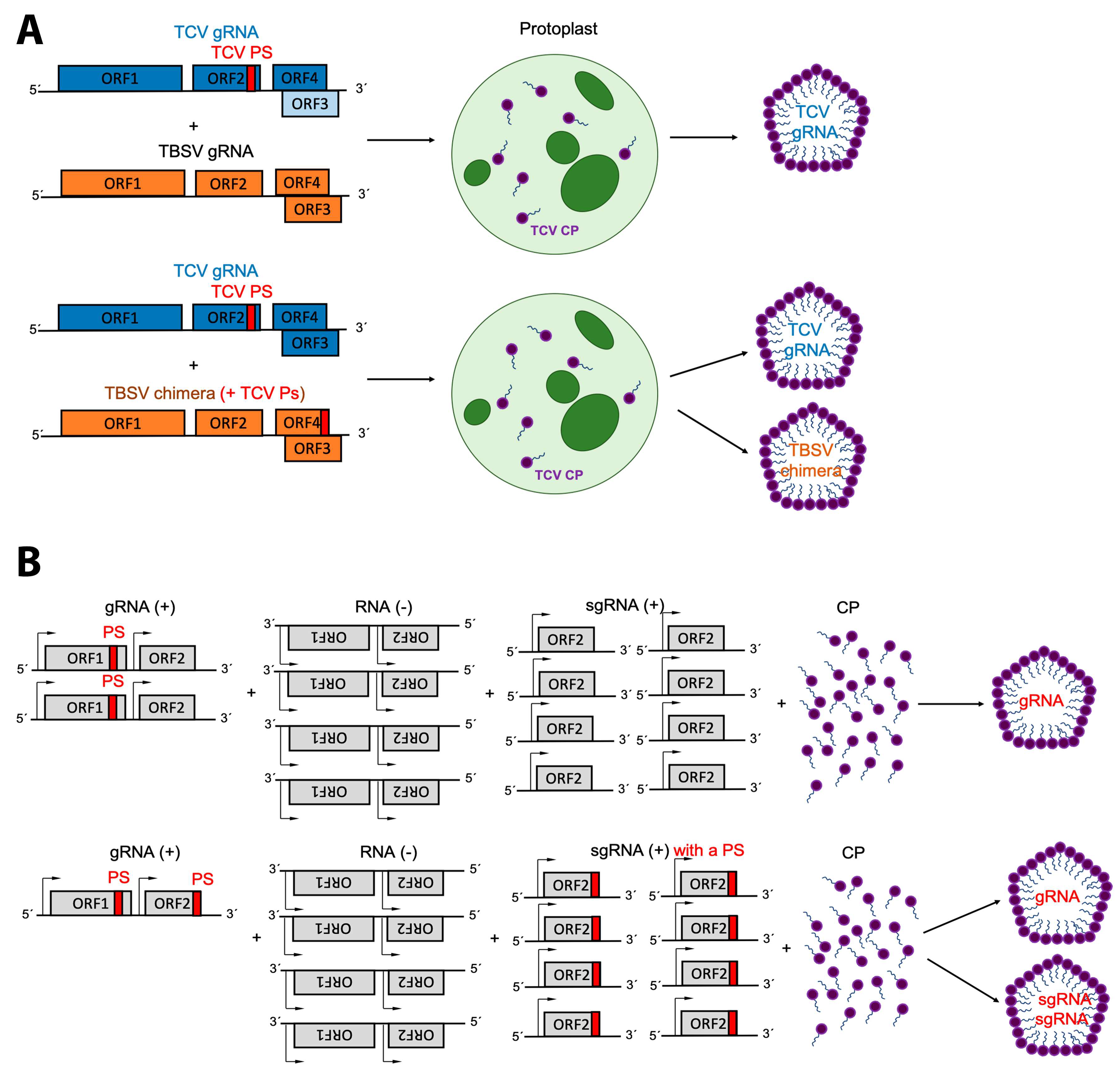
3. Packaging Signals
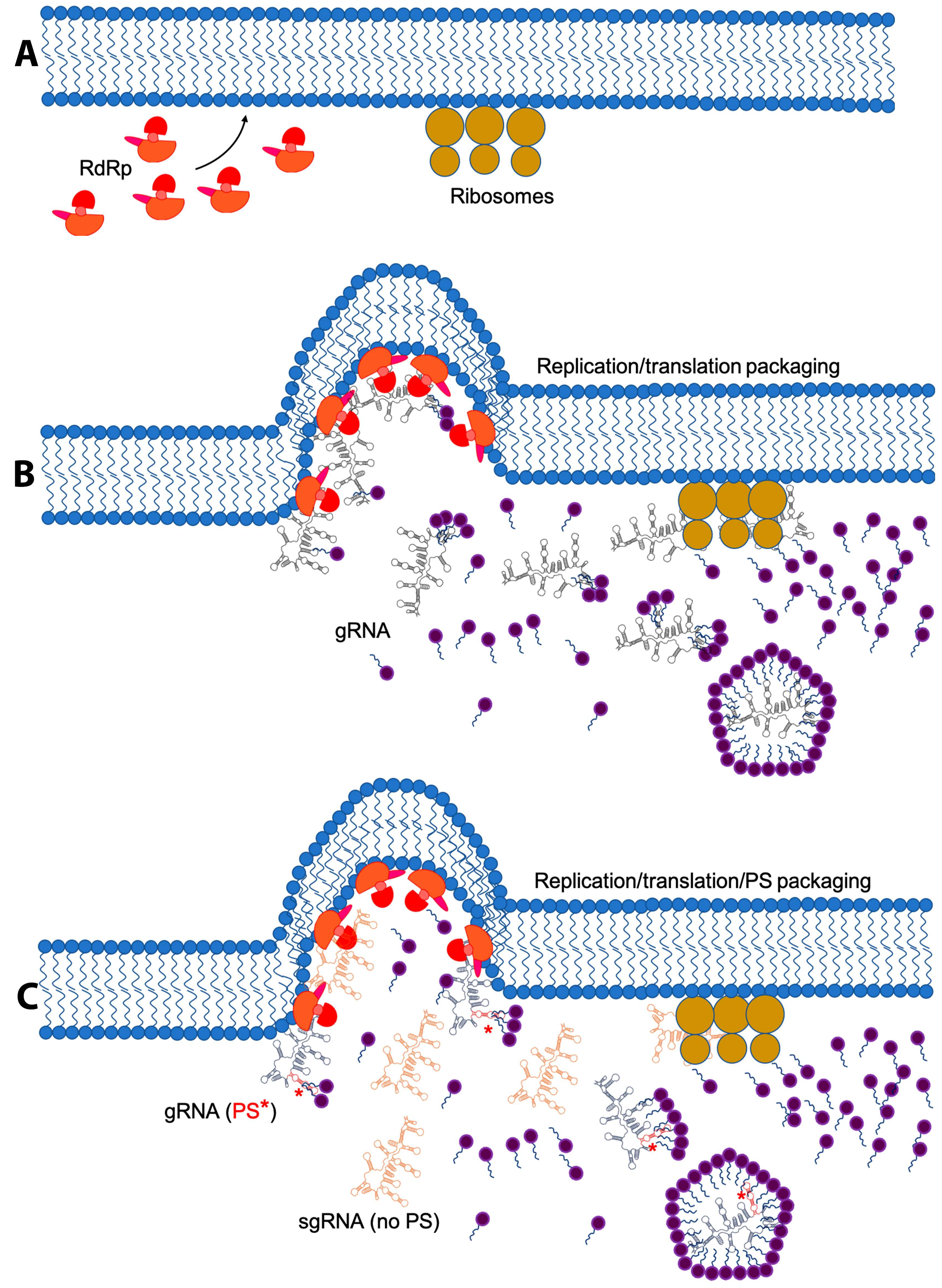
4. Replication and Translation Contribute to Packaging Specificity
5. Membrane Re-Arrangement and Viral Factories
6. Conclusions: Finding a Common Ground
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, Y.; Wang, C.; Mueller, S.; Paul, A.V.; Wimmer, E.; Jiang, P. Direct interaction between two viral proteins, the nonstructural protein 2c and the capsid protein vp3, is required for enterovirus morphogenesis. PLoS Pathog. 2010, 6, e1001066. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ren, J.; Gao, Q.; Hu, Z.; Sun, Y.; Li, X.; Rowlands, D.J.; Yin, W.; Wang, J.; Stuart, D.I.; et al. Hepatitis a virus and the origins of picornaviruses. Nature 2015, 517, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Bishop, N.E.; Hugo, D.L.; Borovec, S.V.; Anderson, D.A. Rapid and efficient purification of hepatitis a virus from cell culture. J. Virol. Methods 1994, 47, 203–216. [Google Scholar] [CrossRef]
- Lomonossoff, G.P.; Johnson, J.E. The synthesis and structure of comovirus capsids. Prog. Biophys. Mol. Biol. 1991, 55, 107–137. [Google Scholar] [CrossRef]
- Cadena-Nava, R.D.; Comas-Garcia, M.; Garmann, R.F.; Rao, A.L.; Knobler, C.M.; Gelbart, W.M. Self-assembly of viral capsid protein and RNA molecules of different sizes: Requirement for a specific high protein/RNA mass ratio. J. Virol. 2012, 86, 3318–3326. [Google Scholar] [CrossRef]
- Comas-Garcia, M.; Garmann, R.F.; Singaram, S.W.; Ben-Shaul, A.; Knobler, C.M.; Gelbart, W.M. Characterization of viral capsid protein self-assembly around short single-stranded RNA. J. Phys. Chem. B 2014, 118, 7510–7519. [Google Scholar] [CrossRef]
- Garmann, R.F.; Comas-Garcia, M.; Gopal, A.; Knobler, C.M.; Gelbart, W.M. The assembly pathway of an icosahedral single-stranded RNA virus depends on the strength of inter-subunit attractions. J. Mol. Biol. 2014, 426, 1050–1060. [Google Scholar] [CrossRef]
- Garmann, R.F.; Comas-Garcia, M.; Knobler, C.M.; Gelbart, W.M. Physical principles in the self-assembly of a simple spherical virus. Acc. Chem. Res. 2016, 49, 48–55. [Google Scholar] [CrossRef]
- Dykeman, E.C.; Stockley, P.G.; Twarock, R. Building a viral capsid in the presence of genomic RNA. Phys. Rev. E 2013, 87, 022717. [Google Scholar] [CrossRef]
- Johnson, J.M.; Willits, D.A.; Young, M.J.; Zlotnick, A. Interaction with capsid protein alters RNA structure and the pathway for in vitro assembly of cowpea chlorotic mottle virus. J. Mol. Biol. 2004, 335, 455–464. [Google Scholar] [CrossRef]
- Zlotnick, A. Are weak protein-protein interactions the general rule in capsid assembly? Virology 2003, 315, 269–274. [Google Scholar] [CrossRef]
- Zlotnick, A. To build a virus capsid: An equilibrium model of the self assembly of polyhedral protein complexes. J. Mol. Biol. 1994, 241, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Zlotnick, A. Distinguishing reversible from irreversible virus capsid assembly. J. Mol. Biol. 2007, 366, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Zlotnick, A.; Aldrich, R.; Johnson, J.E.; Ceres, P.; Young, M.J. Mechanism of capsid assembly for an icosahedral plant virus. Virology 2000, 277, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Zlotnick, A.; Porterfield, J.Z.; Wang, J.C.-Y. To build a virus on a nucleic acid substrate. Biophys. J. 2013, 104, 1595–1604. [Google Scholar] [CrossRef]
- Bruinsma, R.F.; Comas-Garcia, M.; Garmann, R.F.; Grosberg, A.Y. Equilibrium self-assembly of small RNA viruses. Phys. Rev. E 2016, 93, 032405. [Google Scholar] [CrossRef]
- Hagan, M.F. Controlling viral capsid assembly with templating. Phys. Rev. E 2008, 77, 051904. [Google Scholar] [CrossRef] [PubMed]
- Perlmutter, J.D.; Hagan, M.F. Mechanisms of virus assembly. Phys. Chem. 2015, 66, 217–239. [Google Scholar] [CrossRef]
- Perlmutter, J.D.; Perkett, M.R.; Hagan, M.F. Pathways for virus assembly around nucleic acids. J. Mol. Biol. 2014, 426, 3148–3165. [Google Scholar] [CrossRef] [PubMed]
- Erdemci-Tandogan, G.; Wagner, J.; van der Schoot, P.; Podgornik, R.; Zandi, R. Rna topology remolds electrostatic stabilization of viruses. Phys. Rev. E 2014, 89, 032707. [Google Scholar] [CrossRef]
- Elrad, O.M.; Hagan, M.F. Mechanisms of size control and polymorphism in viral capsid assembly. Nano Lett. 2008, 8, 3850–3857. [Google Scholar] [CrossRef] [PubMed]
- Gaussier, H.; Yang, Q.; Catalano, C.E. Building a virus from scratch: Assembly of an infectious virus using purified components in a rigorously defined biochemical assay system. J. Mol. Biol. 2006, 357, 1154–1166. [Google Scholar] [CrossRef] [PubMed]
- Aksyuk, A.A.; Rossmann, M.G. Bacteriophage assembly. Viruses 2011, 3, 172–203. [Google Scholar] [CrossRef] [PubMed]
- Kainov, D.E.; Tuma, R.; Mancini, E.J. Hexameric molecular motors: P4 packaging atpase unravels the mechanism. Cell. Mol. Life Sci. 2006, 63, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Olson, N.H.; Van Etten, J.L.; Bergoin, M.; Rossmann, M.G.; Baker, T.S. Structure and assembly of large lipid-containing dsDNA viruses. Nat. Struct. Mol. Biol. 2000, 7, 101–103. [Google Scholar]
- Hohn, T. Packaging of genomes in bacteriophages: A comparison of ssRNA bacteriophages and dsDNA bactgeriophages. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1976, 276, 143–150. [Google Scholar] [CrossRef]
- Homa, F.L.; Brown, J.C. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 1997, 7, 107–122. [Google Scholar] [CrossRef]
- Newcomb, W.; Thomsen, D.R.; Homa, F.L.; Brown, J.C. Assembly of the herpes simplex virus capsid: Identification of soluble scaffold-portal complexes and their role in formation of portal-containing capsids. J. Virol. 2003, 77, 9862–9871. [Google Scholar] [CrossRef]
- Griffith, J.; Dieckmann, M.; Berg, P. Electron microscope localization of a protein bound near the origin of simian virus 40 DNA replication. J. Virol. 1975, 15, 167–172. [Google Scholar]
- Varshavsky, A.J.; Bakayev, V.V.; Chumackov, P.M.; Georgiev, G.P. Minichromosome of simian virus 40: Presence of histone hi. Nucleic Acids Res. 1976, 3, 2101–2113. [Google Scholar] [CrossRef]
- Kler, S.; Asor, R.; Li, C.; Ginsburg, A.; Harries, D.; Oppenheim, A.; Zlotnick, A.; Raviv, U. Rna encapsidation by sv40-derived nanoparticles follows a rapid two-state mechanism. J. Am. Chem. Soc. 2012, 134, 8823–8830. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Abd-El-Latif, M.; Bronstein, M.; Ben-nun-Shaul, O.; Kler, S.; Oppenheim, A. High cooperativity of the sv40 major capsid protein vp1 in virus assembly. PLoS ONE 2007, 2, e765. [Google Scholar] [CrossRef]
- Van Rosmalen, M.G.M.; Li, C.; Zlotnick, A.; Wuite, G.J.L.; Roos, W.H. Effect of dsDNA on the assembly pathway and mechanical strength of sv40 vp1 virus-like particles. Biophys. J. 2018, 115, 1656–1665. [Google Scholar] [CrossRef] [PubMed]
- Borodavka, A.; Tuma, R.; Stockley, P.G. Evidence that viral RNAs have evolved for efficient, two-stage packaging. Proc. Natl. Acad. Sci. USA 2012, 109, 15769–15774. [Google Scholar] [CrossRef] [PubMed]
- Ford, R.J.; Barker, A.M.; Bakker, S.E.; Coutts, R.H.; Ranson, N.A.; Phillips, S.E.; Pearson, A.R.; Stockley, P.G. Sequence-specific, RNA-protein interactions overcome electrostatic barriers preventing assembly of satellite tobacco necrosis virus coat protein. J. Mol. Biol. 2013, 425, 1050–1064. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.; White, S.J.; Thompson, R.F.; Bingham, R.; Weiss, E.U.; Maskell, D.P.; Zlotnick, A.; Dykeman, E.; Tuma, R.; Twarock, R.; et al. HBV RNA pre-genome encodes specific motifs that mediate interactions with the viral core protein that promote nucleocapsid assembly. Nat. Microbiol. 2017, 2, 17098. [Google Scholar] [CrossRef] [PubMed]
- Sorger, P.; Stockley, P.G.; Harrison, S.C. Structure and assembly of turnip crinkle virus II. Mechanism of reassembly in vitro. J. Mol. Biol. 1986, 191, 639–658. [Google Scholar] [CrossRef]
- Chaturvedi, S.; Rao, A.L. Live cell imaging of interactions between replicase and capsid protein of brome mosaic virus using bimolecular fluorescence complementation: Implications for replication and genome packaging. Virology 2014, 464–465, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Volkova, E.; Gorchakov, R.; Frolov, I. The efficient packaging of venezuelan equine encephalitis virus-specific RNAs into viral particles is determined by nsp1-3 synthesis. Virology 2006, 344, 315–327. [Google Scholar] [CrossRef]
- Qu, F.; Morris, T.J. Encapsidation of turnip crinkle virus is defined by a specific packaging signal and RNA size. J. Virol. 1997, 71, 1428–1435. [Google Scholar] [PubMed]
- Venter, P.A.; Krishna, N.K.; Schneemann, A. Capsid protein synthesis from replicating RNA directs specific packaging of the genome of a multipartite, positive-strand RNA virus. J. Virol. 2005, 79, 6239–6248. [Google Scholar] [CrossRef]
- Khromykh, A.A.; Varnavski, A.N.; Sedlak, P.L.; Westaway, E.G. Coupling between replication and packaging of flavivirus RNA: Evidence derived from the use of DNA-based full-length cDNA clones of kunjin virus. J. Virol. 2001, 75, 4633–4640. [Google Scholar] [CrossRef]
- Nugent, C.I.; Johnson, K.L.; Sarnow, P.; Kirkegaard, K. Functional coupling between replication and packaging of poliovirus replicon RNA. J. Virol. 1999, 73, 427–435. [Google Scholar] [PubMed]
- Annamalai, P.; Rao, A.L.N. Packaging of brome mosaic virus subgenomic RNA is functionally coupled to replication-dependent transcription and translation of coat protein. J. Virol. 2006, 80, 10096–10108. [Google Scholar] [CrossRef] [PubMed]
- Bamunusinghe, D.; Seo, J.K.; Rao, A.L. Subcellular localization and rearrangement of endoplasmic reticulum by brome mosaic virus capsid protein. J. Virol. 2011, 85, 2953–2963. [Google Scholar] [CrossRef] [PubMed]
- Romero-Brey, I.; Bartenschlager, R. Membranous replication factories induced by plus-strand RNA viruses. Viruses 2014, 6, 2826–2857. [Google Scholar] [CrossRef] [PubMed]
- Laliberte, J.F.; Sanfacon, H. Cellular remodeling during plant virus infection. Annu. Rev. Phytopathol. 2010, 48, 69–91. [Google Scholar] [CrossRef]
- Ahlquist, P. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol. 2006, 4, 371–382. [Google Scholar] [CrossRef]
- Den Boon, J.A.; Ahlquist, P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 2010, 64, 241–256. [Google Scholar] [CrossRef] [PubMed]
- Diaz, A.; Wang, X.; Ahlquist, P. Membrane-shaping host reticulon proteins play crucial roles in viral RNA replication compartment formation and function. Proc. Natl. Acad. Sci. USA 2010, 107, 16291–16296. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.J.; Schwartz, M.D.; Ahlquist, P. Flock house virus RNA replicates on outer mitochondrial membranes in drosophila cells. J. Virol. 2001, 75, 11664–11676. [Google Scholar] [CrossRef] [PubMed]
- Noueiry, A.O.; Ahlquist, P. Brome mosaic virus RNA replication: Revealing the role of the host in RNA virus replication. Annu. Rev. Phytopathol. 2003, 41, 77–98. [Google Scholar] [CrossRef]
- Rao, A.L. Genome packaging by spherical plant RNA viruses. Annu. Rev. Phytopathol. 2006, 44, 61–87. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Li, Z.; Lai, M.; Shu, S.; Du, Y.; Zhou, Z.H.; Sun, R. In situ structures of the genome and genome-delivery apparatus in a single-stranded RNA virus. Nature 2017, 541, 112–116. [Google Scholar] [CrossRef]
- Stockley, P.G.; Rolfsson, O.; Thompson, G.S.; Basnak, G.; Francese, S.; Stonehouse, N.J.; Homans, S.W.; Ashcroft, A.E. A simple, RNA-mediated allosteric switch controls the pathway to formation of a t = 3 viral capsid. J. Mol. Biol. 2007, 369, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Mastico, R.A.; Talbot, S.J.; Stockley, P.G. Multiple presentation of foreign peptides on the surface of an RNA-free spherical bacteriophage capsid. J. Gen. Virol. 1993, 74 Pt 4, 541–548. [Google Scholar] [CrossRef]
- Rossmann, M.G.; Johnson, J.E. Icosahedral RNA virus structure. Annu. Rev. Biochem. 1989, 58, 533–573. [Google Scholar] [CrossRef]
- Baker, T.S.; Olson, N.H.; Fuller, S.D. Adding the third dimension to virus life cycles: Three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev. 1999, 63, 862. [Google Scholar] [CrossRef]
- White, J.M. Viral and cellular membrane fusion proteins. Annu. Rev. Physiol. 1990, 52, 675–697. [Google Scholar] [CrossRef]
- Hardy, M.E. Norovirus protein structure and function. FEMS Microbiol. Lett. 2005, 253, 1–8. [Google Scholar] [CrossRef]
- Ng, K.K.; Pendas-Franco, N.; Rojo, J.; Boga, J.A.; Machin, A.; Alonso, J.M.; Parra, F. Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft. J. Biol. Chem. 2004, 279, 16638–16645. [Google Scholar] [CrossRef] [PubMed]
- Hogle, J.M.; Chow, M.; Filman, D.J. Three-dimensional structure of poliovirus at 2.9 a resolution. Science 1985, 229, 1358–1365. [Google Scholar] [CrossRef] [PubMed]
- Salonen, A.; Ahola, T.; Kaariainen, L. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 2005, 285, 139–173. [Google Scholar] [PubMed]
- Adolph, K.W.; Butler, P.J.G. Studies on the assembly of a spherical plant virus+:: Iii. Reassembly of infectious virus under mild conditions. J. Mol. Biol. 1977, 109, 345–357. [Google Scholar] [CrossRef]
- Adolph, K.W.; Butler, P.J. Assembly of a spherical plant virus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1976, 276, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Bancroft, J.B. A virus made from parts of the genomes of brome mosaic and cowpea chlorotic mottle viruses. J. Gen. Virol. 1972, 14, 223–228. [Google Scholar] [CrossRef]
- Fraenkel-Conrat, H.; Williams, R.C. Reconstitution of active tobacco mosaic virus from its inactive protein and nucleic acid components. Biochim. Biophys. Acta 1955, 41, 690–698. [Google Scholar] [CrossRef]
- Comas-Garcia, M.; Cadena-Nava, R.D.; Rao, A.L.; Knobler, C.M.; Gelbart, W.M. In vitro quantification of the relative packaging efficiencies of single-stranded RNA molecules by viral capsid protein. J. Virol. 2012, 86, 12271–12282. [Google Scholar] [CrossRef] [PubMed]
- Garmann, R.F.; Comas-Garcia, M.; Koay, M.S.; Cornelissen, J.J.; Knobler, C.M.; Gelbart, W.M. Role of electrostatics in the assembly pathway of a single-stranded RNA virus. J. Virol. 2014, 88, 10472–10479. [Google Scholar] [CrossRef]
- Endres, D.; Zlotnick, A. Model-based analysis of assembly kinetics for virus capsids or other spherical polymers. Biophys. J. 2002, 83, 1217–1230. [Google Scholar] [CrossRef]
- Johnson, J.E.; Tang, J.; Nyame, Y.; Willits, D.; Young, M.J.; Zlotnick, A. Regulating self-assembly of spherical oligomers. Nano Lett. 2005, 5, 765–770. [Google Scholar] [CrossRef]
- Porterfield, J.Z.; Dhason, M.S.; Loeb, D.D.; Nassal, M.M.; Stray, S.J.; Zlotnick, A. Full-length hepatitis b virus core protein packages viral and heterologous RNA with similarly high levels of cooperativity. J. Virol. 2010, 84, 7174–7184. [Google Scholar] [CrossRef] [PubMed]
- Dhason, M.S.; Wang, J.C.-Y.; Hagan, M.F.; Zlotnick, A. Differential assembly of hepatitis b virus core protein on single- and double-stranded nucleic acid suggest the dsDNA-filled core is spring-loaded. Virology 2012, 430, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Rayaprolu, V.; Moore, A.; Wang, J.C.; Goh, B.C.; Perilla, J.R.; Zlotnick, A.; Mukhopadhyay, S. Length of encapsidated cargo impacts stability and structure of in vitro assembled alphavirus core-like particles. J. Phys. Condens. Matter 2017, 29, 484003. [Google Scholar] [CrossRef] [PubMed]
- Adolph, K.W.; Butler, P.J.G. Studies on the assembly of a spherical plant virus* 1:: I. States of aggregation of the isolated protein. J. Mol. Biol. 1974, 88, 327–338. [Google Scholar] [CrossRef]
- Adolph, K.W. Conformational features of cowpea chlorotic mottle virus RNA and the stability of the virus. J. Gen. Virol. 1975, 28, 137–145. [Google Scholar] [CrossRef]
- Adolph, K.W. Structural transitions of cowpea chlorotic mottle virus. J. Gen. Virol. 1975, 28, 147–154. [Google Scholar] [CrossRef]
- Chen, C.; Kwak, E.S.; Stein, B.; Kao, C.C.; Dragnea, B.B. Packaging of gold particles in viral capsids. J. Nanosci. Nanotechnol. 2005, 5, 2029–2033. [Google Scholar] [CrossRef]
- Cheng, F.; Tsvetkova, I.B.; Khuong, Y.L.; Moore, A.W.; Arnold, R.J.; Goicochea, N.L.; Dragnea, B.; Mukhopadhyay, S. The packaging of different cargo into enveloped viral nanoparticles. Mol. Pharm. 2013, 10, 51–58. [Google Scholar] [CrossRef]
- Chevreuil, M.; Law-Hine, D.; Chen, J.; Bressanelli, S.; Combet, S.; Constantin, D.; Degrouard, J.; Moller, J.; Zeghal, M.; Tresset, G. Nonequilibrium self-assembly dynamics of icosahedral viral capsids packaging genome or polyelectrolyte. Nat. Commun. 2018, 9, 3071. [Google Scholar] [CrossRef]
- Stockley, P.G.; Ashcroft, A.E.; Francese, S.; Thompson, G.S.; Ranson, N.; Smith, A.M.; Homans, S.W.; Stonehouse, N.J. Dissecting the fine details of assembly of a t = 3 phage capsid. J. Theor. Med. 2005, 6, 119–125. [Google Scholar] [CrossRef]
- Basnak, G.; Morton, V.L.; Rolfsson, O.; Stonehouse, N.J.; Ashcroft, A.E.; Stockley, P.G. Viral genomic single-stranded RNA directs the pathway toward a t = 3 capsid. J. Mol. Biol. 2010, 395, 924–936. [Google Scholar] [CrossRef] [PubMed]
- Lane, S.W.; Dennis, C.A.; Lane, C.L.; Trinh, C.H.; Rizkallah, P.J.; Stockley, P.G.; Phillips, S.E.V. Construction and crystal structure of recombinant stnv capsids. J. Mol. Biol. 2011, 413, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Borodavka, A.; Tuma, R.; Stockley, P.G. A two-stage mechanism of viral RNA compaction revealed by single molecule fluorescence. RNA Biol. 2013, 10, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Douglas, T.; Young, M.J. Host–guest encapsulation of materials by assembled virus protein cages. Nature 1998, 393, 152–155. [Google Scholar] [CrossRef]
- Shenton, W.; Douglas, T.; Young, M.J.; Stubbs, G.; Mann, S. Inorganic-organic nanotube composites from template mineralization of tobacco mosaic virus. Adv. Mater. 1999, 11, 253–256. [Google Scholar] [CrossRef]
- Comellas-Aragonès, M.; Engelkamp, H.; Claessen, V.I.; Sommerdijk, N.A.J.M.; Rowan, A.E.; Christianen, P.C.M.; Maan, J.C.; Verduin, B.J.M.; Cornelissen, J.J.L.M.; Nolte, R.J.M. A virus-based single-enzyme nanoreactor. Nat. Nanotechnol. 2007, 2, 635–639. [Google Scholar] [CrossRef] [PubMed]
- Minten, I.J.; Claessen, V.I.; Blank, K.; Rowan, A.E.; Nolte, R.J.M.; Cornelissen, J.J.L.M. Catalytic capsids: The art of confinement. Chem. Sci. 2011, 2, 358–362. [Google Scholar] [CrossRef]
- Douglas, T.; Young, M.J. Virus particles as templates for materials synthesis. Adv. Mater. 1999, 11, 679–681. [Google Scholar] [CrossRef]
- Flynn, C.E.; Lee, S.W.; Peelle, B.R.; Belcher, A.M. Viruses as vehicles for growth, organization and assembly of materials. Acta Mater. 2003, 51, 5867–5880. [Google Scholar] [CrossRef]
- Sánchez-Sánchez, L.; Cadena-Nava, R.D.; Palomares, L.A.; Ruiz-Garcia, J.; Koay, M.S.T.; Cornelissen, J.J.L.M.; Vazquez-Duhalt, R. Chemotherapy pro-drug activation by biocatalytic virus-like nanoparticles containing cytochrome p450. Enzyme Microb. Technol. 2014, 60, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Steinmetz, N.F. Viral nanoparticles as platforms for next-generation therapeutics and imaging devices. Nanomedicine 2010, 6, 634–641. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, I.; Tsvetkova, I.; Wen, A.M.; Shukla, S.; Masarapu, M.H.; Dragnea, B.; Steinmetz, N.F. Engineering of brome mosaic virus for biomedical applications. RSC Adv. 2012, 2, 3670–3677. [Google Scholar] [CrossRef]
- Azizgolshani, O.; Garmann, R.F.; Cadena-Nava, R.; Knobler, C.M.; Gelbart, W.M. Reconstituted plant viral capsids can release genes to mammalian cells. Virology 2013, 441, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Loredo-Tovias, M.; Duran-Meza, A.L.; Villagrana-Escareno, M.V.; Vega-Acosta, R.; Reynaga-Hernandez, E.; Flores-Tandy, L.M.; Valdes-Resendiz, O.E.; Cadena-Nava, R.D.; Alvizo-Paez, E.R.; Ruiz-Garcia, J. Encapsidated ultrasmall nanolipospheres as novel nanocarriers for highly hydrophobic anticancer drugs. Nanoscale 2017, 9, 11625–11631. [Google Scholar] [CrossRef]
- Tapia-Moreno, A.; Juarez-Moreno, K.; Gonzalez-Davis, O.; Cadena-Nava, R.D.; Vazquez-Duhalt, R. Biocatalytic virus capsid as nanovehicle for enzymatic activation of tamoxifen in tumor cells. Biotechnol. J. 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Krishna, N.K.; Marshall, D.; Schneemann, A. Analysis of RNA packaging in wild-type and mosaic protein capsids of flock house virus using recombinant baculovirus vectors. Virology 2003, 305, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Rumenapf, T.; Strauss, E.G.; Strauss, J.H. Subgenomic mRNA of aura alphavirus is packaged into virions. J. Virol. 1994, 68, 56–62. [Google Scholar] [PubMed]
- Witherell, G.W.; Gott, J.M.; Uhlenbeck, O.C. Specific interaction between RNA phage coat proteins and RNA. Prog. Nucleic Acid Res. Mol. Biol. 1991, 40, 185–220. [Google Scholar] [PubMed]
- Lever, A.; Gottlinger, H.; Haseltine, W.; Sodroski, J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type-1 RNA into virions. J. Virol. 1989, 63, 4085–4087. [Google Scholar] [PubMed]
- Weiss, B.; Geigenmuller-Gnirke, U.; Schlesinger, S. Interactions between sindbis virus RNAs and a 68 amino acid derivative of the viral capsid protein further defines the capsid binding site. Nucleic Acids Res. 1994, 22, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Toropova, K.; Basnak, G.; Twarock, R.; Stockley, P.G.; Ranson, N.A. The three-dimensional structure of genomic RNA in bacteriophage ms2: Implications for assembly. J. Mol. Biol. 2008, 375, 824–836. [Google Scholar] [CrossRef] [PubMed]
- Dykeman, E.C.; Grayson, N.E.; Toropova, K.; Ranson, N.A.; Stockley, P.G.; Twarock, R. Simple rules for efficient assembly predict the layout of a packaged viral RNA. J. Mol. Biol. 2011, 408, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Bunka, D.H.; Lane, S.W.; Lane, C.L.; Dykeman, E.C.; Ford, R.J.; Barker, A.M.; Twarock, R.; Phillips, S.E.; Stockley, P.G. Degenerate RNA packaging signals in the genome of satellite tobacco necrosis virus: Implications for the assembly of a t = 1 capsid. J. Mol. Biol. 2011, 413, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Dykeman, E.C.; Stockley, P.G.; Twarock, R. Solving a levinthal’s paradox for virus assembly identifies a unique antiviral strategy. Proc. Natl. Acad. Sci. USA 2014, 111, 5361–5366. [Google Scholar] [CrossRef] [PubMed]
- Bingham, R.J.; Dykeman, E.C.; Twarock, R. RNA virus evolution via a quasispecies-based model reveals a drug target with a high barrier to resistance. Viruses 2017, 9, 347. [Google Scholar] [CrossRef]
- Twarock, R.; Bingham, R.J.; Dykeman, E.C.; Stockley, P.G. A modelling paradigm for RNA virus assembly. Curr. Opin. Virol. 2018, 31, 74–81. [Google Scholar] [CrossRef]
- Stewart, H.; Bingham, R.J.; White, S.J.; Dykeman, E.C.; Zothner, C.; Tuplin, A.K.; Stockley, P.G.; Twarock, R.; Harris, M. Identification of novel RNA secondary structures within the hepatitis c virus genome reveals a cooperative involvement in genome packaging. Sci. Rep. 2016, 6, 22952. [Google Scholar] [CrossRef]
- Comas-Garcia, M.; Davis, S.R.; Rein, A. On the selective packaging of genomic RNA by HIV-1. Viruses 2016, 8, 246. [Google Scholar] [CrossRef]
- Comas-Garcia, M.; Kroupa, T.; Datta, S.A.; Harvin, D.P.; Hu, W.S.; Rein, A. Efficient support of virus-like particle assembly by the HIV-1 packaging signal. eLife 2018, 7, e38438. [Google Scholar] [CrossRef]
- Dilley, K.A.; Nikolaitchik, O.A.; Galli, A.; Burdick, R.C.; Levine, L.; Li, K.; Rein, A.; Pathak, V.K.; Hu, W.S. Interactions between HIV-1 gag and viral RNA genome enhance virion assembly. J. Virol. 2017, 91, e02319-16. [Google Scholar] [CrossRef] [PubMed]
- Zimmern, D. The nucleotide sequence at the origin for assembly on tobacco mosaic virus RNA. Cell 1977, 11, 463–482. [Google Scholar] [CrossRef]
- Zimmern, D.; Butler, P.J. The isolation of tobacco mosaic virus RNA fragments containing the origin for viral assembly. Cell 1977, 11, 455–462. [Google Scholar] [CrossRef]
- Butler, P.J.; Finch, J.T.; Zimmern, D. Configuration of tobacco mosaic virus, RNA during virus assembly. Nature 1977, 265, 217–219. [Google Scholar] [CrossRef]
- Zimmern, D. An extended secondary structure model for the tmv assembly origin, and its correlation with protection studies and an assembly defective mutant. EMBO J. 1983, 2, 1901–1907. [Google Scholar] [CrossRef]
- Knapman, T.W.; Morton, V.L.; Stonehouse, N.J.; Stockley, P.G.; Ashcroft, A.E. Determining the topology of virus assembly intermediates using ion mobility spectrometry-mass spectrometry. Rapid Commun. Mass Spectrom. 2010, 24, 3033–3042. [Google Scholar] [CrossRef] [PubMed]
- Elrad, O.M.; Hagan, M.F. Encapsulation of a polymer by an icosahedral virus. Phys. Biol. 2010, 7, 045003. [Google Scholar] [CrossRef]
- Perlmutter, J.D.; Qiao, C.; Hagan, M.F. Viral genome structures are optimal for capsid assembly. eLife 2013, 2, e00632. [Google Scholar] [CrossRef]
- Perlmutter, J.D.; Hagan, M.F. The role of packaging sites in efficient and specific virus assembly. J. Mol. Biol. 2015, 427, 2451–2467. [Google Scholar] [CrossRef]
- Bruinsma, R.F.; Gelbart, W.M.; Reguera, D.; Rudnick, J.; Zandi, R. Viral self-assembly as a thermodynamic process. Phys. Rev. Lett. 2003, 90, 248101. [Google Scholar] [CrossRef]
- Zandi, R.; van der Schoot, P.; Reguera, D.; Kegel, W.; Reiss, H. Classical nucleation theory of virus capsids. Biophys. J. 2006, 90, 1939–1948. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zandi, R.; Anavitarte, A.; Knobler, C.M.; Gelbart, W.M. Packaging of a polymer by a viral capsid: The interplay between polymer length and capsid size. Biophys. J. 2008, 94, 1428–1436. [Google Scholar] [CrossRef] [PubMed]
- Zandi, R.; van der Schoot, P. Size regulation of ss-RNA viruses. Biophys. J. 2009, 96, 9–20. [Google Scholar] [CrossRef]
- Chatel-Chaix, L.; Bartenschlager, R. Dengue virus- and hepatitis c virus-induced replication and assembly compartments: The enemy inside—Caught in the web. J. Virol. 2014, 88, 5907–5911. [Google Scholar] [CrossRef] [PubMed]
- Diaz, A.; Zhang, J.; Ollwerther, A.; Wang, X.; Ahlquist, P. Host escrt proteins are required for bromovirus RNA replication compartment assembly and function. PLoS Pathog. 2015, 11, e1004742. [Google Scholar] [CrossRef]
- Witherell, G.W.; Wu, H.N.; Uhlenbeck, O.C. Cooperative binding of r17 coat protein to RNA. Biochemistry 1990, 29, 11051–11057. [Google Scholar] [CrossRef]
- Meyers, G.; Wirblich, C.; Thiel, H.J. Genomic and subgenomic RNAs of rabbit hemorrhagic disease virus are both protein-linked and packaged into particles. Virology 1991, 184, 677–686. [Google Scholar] [CrossRef]
- Asanaka, M.; Atmar, R.L.; Ruvolo, V.; Crawford, S.E.; Neill, F.H.; Estes, M.K. Replication and packaging of norwalk virus RNA in cultured mammalian cells. Proc. Natl. Acad. Sci. USA 2005, 102, 10327–10332. [Google Scholar] [CrossRef]
- Kim, D.Y.; Firth, A.E.; Atasheva, S.; Frolova, E.I.; Frolov, I. Conservation of a packaging signal and the viral genome RNA packaging mechanism in alphavirus evolution. J. Virol. 2011, 85, 8022–8036. [Google Scholar] [CrossRef]
- Frolova, E.; Frolov, I.; Schlesinger, S. Packaging signals in alphaviruses. J. Virol. 1997, 71, 248–258. [Google Scholar]
- Frolov, I.; Hoffman, T.A.; Pragai, B.M.; Dryga, S.A.; Huang, H.V.; Schlesinger, S.; Rice, C.M. Alphavirus-based expression vectors: Strategies and applications. Proc. Natl. Acad. Sci. USA 1996, 93, 11371–11377. [Google Scholar] [CrossRef] [PubMed]
- Bredenbeek, P.J.; Frolov, I.; Rice, C.M.; Schlesinger, S. Sindbis virus expression vectors: Packaging of RNA replicons by using defective helper RNAs. J. Virol. 1993, 67, 6439–6446. [Google Scholar] [PubMed]
- White, C.L.; Thomson, M.; Dimmock, N.J. Deletion analysis of a defective interfering semliki forest virus RNA genome defines a region in the nsp2 sequence that is required for efficient packaging of the genome into virus particles. J. Virol. 1998, 72, 4320–4326. [Google Scholar] [PubMed]
- Fosmire, J.A.; Hwang, K.; Makino, S. Identification and characterization of a coronavirus packaging signal. J. Virol. 1992, 66, 3522–3530. [Google Scholar] [PubMed]
- Choi, Y.G.; Rao, A.L.N. Packaging of brome mosaic virus RNA3 is mediated through a bipartite signal. J. Virol. 2003, 77, 9750–9757. [Google Scholar] [CrossRef]
- Annamalai, P.; Rao, A.L. Dispensability of 3′ tRNA-like sequence for packaging cowpea chlorotic mottle virus genomic RNAs. Virology 2005, 332, 650–658. [Google Scholar] [CrossRef]
- Annamalai, P.; Rao, A.L.N. In vivo packaging of brome mosaic virus RNA3, but not RNAs 1 and 2, is dependent on a cis-acting 3′ tRNA-like structure. J. Virol. 2007, 81, 173–181. [Google Scholar] [CrossRef]
- Fraenkel-Conrat, H.; Staehelin, M.; Crawford, L.V. Tobacco mosaic virus reconstitution using inactivated nucleic acid. Proc. Soc. Exp. Biol. Med. 1959, 102, 118–121. [Google Scholar] [CrossRef]
- Fraenkel-Conrat, H.; Singer, B. Reconstitution of tobacco mosaic virus. Iii. Improved methods and the use of mixed nucleic acids. Biochim. Biophys. Acta 1959, 33, 359–370. [Google Scholar] [CrossRef]
- Durham, A.C.; Klug, A. Polymerization of tobacco mosaic virus protein and its control. Nat. New Biol. 1971, 229, 42–46. [Google Scholar] [CrossRef]
- Mandelkow, E.; Holmes, K.C.; Gallwitz, U. A new helical aggregate of tobacco mosaic virus protein. J. Mol. Biol. 1976, 102, 265–285. [Google Scholar] [CrossRef]
- Mandelkow, E.; Stubbs, G.; Warren, S. Structures of the helical aggregates of tobacco mosaic virus protein. J. Mol. Biol. 1981, 152, 375–386. [Google Scholar] [CrossRef]
- Butler, P.J. Structures and roles of the polymorphic forms of tobacco mosaic virus protein. Vi. Assembly of the nucleoprotein rods of tobacco mosaic virus from the protein disks and RNA. J. Mol. Biol. 1972, 72, 25–35. [Google Scholar] [CrossRef]
- Butler, P.J.; Klug, A. Assembly of the particle of tobacco mosaic virus from RNA and disks of protein. Nat. New Biol. 1971, 229, 47–50. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.R.; Butler, P.J. Essential features of the assembly origin of tobacco mosaic virus RNA as studied by directed mutagenesis. Nucleic Acids Res. 1986, 14, 9229–9242. [Google Scholar] [CrossRef]
- Gao, S.; Zhang, R.; Yu, Z.; Xi, Z. Antofine analogues can inhibit tobacco mosaic virus assembly through small-molecule-RNA interactions. ChemBioChem 2012, 13, 1622–1627. [Google Scholar] [CrossRef]
- Butler, P.J. The current picture of the structure and assembly of tobacco mosaic virus. J. Gen. Virol. 1984, 65 Pt 2, 253–279. [Google Scholar] [CrossRef]
- Lehtovaara, P.; Soderlund, H.; Keranen, S.; Pettersson, R.F.; Kaariainen, L. 18s defective interfering RNA of semliki forest virus contains a triplicated linear repeat. Proc. Natl. Acad. Sci. USA 1981, 78, 5353–5357. [Google Scholar] [CrossRef]
- Lehtovaara, P.; Soderlund, H.; Keranen, S.; Pettersson, R.F.; Kaariainen, L. Extreme ends of the genome are conserved and rearranged in the defective interfering RNAs of semliki forest virus. J. Mol. Biol. 1982, 156, 731–748. [Google Scholar] [CrossRef]
- Strauss, J.H.; Strauss, E.G. The alphaviruses: Gene expression, replication, and evolution. Microbiol. Rev. 1994, 58, 491–562. [Google Scholar]
- Mendes, A.; Kuhn, R.J. Alphavirus nucleocapsid packaging and assembly. Viruses 2018, 10, 138. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, R.J.; Griffin, D.E.; Owen, K.E.; Niesters, H.G.M.; Strauss, J.H. Chimeric sindbis ross river viruses to study interactions between alphavirus nonstructural and structural regions. J. Virol. 1996, 70, 7900–7909. [Google Scholar] [PubMed]
- Berkowitz, R.D.; Luban, J.; Goff, S.P. Specific binding of human immunodeficiency virus type 1 gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J. Virol. 1993, 67, 7190–7200. [Google Scholar] [PubMed]
- Comas-Garcia, M.; Datta, S.A.; Baker, L.; Varma, R.; Gudla, P.R.; Rein, A. Dissection of specific binding of HIV-1 gag to the ‘packaging signal’ in viral RNA. eLife 2017, 6, e27055. [Google Scholar] [CrossRef] [PubMed]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. Selex—A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007, 24, 381–403. [Google Scholar] [CrossRef]
- Aldovini, A.; Young, R.A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J. Virol. 1990, 64, 1920–1926. [Google Scholar] [PubMed]
- Wilkinson, K.A.; Gorelick, R.J.; Vasa, S.M.; Guex, N.; Rein, A.; Mathews, D.H.; Giddings, M.C.; Weeks, K.M. High-throughput shape analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008, 6, e96. [Google Scholar] [CrossRef]
- Keane, S.C.; Heng, X.; Lu, K.; Kharytonchyk, S.; Ramakrishnan, V.; Carter, G.; Barton, S.; Hosic, A.; Florwick, A.; Santos, J.; et al. Rna structure. Structure of the HIV-1 RNA packaging signal. Science 2015, 348, 917–921. [Google Scholar] [CrossRef]
- Kotta-Loizou, I.; Peyret, H.; Saunders, K.; Coutts, R.H.A.; Lomonossoff, G.P. Investigating the biological relevance of in vitro identified putative packaging signals at the 5′ terminus of the satellite tobacco necrosis virus-1 genomic RNA. J. Virol. 2019. [Google Scholar] [CrossRef]
- Venter, P.A.; Schneemann, A. Assembly of two independent populations of flock house virus particles with distinct RNA packaging characteristics in the same cell. J. Virol. 2007, 81, 613–619. [Google Scholar] [CrossRef]
- Kim, D.Y.; Atasheva, S.; Frolova, E.I.; Frolov, I. Venezuelan equine encephalitis virus nsp2 protein regulates packaging of the viral genome into infectious virions. J. Virol. 2013, 87, 4202–4213. [Google Scholar] [CrossRef] [PubMed]
- Allison, R.; Janda, M.; Ahlquist, P. Infectious in vitro transcripts from cowpea chlorotic mottle virus cDNA clones and exchange of individual RNA components with brome mosaic virus. J. Virol. 1988, 62, 3581–3588. [Google Scholar] [PubMed]
- Rulli, S.J., Jr.; Hibbert, C.S.; Mirro, J.; Pederson, T.; Biswal, S.; Rein, A. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J. Virol. 2007, 81, 6623–6631. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.-K.; Kwon, S.-J.; Rao, A.L.N. A physical interaction between viral replicase and capsid protein is required for genome-packaging specificity in an RNA virus. J. Virol. 2012, 86, 6210–6221. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.K.; Kwon, S.J.; Rao, A.L.N. Molecular dissection of flock house virus protein b2 reveals that electrostatic interactions between n-terminal domains of b2 monomers are critical for dimerization. Virology 2012, 432, 296–305. [Google Scholar] [CrossRef]
- Annamalai, P.; Rao, A.L. Replication-independent expression of genome components and capsid protein of brome mosaic virus in planta: A functional role for viral replicase in RNA packaging. Virology 2005, 338, 96–111. [Google Scholar] [CrossRef]
- Annamalai, P.; Rofail, F.; Demason, D.A.; Rao, A.L. Replication-coupled packaging mechanism in positive-strand RNA viruses: Synchronized coexpression of functional multigenome RNA components of an animal and a plant virus in nicotiana benthamiana cells by agroinfiltration. J. Virol. 2008, 82, 1484–1495. [Google Scholar] [CrossRef] [PubMed]
- Dreher, T.W.; Rao, A.L.; Hall, T.C. Replication in vivo of mutant brome mosaic virus RNAs defective in aminoacylation. J. Mol. Biol. 1989, 206, 425–438. [Google Scholar] [CrossRef]
- Damayanti, T.A.; Tsukaguchi, S.; Mise, K.; Okuno, T. Cis-acting elements required for efficient packaging of brome mosaic virus RNA3 in barley protoplasts. J. Virol. 2003, 77, 9979–9986. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.G.; Dreher, T.W.; Rao, A.L. tRNA elements mediate the assembly of an icosahedral RNA virus. Proc. Natl. Acad. Sci. USA 2002, 99, 655–660. [Google Scholar] [CrossRef]
- Annamalai, P.; Rao, A.L.N. Delivery and expression of functional viral RNA genomes in planta by agroinfiltration. Curr. Protoc. Microbiol. 2006. [Google Scholar] [CrossRef]
- Kujala, P.; Ikaheimonen, A.; Ehsani, N.; Vihinen, H.; Auvinen, P.; Kaariainen, L. Biogenesis of the semliki forest virus RNA replication complex. J. Virol. 2001, 75, 3873–3884. [Google Scholar] [CrossRef] [PubMed]
- Schlegel, A.; Giddings, T.H., Jr.; Ladinsky, M.S.; Kirkegaard, K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 1996, 70, 6576–6588. [Google Scholar] [PubMed]
- Jones, D.M.; McLauchlan, J. Hepatitis c virus: Assembly and release of virus particles. J. Biol. Chem. 2010, 285, 22733–22739. [Google Scholar] [CrossRef] [PubMed]
- Romero-Brey, I.; Merz, A.; Chiramel, A.; Lee, J.Y.; Chlanda, P.; Haselman, U.; Santarella-Mellwig, R.; Habermann, A.; Hoppe, S.; Kallis, S.; et al. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis c virus replication. PLoS Pathog. 2012, 8, e1003056. [Google Scholar] [CrossRef] [PubMed]
- Paul, D.; Bartenschlager, R. Architecture and biogenesis of plus-strand RNA virus replication factories. World J. Virol. 2013, 2, 32–48. [Google Scholar] [CrossRef] [PubMed]
- Magliano, D.; Marshall, J.A.; Bowden, D.S.; Vardaxis, N.; Meanger, J.; Lee, J.Y. Rubella virus replication complexes are virus-modified lysosomes. Virology 1998, 240, 57–63. [Google Scholar] [CrossRef]
- Barajas, D.; Jiang, Y.; Nagy, P.D. A unique role for the host escrt proteins in replication of tomato bushy stunt virus. PLoS Pathog. 2009, 5, e1000705. [Google Scholar] [CrossRef]
- Torrance, L.; Cowan, G.H.; Gillespie, T.; Ziegler, A.; Lacomme, C. Barley stripe mosaic virus-encoded proteins triple-gene block 2 and gammab localize to chloroplasts in virus-infected monocot and dicot plants, revealing hitherto-unknown roles in virus replication. J. Gen. Virol. 2006, 87, 2403–2411. [Google Scholar] [CrossRef]
- Miller, S.; Krijnse-Locker, J. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 2008, 6, 363–374. [Google Scholar] [CrossRef]
- Zhao, H.; Lindqvist, B.; Garoff, H.; von Bonsdorff, C.H.; Liljestrom, P. A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO J. 1994, 13, 4204–4211. [Google Scholar] [CrossRef] [PubMed]
- Frolova, E.I.; Gorchakov, R.; Pereboeva, L.; Atasheva, S.; Frolov, I. Functional sindbis virus replicative complexes are formed at the plasma membrane. J. Virol. 2010, 84, 11679–11695. [Google Scholar] [CrossRef] [PubMed]
- Salonen, A.; Vasiljeva, L.; Merits, A.; Magden, J.; Jokitalo, E.; Kaariainen, L. Properly folded nonstructural polyprotein directs the semliki forest virus replication complex to the endosomal compartment. J. Virol. 2003, 77, 1691–1702. [Google Scholar] [CrossRef]
- Schwartz, M.; Chen, J.; Janda, M.; Sullivan, M.; den Boon, J.; Ahlquist, P. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 2002, 9, 505–514. [Google Scholar] [CrossRef]
- Bamunusinghe, D.; Chaturvedi, S.; Seo, J.K.; Rao, A.L. Mutations in the capsid protein of brome mosaic virus affecting encapsidation eliminate vesicle induction in planta: Implications for virus cell-to-cell spread. J. Virol. 2013, 87, 8982–8992. [Google Scholar] [CrossRef] [PubMed]
- Dykeman, E.C.; Stockley, P.G.; Twarock, R. Packaging signals in two single-stranded RNA viruses imply a conserved assembly mechanism and geometry of the packaged genome. J. Mol. Biol. 2013, 425, 3235–3249. [Google Scholar] [CrossRef]
- Abd El-Wahab, E.W.; Smyth, R.P.; Mailler, E.; Bernacchi, S.; Vivet-Boudou, V.; Hijnen, M.; Jossinet, F.; Mak, J.; Paillart, J.C.; Marquet, R. Specific recognition of the HIV-1 genomic RNA by the gag precursor. Nat. Commun. 2014, 5, 4304. [Google Scholar] [CrossRef]
- Bernacchi, S.; Abd El-Wahab, E.W.; Dubois, N.; Hijnen, M.; Smyth, R.P.; Mak, J.; Marquet, R.; Paillart, J.C. Hiv-1 pr55gag binds genomic and spliced RNAs with different affinity and stoichiometry. RNA Biol. 2017, 14, 90–103. [Google Scholar] [CrossRef]
- Webb, J.A.; Jones, C.P.; Parent, L.J.; Rouzina, I.; Musier-Forsyth, K. Distinct binding interactions of HIV-1 gag to psi and non-psi RNAs: Implications for viral genomic RNA packaging. RNA 2013, 19, 1078–1088. [Google Scholar] [CrossRef]
- Morton, V.L.; Stockley, P.G.; Stonehouse, N.J.; Ashcroft, A.E. Insights into virus capsid assembly from non-covalent mass spectrometry. Mass Spectrom. Rev. 2008, 27, 575–595. [Google Scholar] [CrossRef]
- Dykeman, E.C.; Stockley, P.G.; Twarock, R. Dynamic allostery controls coat protein conformer switching during ms2 phage assembly. J. Mol. Biol. 2010, 395, 916–923. [Google Scholar] [CrossRef] [PubMed]
- Morton, V.L.; Dykeman, E.C.; Stonehouse, N.J.; Ashcroft, A.E.; Twarock, R.; Stockley, P.G. The impact of viral RNA on assembly pathway selection. J. Mol. Biol. 2010, 401, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Belyi, V.A.; Muthukumar, M. Electrostatic origin of the genome packing in viruses. Proc. Natl. Acad. Sci. USA 2006, 103, 17174–17178. [Google Scholar] [CrossRef] [PubMed]
| Family | Genus/Clade | Species | Packaging Signal-Mediated Packaging | Replication-Mediated Packaging* | Assembly/Replication Sites | Length-Dependence |
|---|---|---|---|---|---|---|
| Togaviridae | Alphavirus/SINV | Sindbis (SINV) | One (nsP1 *) | N.D. | CPV | Yes * |
| Alphavirus/SINV | Venezuelan equine encephalitis virus (VEEV) | One (nsP1 *) | Yes | CPV | N.D. | |
| Alphavirus/SFV | Semliki forest virus (SFV) | One (nsP2 *) | No | CPV | N.D. | |
| Alphavirus/SFV | Ross river virus (RRV) | Three (nsP2 *) | N.D. | CPV | N.D. | |
| Bromoviridae | Bromovirus | Brome mosaic virus (BMV) | One (3′UTR **) | Yes | ER | N.D. |
| Bromovirus | Cowpea chlorotic virus (CCMV) | None ** | Yes | N.D. (Probably ER) | Yes ** | |
| Nodaviridae | Alphanodavirus | Flock house virus (FHV) | N.D. | Yes | ER | Yes * |
| Tombusviridae | Betacarmovirus | Turnip crinkle virus (TCV) | One (CP */RdRp **) | N.D. | N.D. | Yes * |
| Picornaviridae | Enterovirus | Poliovurus | N.D. | Yes | Golgi | N.D. |
| Flaviviridae | Flavivirus | Kunjin virus (KUNV) | N.D. | Yes | ER | N.D. |
| Flaviviridae | Hepacivirus | Hepatitis C (HCV) | Multiple ** | N.D. | ER | N.D. |
| Hepadnaviridae | Orthohepadnavirus | Hepatitis B (HBV) | Multiple ** | N.D. | N.D. | No ** |
| Leviviridae | Levivirus | Enterobacteria phage MS2 | Multiple ** | N.D. | No | No |
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Comas-Garcia, M. Packaging of Genomic RNA in Positive-Sense Single-Stranded RNA Viruses: A Complex Story. Viruses 2019, 11, 253. https://doi.org/10.3390/v11030253
Comas-Garcia M. Packaging of Genomic RNA in Positive-Sense Single-Stranded RNA Viruses: A Complex Story. Viruses. 2019; 11(3):253. https://doi.org/10.3390/v11030253
Chicago/Turabian StyleComas-Garcia, Mauricio. 2019. "Packaging of Genomic RNA in Positive-Sense Single-Stranded RNA Viruses: A Complex Story" Viruses 11, no. 3: 253. https://doi.org/10.3390/v11030253
APA StyleComas-Garcia, M. (2019). Packaging of Genomic RNA in Positive-Sense Single-Stranded RNA Viruses: A Complex Story. Viruses, 11(3), 253. https://doi.org/10.3390/v11030253





