Cytomegalovirus-Driven Adaption of Natural Killer Cells in NKG2Cnull Human Immunodeficiency Virus-Infected Individuals
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Peripheral Blood Mononuclear Cell Isolation
2.2. Identification of NKG2Cnull Individuals and a Matched Control Group
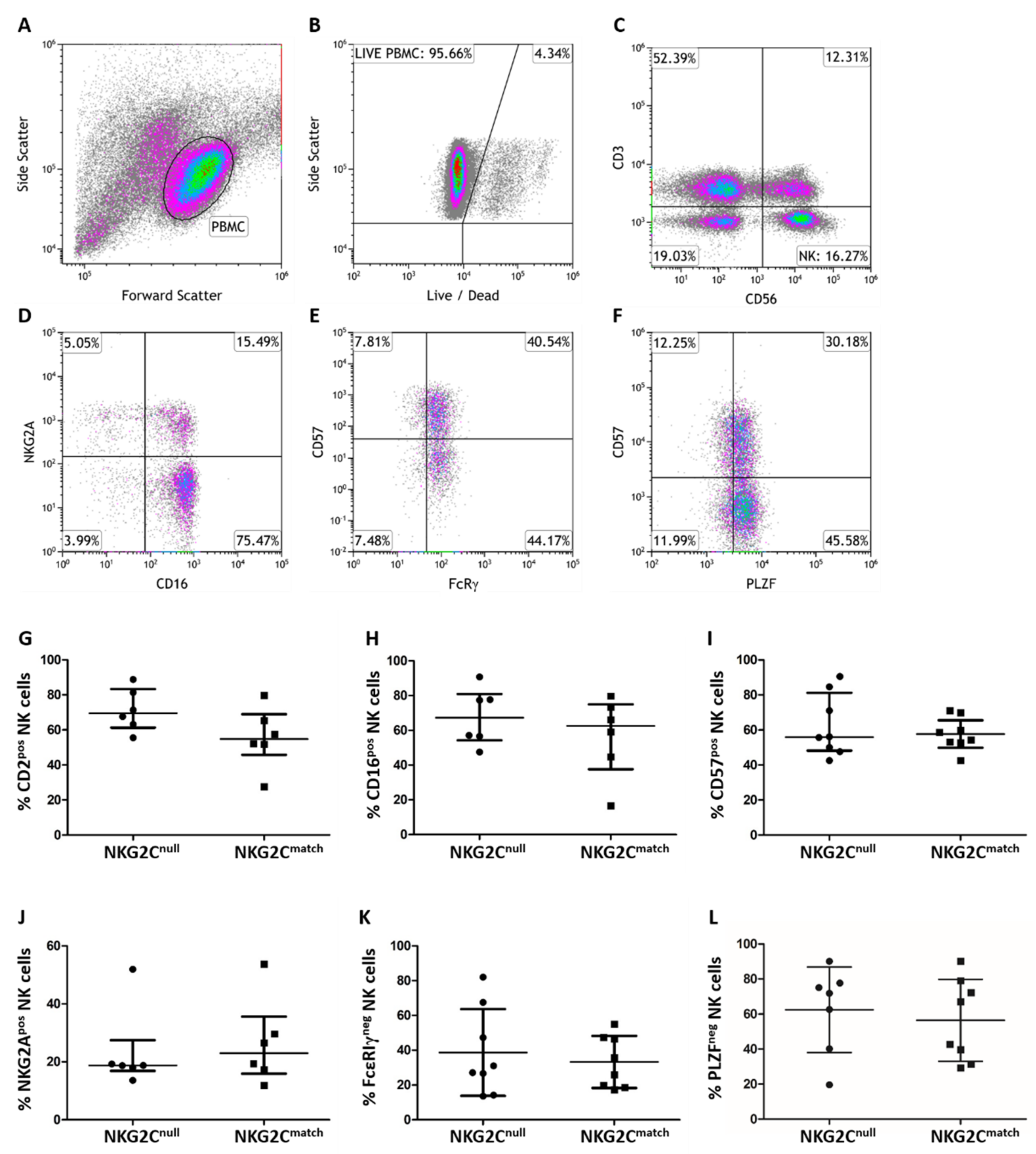
2.3. Flow Cytometry
2.4. NK Cell Stimulations
2.5. Cytotoxicity Assays
2.6. Statistical Analysis
3. Results
3.1. General Characteristics of NKG2Cnull and NKG2Cmatch Groups
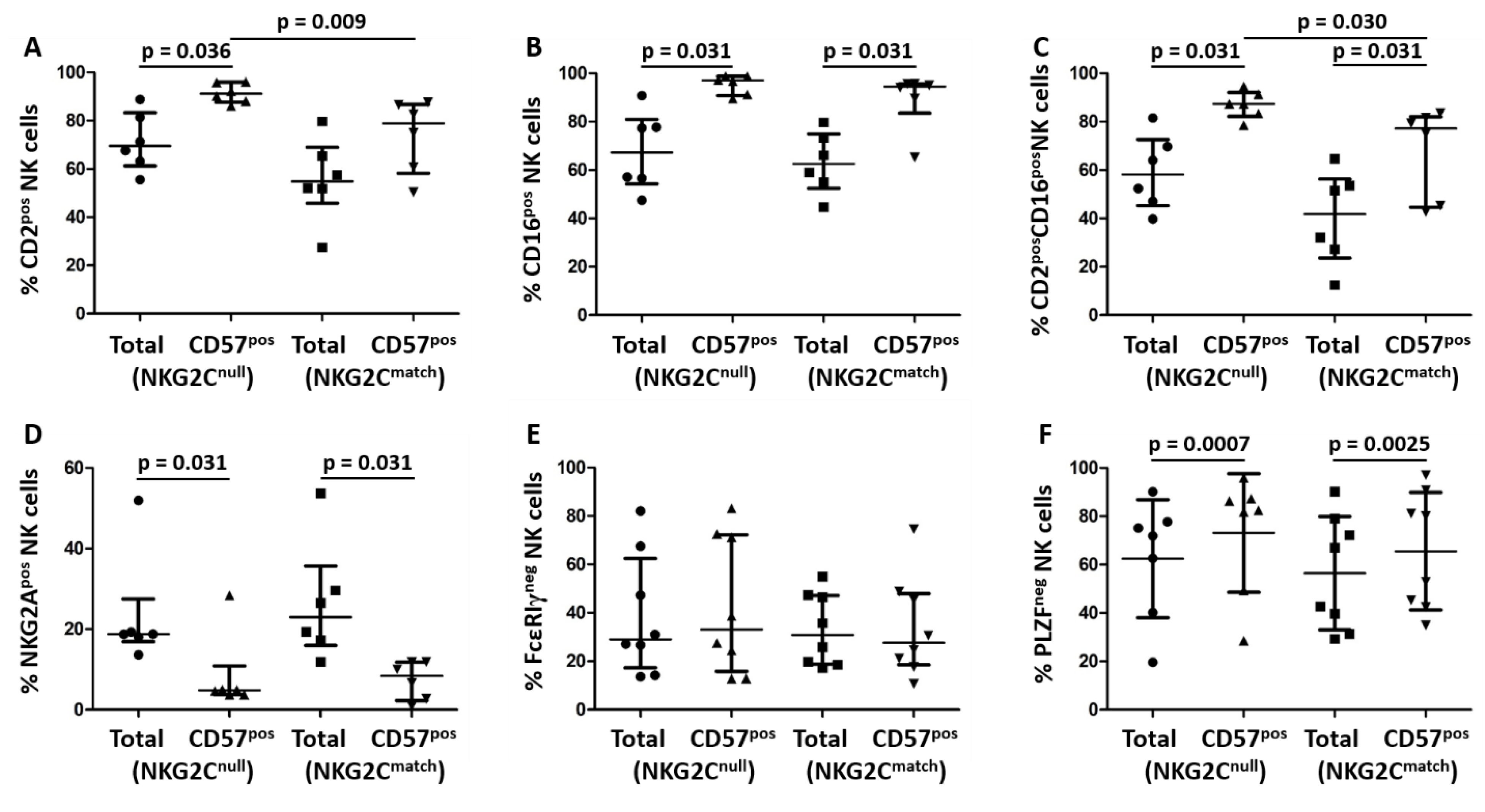
3.2. Phenotypic Comparison of NK Cells from NKG2Cnull and a Matched Control Group
3.3. Comparison of Cytokine Production by NK Cells from NKG2Cnull and NKG2Cmatch Groups
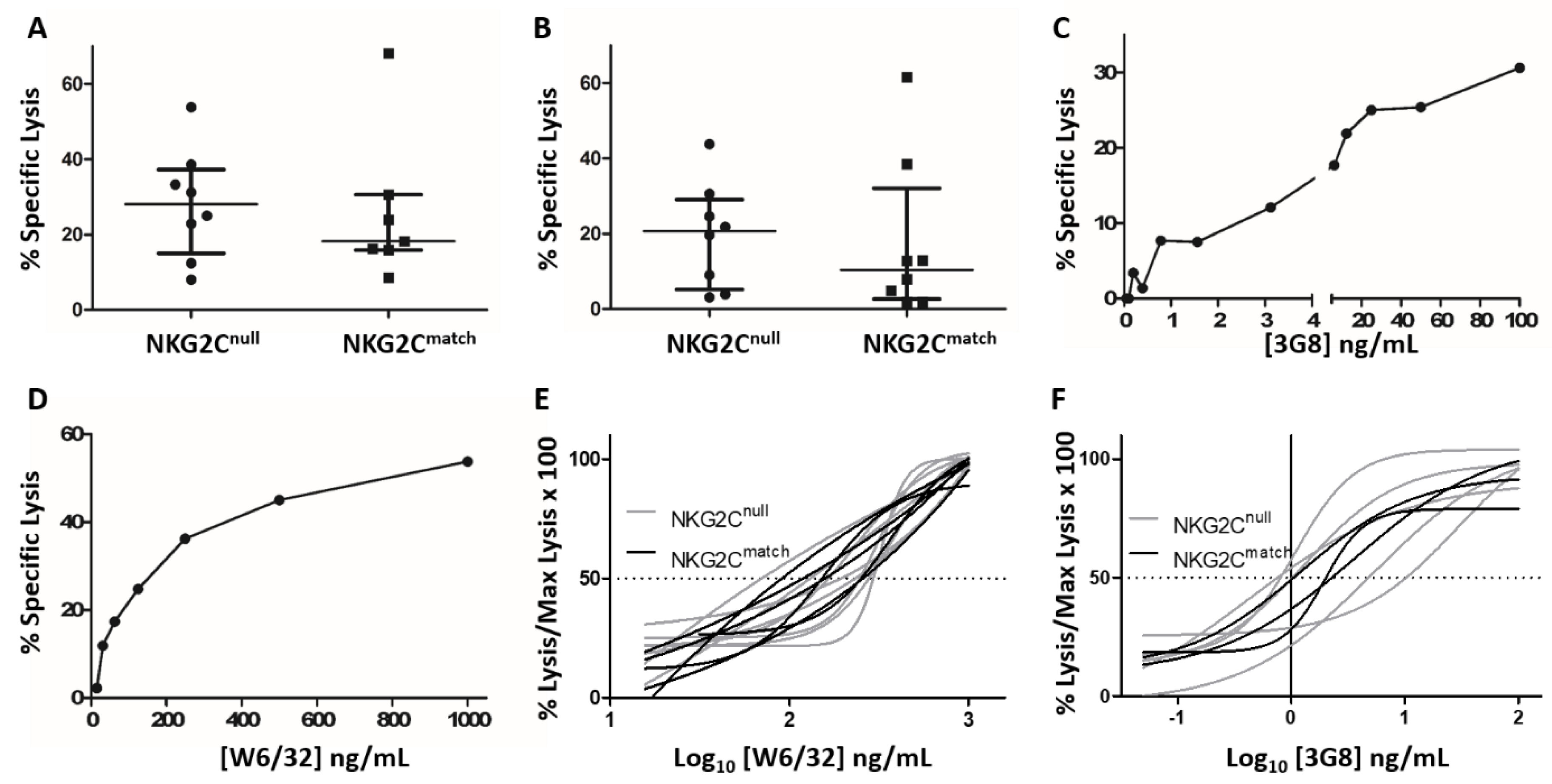
3.4. Comparison of ADCC by NK Cells from the NKG2Cnull and Matched Control Groups
4. Discussion
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Kinney, J.S.; Onorato, I.M.; Stewart, J.A.; Pass, R.F.; Stagno, S.; Cheeseman, S.H.; Chin, J.; Kumar, M.L.; Yaeger, A.S.; Herrmann, K.L.; et al. Cytomegaloviral infection and disease. J. Infect. Dis. 1985, 151, 772–774. [Google Scholar] [CrossRef] [PubMed]
- Onorato, I.M.; Morens, D.M.; Martone, W.J.; Stansfield, S.K. Epidemiology of cytomegaloviral infections: Recommendations for prevention and control. Rev. Infect. Dis. 1985, 7, 479–497. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, J. Human cytomegalovirus: Latency and reactivation in the myeloid lineage. J. Clin. Virol. 2008, 41, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Lang, A.; Brien, J.D.; Nikolich-Zugich, J. Inflation and long-term maintenance of cd8 t cells responding to a latent herpesvirus depend upon establishment of latency and presence of viral antigens. J. Immunol. 2009, 183, 8077–8087. [Google Scholar] [CrossRef] [PubMed]
- Heath, J.; Newhook, N.; Comeau, E.; Gallant, M.; Fudge, N.; Grant, M. Nkg2c(+)cd57(+) natural killer cell expansion parallels cytomegalovirus-specific cd8(+) t cell evolution towards senescence. J. Immunol. Res. 2016, 2016, 7470124. [Google Scholar] [CrossRef] [PubMed]
- Guma, M.; Angulo, A.; Vilches, C.; Gomez-Lozano, N.; Malats, N.; Lopez-Botet, M. Imprint of human cytomegalovirus infection on the nk cell receptor repertoire. Blood 2004, 104, 3664–3671. [Google Scholar] [CrossRef] [PubMed]
- Guma, M.; Budt, M.; Saez, A.; Brckalo, T.; Hengel, H.; Angulo, A.; Lopez-Botet, M. Expansion of cd94/nkg2c+ nk cells in response to human cytomegalovirus-infected fibroblasts. Blood 2006, 107, 3624–3631. [Google Scholar] [CrossRef]
- Kuijpers, T.W.; Baars, P.A.; Dantin, C.; van den Burg, M.; van Lier, R.A.; Roosnek, E. Human nk cells can control cmv infection in the absence of t cells. Blood 2008, 112, 914–915. [Google Scholar] [CrossRef]
- Della Chiesa, M.; Falco, M.; Podesta, M.; Locatelli, F.; Moretta, L.; Frassoni, F.; Moretta, A. Phenotypic and functional heterogeneity of human nk cells developing after umbilical cord blood transplantation: A role for human cytomegalovirus? Blood 2012, 119, 399–410. [Google Scholar] [CrossRef]
- Foley, B.; Cooley, S.; Verneris, M.R.; Pitt, M.; Curtsinger, J.; Luo, X.; Lopez-Verges, S.; Lanier, L.L.; Weisdorf, D.; Miller, J.S. Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated nkg2c+ natural killer cells with potent function. Blood 2012, 119, 2665–2674. [Google Scholar] [CrossRef]
- Lopez-Verges, S.; Milush, J.M.; Schwartz, B.S.; Pando, M.J.; Jarjoura, J.; York, V.A.; Houchins, J.P.; Miller, S.; Kang, S.M.; Norris, P.J.; et al. Expansion of a unique cd57(+)nkg2chi natural killer cell subset during acute human cytomegalovirus infection. Proc. Natl. Acad. Sci. USA 2011, 108, 14725–14732. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Verges, S.; Milush, J.M.; Pandey, S.; York, V.A.; Arakawa-Hoyt, J.; Pircher, H.; Norris, P.J.; Nixon, D.F.; Lanier, L.L. Cd57 defines a functionally distinct population of mature nk cells in the human cd56dimcd16+ nk-cell subset. Blood 2010, 116, 3865–3874. [Google Scholar] [CrossRef] [PubMed]
- Brunetta, E.; Fogli, M.; Varchetta, S.; Bozzo, L.; Hudspeth, K.L.; Marcenaro, E.; Moretta, A.; Mavilio, D. Chronic hiv-1 viremia reverses nkg2a/nkg2c ratio on natural killer cells in patients with human cytomegalovirus co-infection. AIDS 2010, 24, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Sinzger, C.; Frascaroli, G.; Reichel, J.; Bayer, C.; Wang, L.; Schirmbeck, R.; Mertens, T. Human cytomegalovirus-induced nkg2c(hi) cd57(hi) natural killer cells are effectors dependent on humoral antiviral immunity. J. Virol. 2013, 87, 7717–7725. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Botet, M.; Muntasell, A.; Martinez-Rodriguez, J.E.; Lopez-Montanes, M.; Costa-Garcia, M.; Pupuleku, A. Development of the adaptive nk cell response to human cytomegalovirus in the context of aging. Mech. Ageing Dev. 2016, 158, 23–26. [Google Scholar] [CrossRef] [PubMed]
- Luetke-Eversloh, M.; Cicek, B.B.; Siracusa, F.; Thom, J.T.; Hamann, A.; Frischbutter, S.; Baumgrass, R.; Chang, H.D.; Thiel, A.; Dong, J.; et al. Nk cells gain higher ifn-gamma competence during terminal differentiation. Eur. J. Immunol. 2014, 44, 2074–2084. [Google Scholar] [CrossRef] [PubMed]
- Luetke-Eversloh, M.; Hammer, Q.; Durek, P.; Nordstrom, K.; Gasparoni, G.; Pink, M.; Hamann, A.; Walter, J.; Chang, H.D.; Dong, J.; et al. Human cytomegalovirus drives epigenetic imprinting of the ifng locus in nkg2chi natural killer cells. PLoS Pathog. 2014, 10, e1004441. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.; Zhang, T.; Scott, J.M.; Kim, A.R.; Lee, T.; Kakarla, T.; Kim, A.; Sunwoo, J.B.; Kim, S. Identification of human nk cells that are deficient for signaling adaptor fcrgamma and specialized for antibody-dependent immune functions. Int. Immunol. 2012, 24, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Scott, J.M.; Hwang, I.; Kim, S. Cutting edge: Antibody-dependent memory-like nk cells distinguished by fcrgamma deficiency. J. Immunol. 2013, 190, 1402–1406. [Google Scholar] [CrossRef] [PubMed]
- Schlums, H.; Cichocki, F.; Tesi, B.; Theorell, J.; Beziat, V.; Holmes, T.D.; Han, H.; Chiang, S.C.; Foley, B.; Mattsson, K.; et al. Cytomegalovirus infection drives adaptive epigenetic diversification of nk cells with altered signaling and effector function. Immunity 2015, 42, 443–456. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Zhang, T.; Hwang, I.; Kim, A.; Nitschke, L.; Kim, M.; Scott, J.M.; Kamimura, Y.; Lanier, L.L.; Kim, S. Epigenetic modification and antibody-dependent expansion of memory-like nk cells in human cytomegalovirus-infected individuals. Immunity 2015, 42, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Mela, C.M.; Burton, C.T.; Imami, N.; Nelson, M.; Steel, A.; Gazzard, B.G.; Gotch, F.M.; Goodier, M.R. Switch from inhibitory to activating nkg2 receptor expression in hiv-1 infection: Lack of reversion with highly active antiretroviral therapy. AIDS 2005, 19, 1761–1769. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.V.; Manickam, C.; Ram, D.R.; Kroll, K.; Itell, H.; Permar, S.R.; Barouch, D.H.; Klatt, N.R.; Reeves, R.K. Cmv primes functional alternative signaling in adaptive deltag nk cells but is subverted by lentivirus infection in rhesus macaques. Cell Rep. 2018, 25, 2766–2774.e3. [Google Scholar] [CrossRef] [PubMed]
- Peppa, D.; Pedroza-Pacheco, I.; Pellegrino, P.; Williams, I.; Maini, M.K.; Borrow, P. Adaptive reconfiguration of natural killer cells in hiv-1 infection. Front. Immunol. 2018, 9, 474. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, R.; Tsuchiya, N.; Hikami, K.; Kuroki, K.; Fukazawa, T.; Bijl, M.; Kallenberg, C.G.; Hashimoto, H.; Yabe, T.; Tokunaga, K. Molecular genetic analyses of human nkg2c (klrc2) gene deletion. Int. Immunol. 2004, 16, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.; Low, H.Z.; Kniesch, K.; Jacobs, R.; Schmidt, R.E.; Witte, T. Nkg2c deletion is a risk factor of hiv infection. AIDS Res. Hum. Retrovir. 2012, 28, 844–851. [Google Scholar] [CrossRef] [PubMed]
- Gondois-Rey, F.; Cheret, A.; Granjeaud, S.; Mallet, F.; Bidaut, G.; Lecuroux, C.; Ploquin, M.; Muller-Trutwin, M.; Rouzioux, C.; Avettand-Fenoel, V.; et al. Nkg2c(+) memory-like nk cells contribute to the control of hiv viremia during primary infection: Optiprim-anrs 147. Clin. Transl. Immunol. 2017, 6, e150. [Google Scholar] [CrossRef]
- Ma, M.; Wang, Z.; Chen, X.; Tao, A.; He, L.; Fu, S.; Zhang, Z.; Fu, Y.; Guo, C.; Liu, J.; et al. Nkg2c(+)nkg2a(-) natural killer cells are associated with a lower viral set point and may predict disease progression in individuals with primary hiv infection. Front. Immunol. 2017, 8, 1176. [Google Scholar] [CrossRef]
- Goodier, M.R.; White, M.J.; Darboe, A.; Nielsen, C.M.; Goncalves, A.; Bottomley, C.; Moore, S.E.; Riley, E.M. Rapid nk cell differentiation in a population with near-universal human cytomegalovirus infection is attenuated by nkg2c deletions. Blood 2014, 124, 2213–2222. [Google Scholar] [CrossRef]
- Della Chiesa, M.; Falco, M.; Bertaina, A.; Muccio, L.; Alicata, C.; Frassoni, F.; Locatelli, F.; Moretta, L.; Moretta, A. Human cytomegalovirus infection promotes rapid maturation of nk cells expressing activating killer ig–like receptor in patients transplanted with nkg2c−/−umbilical cord blood. J. Immunol. 2014, 192, 1471–1479. [Google Scholar] [CrossRef]
- Liu, L.L.; Landskron, J.; Ask, E.H.; Enqvist, M.; Sohlberg, E.; Traherne, J.A.; Hammer, Q.; Goodridge, J.P.; Larsson, S.; Jayaraman, J.; et al. Critical role of cd2 co-stimulation in adaptive natural killer cell responses revealed in nkg2c-deficient humans. Cell Rep. 2016, 15, 1088–1099. [Google Scholar] [CrossRef] [PubMed]
- Muntasell, A.; Pupuleku, A.; Cisneros, E.; Vera, A.; Moraru, M.; Vilches, C.; Lopez-Botet, M. Relationship of nkg2c copy number with the distribution of distinct cytomegalovirus-induced adaptive nk cell subsets. J. Immunol. 2016, 196, 3818–3827. [Google Scholar] [CrossRef] [PubMed]
- Barrett, L.; Fudge, N.J.; Heath, J.J.; Grant, M.D. Cytomegalovirus immunity and exhaustive cd8+ t cell proliferation in treated human immunodeficiency virus infection. Clin. Infect. Dis. 2016, 62, 1467–1468. [Google Scholar] [CrossRef] [PubMed]
- Parsons, M.S.; Zipperlen, K.; Gallant, M.; Grant, M. Killer cell immunoglobulin-like receptor 3dl1 licenses cd16-mediated effector functions of natural killer cells. J. Leukoc. Biol. 2010, 88, 905–912. [Google Scholar] [CrossRef] [PubMed]
- Holder, K.A.; Comeau, E.M.; Grant, M.D. Origins of natural killer cell memory: Special creation or adaptive evolution. Immunology 2018, 154, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Braud, V.M.; Allan, D.S.; O’Callaghan, C.A.; Soderstrom, K.; D’Andrea, A.; Ogg, G.S.; Lazetic, S.; Young, N.T.; Bell, J.I.; Phillips, J.H.; et al. Hla-e binds to natural killer cell receptors cd94/nkg2a, b and c. Nature 1998, 391, 795–799. [Google Scholar] [CrossRef] [PubMed]
- Prod’homme, V.; Sugrue, D.M.; Stanton, R.J.; Nomoto, A.; Davies, J.; Rickards, C.R.; Cochrane, D.; Moore, M.; Wilkinson, G.W.; Tomasec, P. Human cytomegalovirus ul141 promotes efficient downregulation of the natural killer cell activating ligand cd112. J. Gen. Virol. 2010, 91, 2034–2039. [Google Scholar] [CrossRef] [PubMed]
- Prod’homme, V.; Tomasec, P.; Cunningham, C.; Lemberg, M.K.; Stanton, R.J.; McSharry, B.P.; Wang, E.C.; Cuff, S.; Martoglio, B.; Davison, A.J.; et al. Human cytomegalovirus ul40 signal peptide regulates cell surface expression of the nk cell ligands hla-e and gpul18. J. Immunol. 2012, 188, 2794–2804. [Google Scholar] [CrossRef] [PubMed]
- Rolle, A.; Pollmann, J.; Ewen, E.M.; Le, V.T.; Halenius, A.; Hengel, H.; Cerwenka, A. Il-12-producing monocytes and hla-e control hcmv-driven nkg2c+ nk cell expansion. J. Clin. Investig. 2014, 124, 5305–5316. [Google Scholar] [CrossRef]
- Hammer, Q.; Ruckert, T.; Borst, E.M.; Dunst, J.; Haubner, A.; Durek, P.; Heinrich, F.; Gasparoni, G.; Babic, M.; Tomic, A.; et al. Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat. Immunol. 2018, 19, 453–463. [Google Scholar] [CrossRef]
- Rolle, A.; Meyer, M.; Calderazzo, S.; Jager, D.; Momburg, F. Distinct hla-e peptide complexes modify antibody-driven effector functions of adaptive nk cells. Cell Rep. 2018, 24, 1967–1976.e4. [Google Scholar] [CrossRef] [PubMed]
- Newhook, N.; Fudge, N.; Grant, M. Nk cells generate memory-type responses to human cytomegalovirus-infected fibroblasts. Eur. J. Immunol. 2017, 47, 1032–1039. [Google Scholar] [CrossRef] [PubMed]
- Malone, D.F.G.; Lunemann, S.; Hengst, J.; Ljunggren, H.G.; Manns, M.P.; Sandberg, J.K.; Cornberg, M.; Wedemeyer, H.; Bjorkstrom, N.K. Cytomegalovirus-driven adaptive-like natural killer cell expansions are unaffected by concurrent chronic hepatitis virus infections. Front. Immunol. 2017, 8, 525. [Google Scholar] [CrossRef] [PubMed]
- Rolle, A.; Halenius, A.; Ewen, E.M.; Cerwenka, A.; Hengel, H.; Momburg, F. Cd2-cd58 interactions are pivotal for the activation and function of adaptive natural killer cells in human cytomegalovirus infection. Eur. J. Immunol. 2016, 46, 2420–2425. [Google Scholar] [CrossRef] [PubMed]
- Beziat, V.; Liu, L.L.; Malmberg, J.A.; Ivarsson, M.A.; Sohlberg, E.; Bjorklund, A.T.; Retiere, C.; Sverremark-Ekstrom, E.; Traherne, J.; Ljungman, P.; et al. Nk cell responses to cytomegalovirus infection lead to stable imprints in the human kir repertoire and involve activating kirs. Blood 2013, 121, 2678–2688. [Google Scholar] [CrossRef]
- Wang, E.C.Y.; Pjechova, M.; Nightingale, K.; Vlahava, V.M.; Patel, M.; Ruckova, E.; Forbes, S.K.; Nobre, L.; Antrobus, R.; Roberts, D.; et al. Suppression of costimulation by human cytomegalovirus promotes evasion of cellular immune defenses. Proc. Natl. Acad. Sci. USA 2018, 115, 4998–5003. [Google Scholar] [CrossRef] [PubMed]
- Fausther-Bovendo, H.; Wauquier, N.; Cherfils-Vicini, J.; Cremer, I.; Debre, P.; Vieillard, V. Nkg2c is a major triggering receptor involved in the v[delta]1 t cell-mediated cytotoxicity against hiv-infected cd4 t cells. AIDS 2008, 22, 217–226. [Google Scholar] [CrossRef]
- Naeger, D.M.; Martin, J.N.; Sinclair, E.; Hunt, P.W.; Bangsberg, D.R.; Hecht, F.; Hsue, P.; McCune, J.M.; Deeks, S.G. Cytomegalovirus-specific t cells persist at very high levels during long-term antiretroviral treatment of hiv disease. PLoS ONE 2010, 5, e8886. [Google Scholar] [CrossRef]

| NKG2Cnull | NKG2Cmatch | p Value | |
|---|---|---|---|
| Age in years (mean ± SD) | 50.1 ± 9.6 | 48.3 ± 10.0 | ns |
| Sex | 7 ♂, 1 ♀ | 7 ♂, 1 ♀ | ns |
| CD4pos T cell nadir (mean ± SD) a | 295 ± 223 | 240 ± 208 | ns |
| % NK cells (mean ± SD) b | 8.7 ± 4.7 | 11.2 ± 5.4 | ns |
| Anti-CMV IgG (mean ± SD) c | 1.34 ± 0.41 | 1.10 ± 0.37 | ns |
| % HCMV-specific CD8pos T cells (median with IQR) d | 7.4 (2.7–11.4) | 1.6 (0.3–4.5) | 0.05 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Comeau, E.M.; Holder, K.A.; Fudge, N.J.; Grant, M.D. Cytomegalovirus-Driven Adaption of Natural Killer Cells in NKG2Cnull Human Immunodeficiency Virus-Infected Individuals. Viruses 2019, 11, 239. https://doi.org/10.3390/v11030239
Comeau EM, Holder KA, Fudge NJ, Grant MD. Cytomegalovirus-Driven Adaption of Natural Killer Cells in NKG2Cnull Human Immunodeficiency Virus-Infected Individuals. Viruses. 2019; 11(3):239. https://doi.org/10.3390/v11030239
Chicago/Turabian StyleComeau, Emilie M., Kayla A. Holder, Neva J. Fudge, and Michael D. Grant. 2019. "Cytomegalovirus-Driven Adaption of Natural Killer Cells in NKG2Cnull Human Immunodeficiency Virus-Infected Individuals" Viruses 11, no. 3: 239. https://doi.org/10.3390/v11030239
APA StyleComeau, E. M., Holder, K. A., Fudge, N. J., & Grant, M. D. (2019). Cytomegalovirus-Driven Adaption of Natural Killer Cells in NKG2Cnull Human Immunodeficiency Virus-Infected Individuals. Viruses, 11(3), 239. https://doi.org/10.3390/v11030239





