Retrospective Detection and Genetic Characterization of Porcine circovirus 3 (PCV3) Strains Identified between 2006 and 2007 in Brazil
Abstract
:1. Introduction
2. Materials and Methods
2.1. PCV3 Detection
2.2. Sequence Assembly and Analysis
3. Results
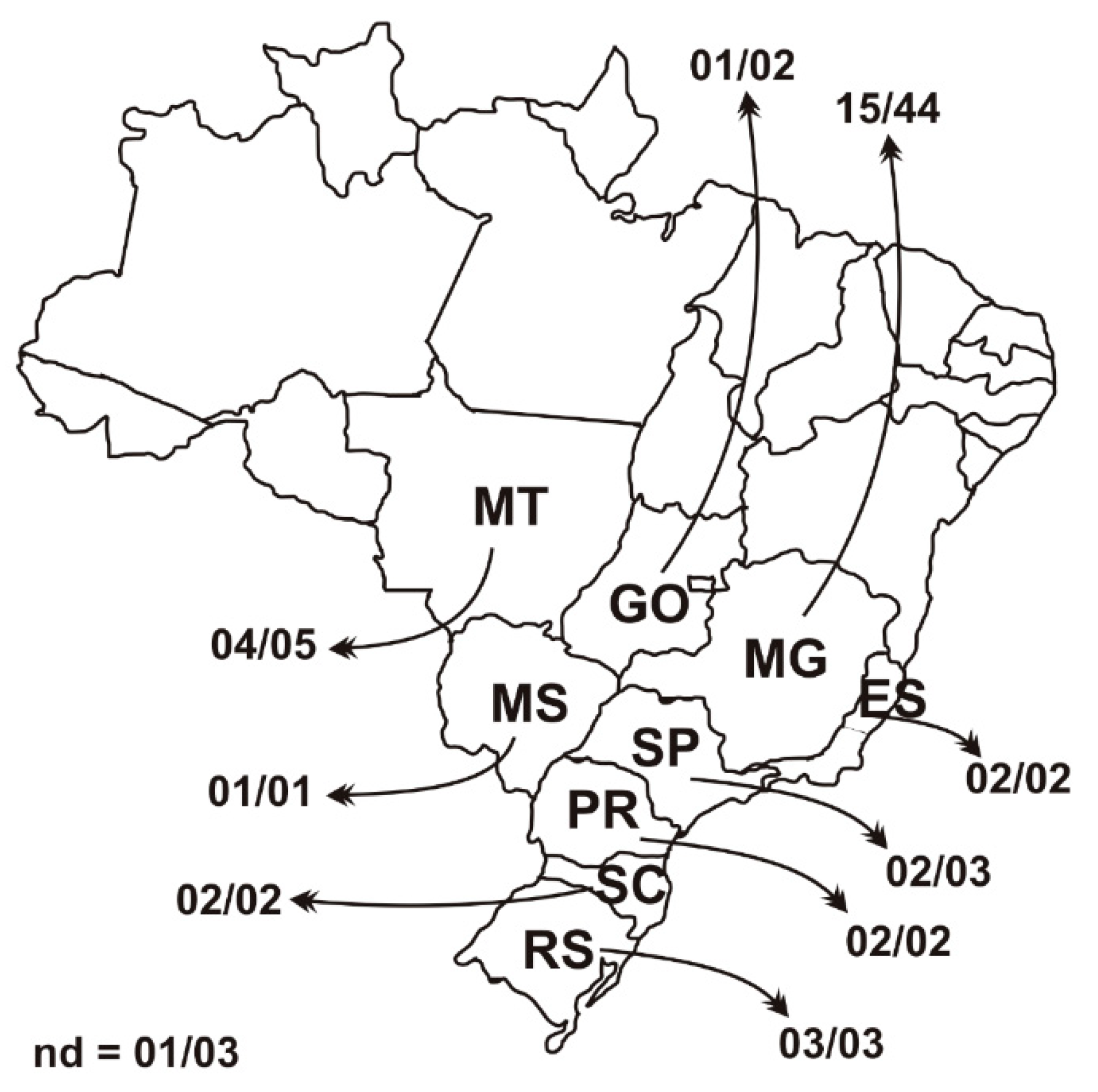
3.1. PCV3 Epidemiology Back in 2006 and 2007
3.2. Polymorphism Analysis and Estimation of Selection Pressures
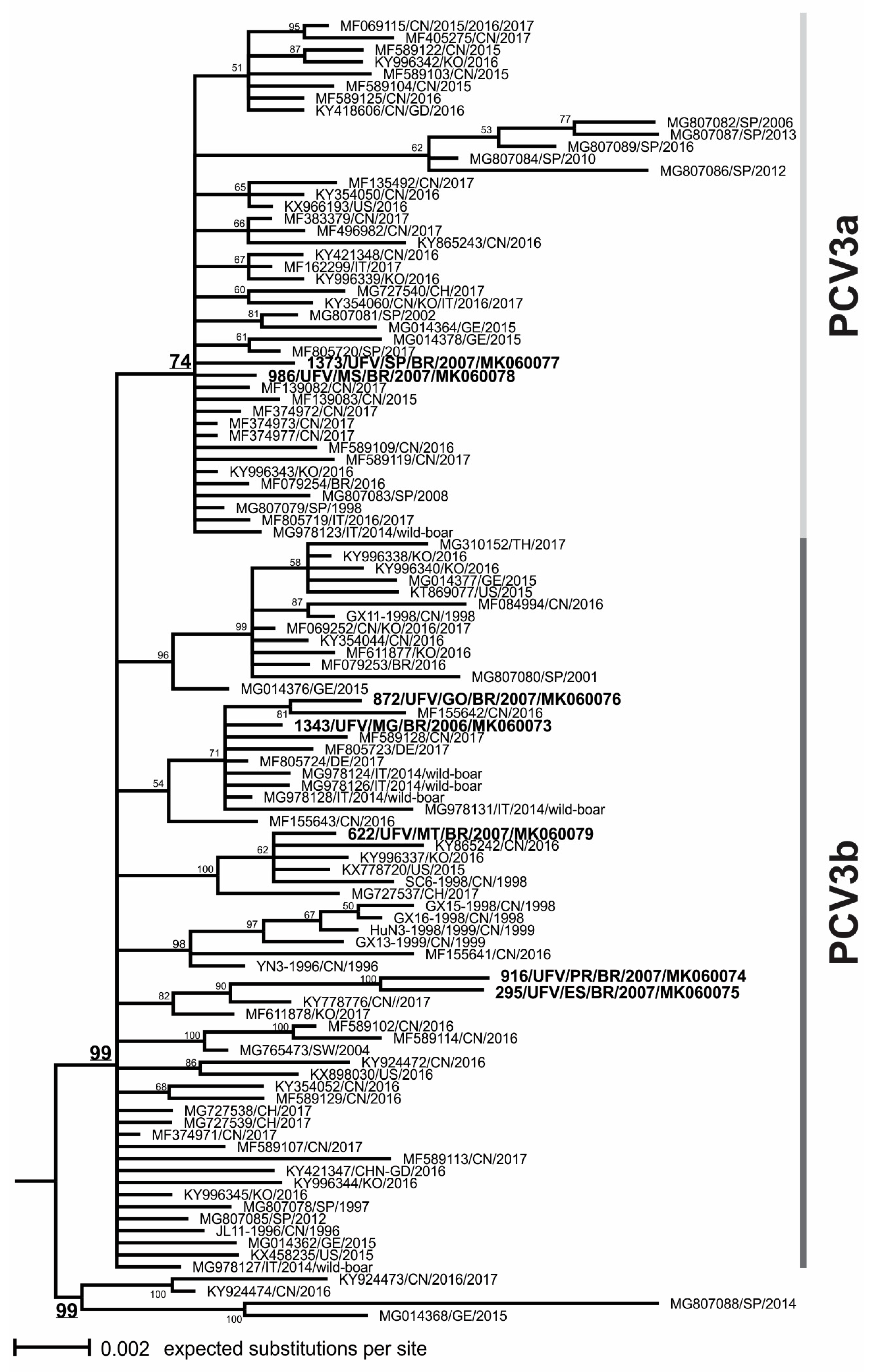
3.3. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Mankertz, A.; Çaliskan, R.; Hattermann, K.; Hillenbrand, B.; Kurzendoerfer, P.; Mueller, B.; Schmitt, C.; Steinfeldt, T.; Finsterbusch, T. Molecular biology of Porcine circovirus: Analyses of gene expression and viral replication. Vet. Microbiol. 2004, 98, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Tischer, I.; Mields, W.; Wolff, D.; Vagt, M.; Griem, W. Studies on epidemiology and pathogenicity of porcine circovirus. Arch. Virol. 1986, 91, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Tischer, I.; Rasch, R.; Tochtermann, G. Characterization of papovavirus-and picornavirus-like particles in permanent pig kidney cell lines. Zentralbl. Bakteriol. Orig. A 1974, 226, 153–167. [Google Scholar] [PubMed]
- Grau-Roma, L.; Fraile, L.; Segales, J. Recent advances in the epidemiology, diagnosis and control of diseases caused by porcine circovirus type 2. Vet. J. 2011, 187, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Madec, F.; Rose, N.; Grasland, B.; Cariolet, R.; Jestin, A. Post-weaning multisystemic wasting syndrome and other PCV2-related problems in pigs: A 12-year experience. Transbound. Emerg. Dis. 2008, 55, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Opriessnig, T.; Meng, X.-J.; Halbur, P.G. Porcine circovirus type 2 associated disease: Update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J. Vet. Diagn. Investig. 2007, 19, 591–615. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthy, S.; Meng, X.-J. Porcine circoviruses: A minuscule yet mammoth paradox. Anim. Health Res. Rev. 2009, 10, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Palinski, R.; Piñeyro, P.; Shang, P.; Yuan, F.; Guo, R.; Fang, Y.; Byers, E.; Hause, B.M. A Novel Porcine Circovirus Distantly Related to Known Circoviruses Is Associated with Porcine Dermatitis and Nephropathy Syndrome and Reproductive Failure. J. Virol. 2017, 91, e01879-16. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Fang, B.; Ma, J.; Liu, Y.; Bu, D.; Zhou, P.; Wang, H.; Jia, K.; Zhang, G. Insights into the epidemic characteristics and evolutionary history of the novel porcine circovirus type 3 in southern China. Transbound. Emerg. Dis. 2018, 65, e296–e303. [Google Scholar] [CrossRef] [PubMed]
- Ku, X.; Chen, F.; Li, P.; Wang, Y.; Yu, X.; Fan, S.; Qian, P.; Wu, M.; He, Q. Identification and genetic characterization of porcine circovirus type 3 in China. Transbound. Emerg. Dis. 2017, 64, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Chen, S.; Meng, F.; Chen, R.; Zhang, Z.; Du, E.; Xue, Q. Full-Length Genome Sequences of Two Chinese Porcine Circovirus Type 3 Strains, NWHEB21 and NWHUN2. Genome Announc. 2018, 6, e00062-18. [Google Scholar] [CrossRef] [PubMed]
- Phan, T.G.; Giannitti, F.; Rossow, S.; Marthaler, D.; Knutson, T.P.; Li, L.; Deng, X.; Resende, T.; Vannucci, F.; Delwart, E. Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol. J. 2016, 13, 184. [Google Scholar] [CrossRef] [PubMed]
- Tochetto, C.; Lima, D.A.; Varela, A.P.M.; Loiko, M.R.; Paim, W.P.; Scheffer, C.M.; Herpich, J.I.; Cerva, C.; Schmitd, C.; Cibulski, S.P.; et al. Full-Genome Sequence of Porcine Circovirus type 3 recovered from serum of sows with stillbirths in Brazil. Transbound. Emerg. Dis. 2018, 65, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Kedkovid, R.; Woonwong, Y.; Arunorat, J.; Sirisereewan, C.; Sangpratum, N.; Lumyai, M.; Kesdangsakonwut, S.; Teankum, K.; Jittimanee, S.; Thanawongnuwech, R. Porcine circovirus type 3 (PCV3) infection in grower pigs from a Thai farm suffering from porcine respiratory disease complex (PRDC). Vet. Microbiol. 2018, 215, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Liu, X.; Zhang, P.; Wang, L.; Liu, Y.; Zhang, L.; Liang, P.; Song, C. Genome characterization of a porcine circovirus type 3 in South China. Transbound. Emerg. Dis. 2018, 65, 264–266. [Google Scholar] [CrossRef] [PubMed]
- Zhai, S.-L.L.; Zhou, X.; Zhang, H.; Hause, B.M.; Lin, T.; Liu, R.; Chen, Q.-L.L.; Wei, W.-K.K.; Lv, D.-H.H.; Wen, X.-H.H.; et al. Comparative epidemiology of porcine circovirus type 3 in pigs with different clinical presentations. Virol. J. 2017, 14, 222. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.H.; Mai, K.J.; Zhou, L.; Wu, R.T.; Tang, X.Y.; Wu, J.L.; He, L.L.; Lan, T.; Xie, Q.M.; Sun, Y.; et al. Detection and genome sequencing of porcine circovirus 3 in neonatal pigs with congenital tremors in South China. Transbound. Emerg. Dis. 2017, 64, 1650–1654. [Google Scholar] [CrossRef] [PubMed]
- Stadejek, T.; Woźniak, A.; Miłek, D.; Biernacka, K.; Wo, A.; Mi, D. First detection of porcine circovirus type 3 on commercial pig farms in Poland. Transbound. Emerg. Dis. 2017, 64, 1350–1353. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Berg, M.; Fossum, C.; Wallgren, P.; Blomström, A.-L.L. Detection and genetic characterisation of porcine circovirus 3 from pigs in Sweden. Virus Genes 2018, 54, 466–469. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Wu, X.; Zhang, L.; Xin, C.; Liu, Y.; Shi, J.; Peng, Z.; Xu, S.; Fu, F.; Yu, J.; et al. The occurrence of porcine circovirus 3 without clinical infection signs in Shandong Province. Transbound. Emerg. Dis. 2017, 64, 1337–1341. [Google Scholar] [CrossRef] [PubMed]
- Franzo, G.; Tucciarone, C.M.; Drigo, M.; Cecchinato, M.; Martini, M.; Mondin, A.; Menandro, M.L. First report of wild boar susceptibility to Porcine circovirus type 3: High prevalence in the Colli Euganei Regional Park (Italy) in the absence of clinical signs. Transbound. Emerg. Dis. 2018, 65, 957–962. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Wei, L.; Lu, Z.; Mi, S.; Bao, F.; Guo, H.; Tu, C.; Zhu, Y.; Gong, W. Retrospective study of porcine circovirus 3 infection in China. Transbound. Emerg. Dis. 2018, 65, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Collins, P.J.; McKillen, J.; Allan, G. Porcine circovirus type 3 in the UK. Vet. Rec. 2017, 181, 599. [Google Scholar] [CrossRef] [PubMed]
- Klaumann, F.; Franzo, G.; Sohrmann, M.; Correa-Fiz, F.; Drigo, M.; Núñez, J.I.; Sibila, M.; Segalés, J. Retrospective detection of Porcine circovirus 3 (PCV-3) in pig serum samples from Spain. Transbound. Emerg. Dis. 2018, 65, 1290–1296. [Google Scholar] [CrossRef] [PubMed]
- Saraiva, G.L.; Vidigal, P.M.P.; Fietto, J.L.R.; Bressan, G.C.; Silva Júnior, A.; de Almeida, M.R. Evolutionary analysis of Porcine circovirus 3 (PCV3) indicates an ancient origin for its current strains and a worldwide dispersion. Virus Genes 2018, 54, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Franzo, G.; Cortey, M.; Segalés, J.; Hughes, J.; Drigo, M. Phylodynamic analysis of porcine circovirus type 2 reveals global waves of emerging genotypes and the circulation of recombinant forms. Mol. Phylogenet. Evol. 2016, 100, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Nawagitgul, P.; Morozov, I.; Bolin, S.R.; Harms, P.A.; Sorden, S.D.; Paul, P.S. Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J. Gen. Virol. 2000, 81, 2281–2287. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [PubMed]
- Fu, Y.X.; Li, W.H. Statistical tests of neutrality of mutations. Genetics 1993, 133, 693–709. [Google Scholar] [PubMed]
- Kosakovsky Pond, S.L.; Frost, S.D.W. Not so different after all: A comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 2005, 22, 1208–1222. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.; Shank, S.D.; Spielman, S.J.; Li, M.; Muse, S.V.; Kosakovsky Pond, S.L. Datamonkey 2.0: A modern web application for characterizing selective and other evolutionary processes. Mol. Biol. Evol. 2018, 35, 773–777. [Google Scholar] [CrossRef] [PubMed]
- Kwon, T.; Yoo, S.J.; Park, C.K.; Lyoo, Y.S. Prevalence of novel porcine circovirus 3 in Korean pig populations. Vet. Microbiol. 2017, 207, 178–180. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Luo, Y.; Liang, L.; Li, J.; Cui, S. Phylogenetic analysis of porcine circovirus type 3 and porcine circovirus type 2 in China detected by duplex nanoparticle-assisted PCR. Infect. Genet. Evol. 2018, 60, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Sun, W.; Li, Z.; Zhuang, X.; Zhao, G.; Xie, C.; Zheng, M.; Jing, J.; Xiao, P.; Wang, M.; et al. The detection of porcine circovirus 3 in Guangxi, China. Transbound. Emerg. Dis. 2018, 65, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Shi, J.; Wu, X.; Peng, Z.; Xin, C.; Zhang, L.; Liu, Y.; Gao, M.; Xu, S.; Han, H.; et al. Presence of Torque teno sus virus 1 and 2 in porcine circovirus 3-positive pigs. Transbound. Emerg. Dis. 2018, 65, 327–330. [Google Scholar] [CrossRef] [PubMed]
- Fux, R.; Söckler, C.; Link, E.K.; Renken, C.; Krejci, R.; Sutter, G.; Ritzmann, M.; Eddicks, M. Full genome characterization of porcine circovirus type 3 isolates reveals the existence of two distinct groups of virus strains. Virol. J. 2018, 15, 25. [Google Scholar] [CrossRef] [PubMed]
| GenBank Accession/Year | 24 | 27 | 75 | 77 | 100 | 104 | 150 |
|---|---|---|---|---|---|---|---|
| MF079253/2016 | A | R | A | S | T | F | L |
| MF079254/2016 | V | K | A | S | T | F | I |
| MK060073/2006 † | A | R | A | G | T | Y | I |
| MK060074/2007 | A | R | S | S | S | F | L |
| MK060075/2007 † | - | - | A | S | T | F | L |
| MK060076/2007 | - | - | A | S | T | Y | I |
| MK060077/2007 | - | - | A | S | T | F | I |
| MK060078/2007 † | - | - | A | S | T | F | I |
| MK060079/2007 | - | - | A | S | T | F | I |
| Tajima’s D | p * | Fu and Li’s D | p * | Fu and Li’s F | p * | |
|---|---|---|---|---|---|---|
| ORF2 | −2.25128 | < 0.01 | −4.76831 | < 0.02 | −4.48081 | < 0.02 |
| Method | Number of Codons | Global dN/dS | Neutral Sites * | Negatives Selected Sites * | Codons |
|---|---|---|---|---|---|
| SLAC | 214 | 0.1549 | 193 (90.19%) | 21 (9.81%) | 14; 21; 41; 49; 54; 55; 57; 70; 73; 82; 85; 95; 96; 129; 134; 147; 165; 192; 203; 212; 213 |
| FEL | 214 | - | 177 (82.71%) | 37 (17.29%) | 9; 11; 14; 16; 21; 26; 32; 38; 41; 49; 53; 54; 55; 57; 70; 73; 82; 85; 95; 99; 106; 118; 129; 134; 147; 162; 165; 166; 171; 176; 190; 192; 199; 203; 211; 212; 213 |
| iFEL | 214 | - | 204 (95.33%) | 10 (4.67%) | 9; 14; 49; 57; 70; 75; 85; 129; 134; 147 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saraiva, G.L.; Vidigal, P.M.P.; Assao, V.S.; Fajardo, M.L.M.; Loreto, A.N.S.; Fietto, J.L.R.; Bressan, G.C.; Lobato, Z.I.P.; Almeida, M.R.d.; Silva-Júnior, A. Retrospective Detection and Genetic Characterization of Porcine circovirus 3 (PCV3) Strains Identified between 2006 and 2007 in Brazil. Viruses 2019, 11, 201. https://doi.org/10.3390/v11030201
Saraiva GL, Vidigal PMP, Assao VS, Fajardo MLM, Loreto ANS, Fietto JLR, Bressan GC, Lobato ZIP, Almeida MRd, Silva-Júnior A. Retrospective Detection and Genetic Characterization of Porcine circovirus 3 (PCV3) Strains Identified between 2006 and 2007 in Brazil. Viruses. 2019; 11(3):201. https://doi.org/10.3390/v11030201
Chicago/Turabian StyleSaraiva, Giuliana Loreto, Pedro Marcus Pereira Vidigal, Viviane Sisdelli Assao, Murilo Leone Miranda Fajardo, Alerrandra Nunes Saraiva Loreto, Juliana Lopes Rangel Fietto, Gustavo Costa Bressan, Zélia Inês Portela Lobato, Márcia Rogéria de Almeida, and Abelardo Silva-Júnior. 2019. "Retrospective Detection and Genetic Characterization of Porcine circovirus 3 (PCV3) Strains Identified between 2006 and 2007 in Brazil" Viruses 11, no. 3: 201. https://doi.org/10.3390/v11030201
APA StyleSaraiva, G. L., Vidigal, P. M. P., Assao, V. S., Fajardo, M. L. M., Loreto, A. N. S., Fietto, J. L. R., Bressan, G. C., Lobato, Z. I. P., Almeida, M. R. d., & Silva-Júnior, A. (2019). Retrospective Detection and Genetic Characterization of Porcine circovirus 3 (PCV3) Strains Identified between 2006 and 2007 in Brazil. Viruses, 11(3), 201. https://doi.org/10.3390/v11030201






