Large Phenotypic and Genetic Diversity of Prophages Induced from the Fish Pathogen Vibrio anguillarum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of Prophages in the Vibrio anguillarum Collection
2.2. Induction and Isolation of Prophages with Mitomycin C
2.3. Spontaneous Induction of Prophages
2.4. Proliferation of Bacteriophages
2.5. Host Range Analysis for Phenotypic Characterization of Induced Phages
2.6. Bacteriophage Purification by CsCl Density Gradient
2.7. Linking Induced Phages to Prophages Using PCR Amplification
2.8. DNA Extraction, Sequencing and Bioinformatic Analysis
2.9. Lysogenization of V. anguillarum Strain BA35 with a Temperate Phage
2.10. Accession Numbers
3. Results
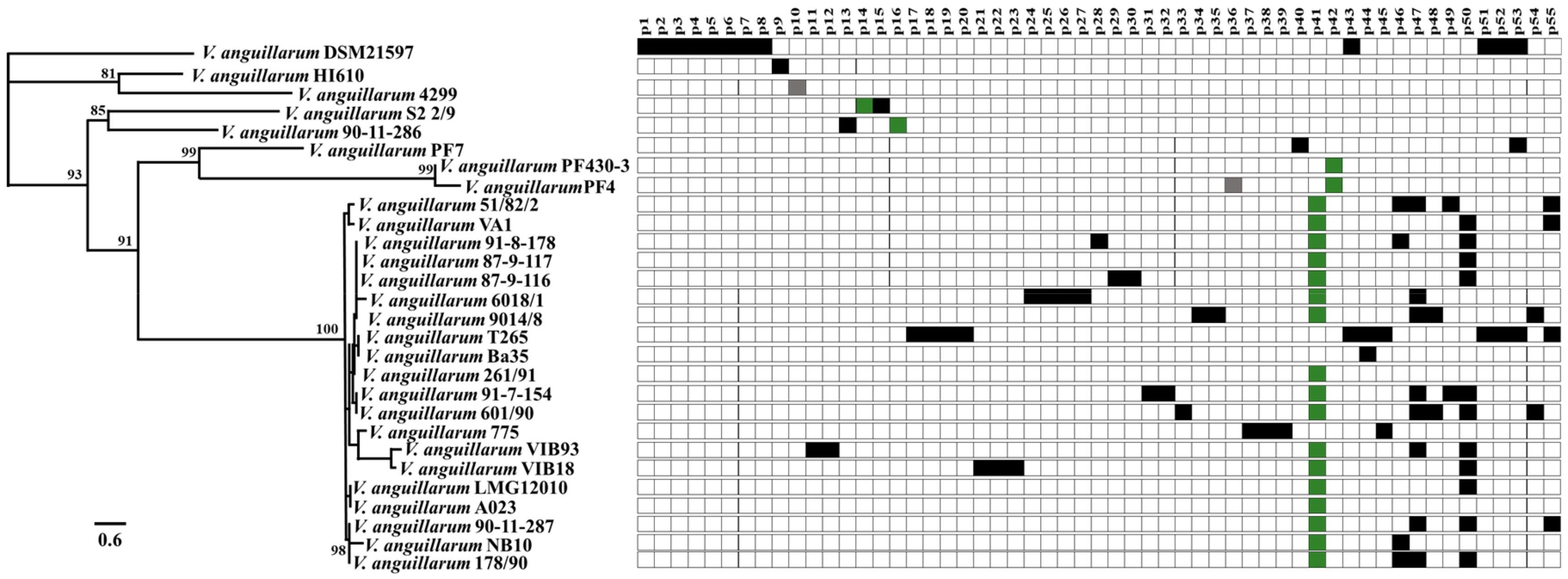
3.1. In Silico Analysis of Prophage-Like Sequences Pool from V. anguillarum Strains
3.2. Mitomycin C and Spontaneous Induction of Vibrio anguillarum Prophages
3.3. Linking Induced Phages to Prophages in Host Genomes
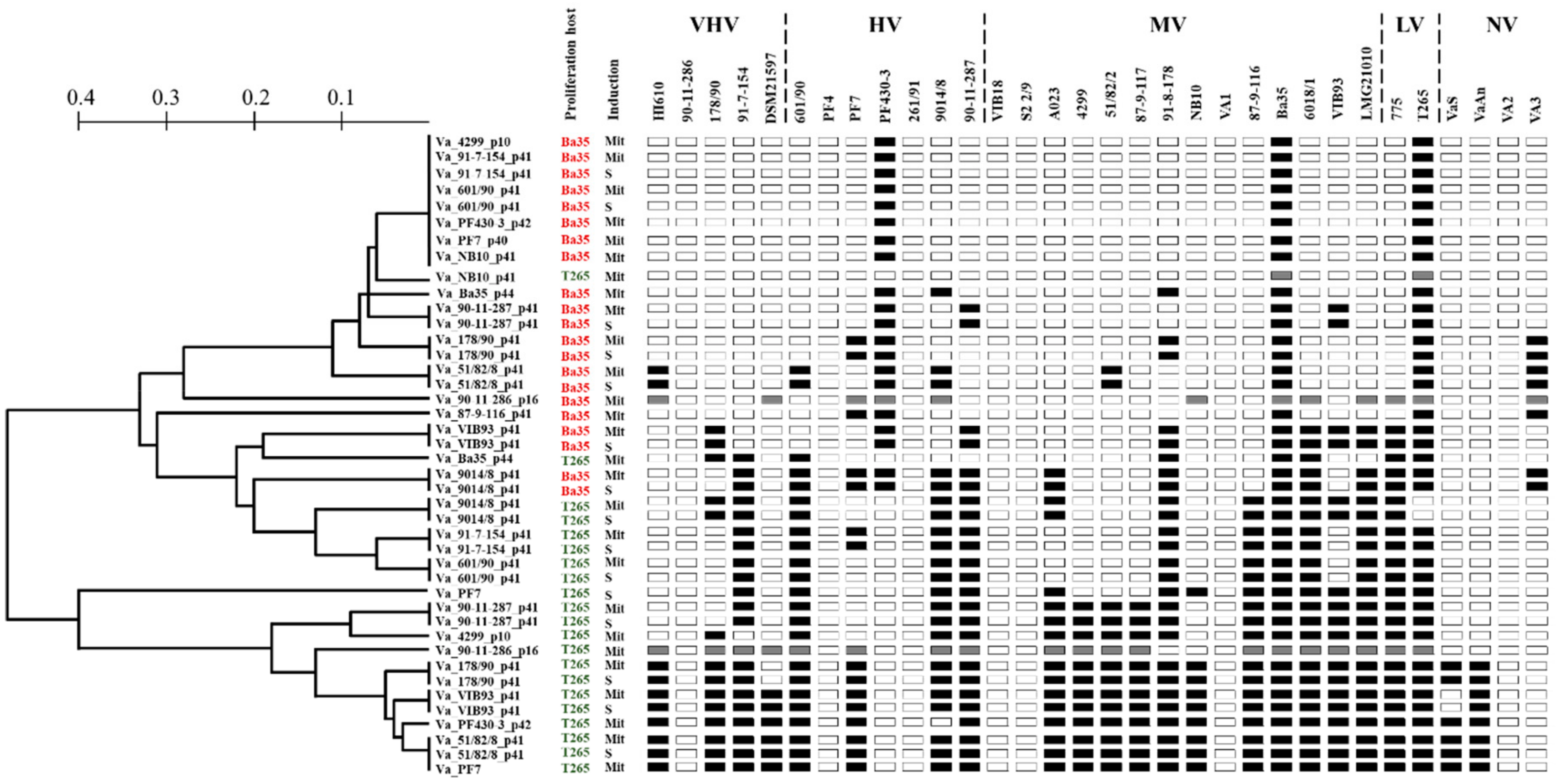
3.4. Phenotypic Characterization of Induced Phages
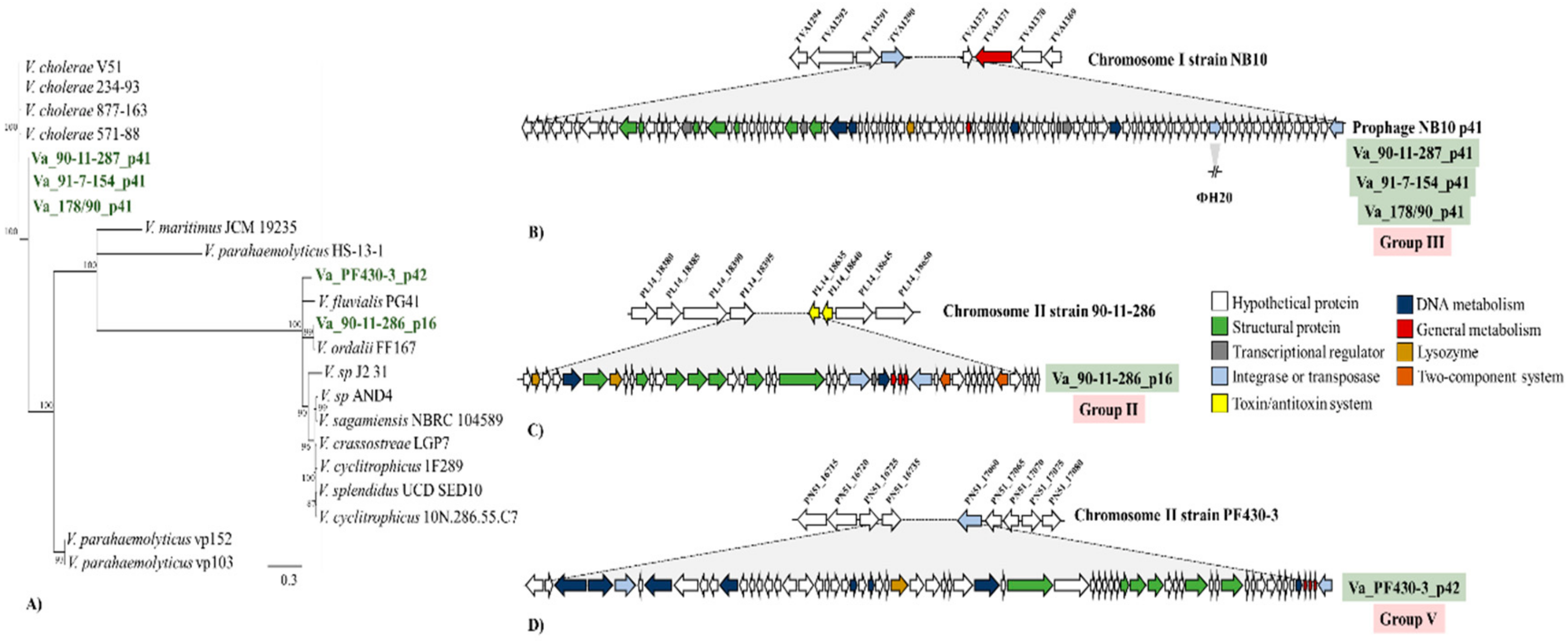
3.5. Sequencing of Induced Vibrio anguillarum Phages
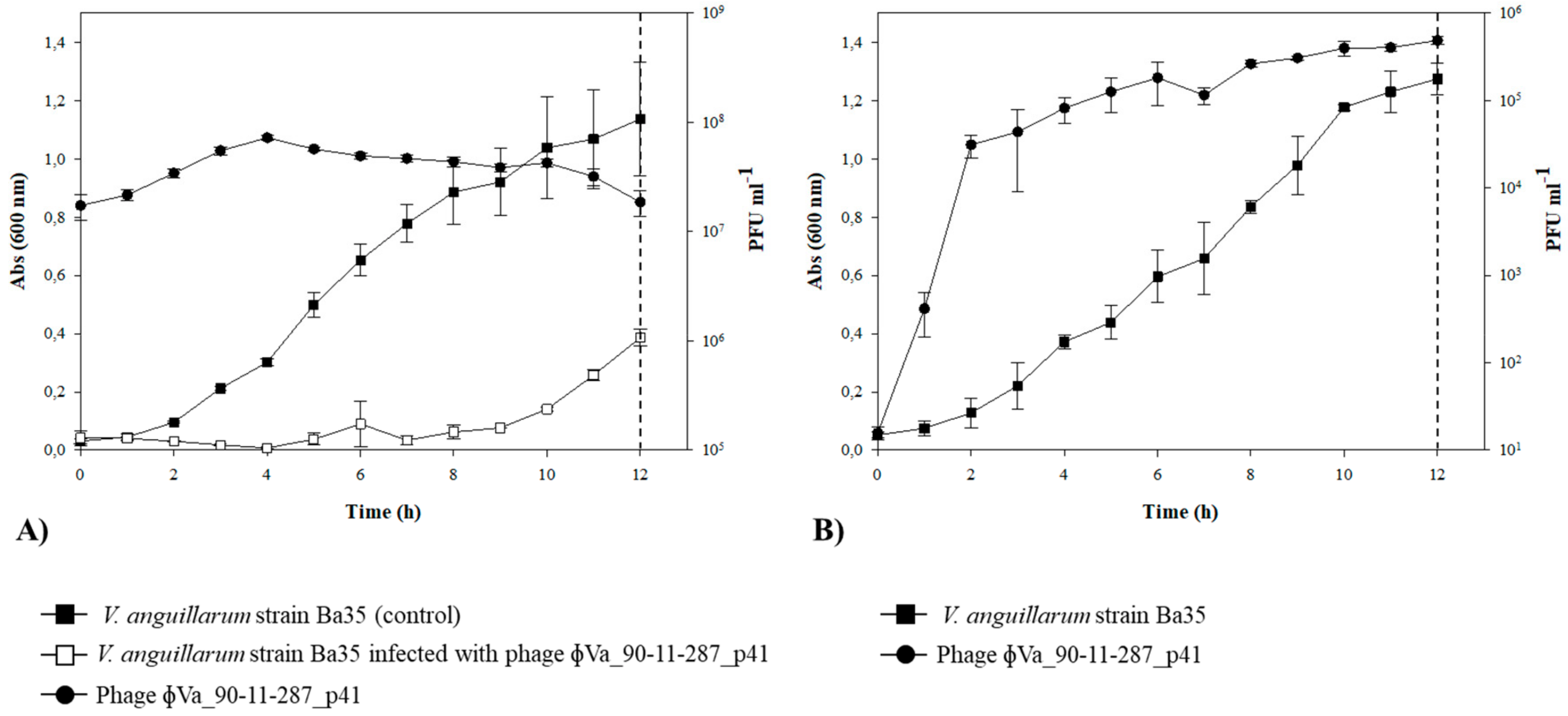
3.6. Lysogenization of V. anguillarum Strain BA35 with Phage ΦVa_90-11-287_p41
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Howard-Varona, C.; Hargreaves, K.R.; Abedon, S.T.; Sullivan, M.B. Lysogeny in nature: Mechanisms, impact and ecology of temperate phages. ISME J. 2017, 11, 1511–1520. [Google Scholar] [CrossRef] [PubMed]
- Casjens, S. Prophages and bacterial genomics: What have we learned so far? Mol. Microbiol. 2003, 49, 277–300. [Google Scholar] [CrossRef] [PubMed]
- Feiner, R.; Argov, T.; Rabinovich, L.; Sigal, N.; Borovok, I.; Herskovits, A.A. A new perspective on lysogeny: Prophages as active regulatory switches of bacteria. Nat. Rev. Microbiol. 2015, 13, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Paul, J.H. Prophages in marine bacteria: Dangerous molecular time bombs or the key to survival in the seas? ISME J. 2008, 2, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Bobay, L.M.; Touchon, M.; Rocha, E.P.C. Pervasive domestication of defective prophages by bacteria. Proc. Natl. Acad. Sci. USA 2014, 111, 12127–12132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takemura, A.F.; Chien, D.M.; Polz, M.F. Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front. Microbiol. 2014, 5, 38. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.A.; Steele, J.A.; Caporaso, J.G.; Steinbrück, L.; Reeder, J.; Temperton, B.; Huse, S.; McHardy, A.C.; Knight, R.; Joint, I.; et al. Defining seasonal marine microbial community dynamics. ISME J. 2012, 6, 298–308. [Google Scholar] [CrossRef]
- Reidl, J.; Klose, K.E. Vibrio cholerae and cholera: Out of the water and into the host. FEMS Microbiol. Rev. 2002, 26, 125–139. [Google Scholar] [CrossRef]
- Stockley, L.; Rangdale, R.; Martinez-Urtaza, J.; Baker-Austin, C.; Martínez-Urtaza, J.; Baker-Austin, C. Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus: A European perspective. Environ. Microbiol. Rep. 2010, 2, 7–18. [Google Scholar]
- Frans, I.; Michiels, C.W.; Bossier, P.; Willems, K.A.; Lievens, B.; Rediers, H.; Michiels, C. Vibrio anguillarum as a fish pathogen: Virulence factors, diagnosis and prevention. J. Fish Dis. 2011, 34, 643–661. [Google Scholar] [CrossRef]
- Austin, B.; Zhang, X.H. Vibrio harveyi: A significant pathogen of marine vertebrates and invertebrates. Lett. Appl. Microbiol. 2006, 43, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Rønneseth, A.; Castillo, D.; D’Alvise, P.; Tønnesen, Ø.; Haugland, G.; Grotkjaer, T.; Engell-Sørensen, K.; Nørremark, L.; Bergh, Ø.; Wergeland, H.I.; et al. Comparative assessment of Vibrio virulence in marine fish larvae. J. Fish Dis. 2017, 40, 1373–1385. [Google Scholar] [CrossRef] [PubMed]
- Castillo, D.; Alvise, P.D.; Xu, R.; Zhang, F.; Middelboe, M.; Gram, L. Comparative Genome Analyses of Vibrio anguillarum Strains Reveal a Link with Pathogenicity Traits. Msystems 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Waldor, M.K.; Mekalanos, J.J. Lysogenic Conversion by a Filamentous Phage Encoding Cholera Toxin. Science 1996, 272, 1910–1914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castillo, D.; Kauffman, K.; Hussain, F.; Kalatzis, P.; Rørbo, N.; Polz, M.F.; Middelboe, M. Widespread distribution of prophage-encoded virulence factors in marine Vibrio communities. Sci. Rep. 2018, 8, 9973. [Google Scholar] [CrossRef] [PubMed]
- Vidgen, M.; Carson, J.; Higgins, M.; Owens, L. Changes to the phenotypic profile of Vibrio harveyi when infected with the Vibrio harveyi myovirus-like (VHML) bacteriophage. J. Appl. Microbiol. 2002, 100, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Williamson, S.J.; Paul, J.H. Environmental factors that influence the transition from lysogenic to lytic existence in the phiHSIC/Listonella pelagia marine phage-host system. Microb. Ecol. 2006, 52, 217–225. [Google Scholar] [CrossRef]
- Munro, J.; Oakey, J.; Bromage, E.; Owens, L. Experimental bacteriophage-mediated virulence in strains of Vibrio harveyi. Dis. Aquat. Org. 2003, 54, 187–194. [Google Scholar] [CrossRef]
- Kalatzis, P.G.; Rørbo, N.I.; Castillo, D.; Mauritzen, J.J.; Jørgensen, J.; Kokkari, C.; Zhang, F.; Katharios, P.; Middelboe, M. Stumbling across the same phage: Comparative genomics of widespread temperate phages infecting the fish pathogen Vibrio anguillarum. Viruses 2017, 9, 122. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, 16–21. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Stenholm, A.R.; Dalsgaard, I.; Middelboe, M. Isolation and Characterization of Bacteriophages Infecting the Fish Pathogen Flavobacterium psychrophilum. Appl. Environ. Microbiol. 2008, 74, 4070–4078. [Google Scholar] [CrossRef] [PubMed]
- Castillo, D.; Christiansen, R.H.; Espejo, R.; Middelboe, M. Diversity and Geographical Distribution of Flavobacterium psychrophilum Isolates and Their Phages: Patterns of Susceptibility to Phage Infection and Phage Host Range. Microb. Ecol. 2014, 67, 748–757. [Google Scholar] [CrossRef] [PubMed]
- De Wachter, R.; Van De Peer, Y. TREECON for Windows: A software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Bioinformatics 1994, 10, 569–570. [Google Scholar]
- Castillo, D.; Higuera, G.; Villa, M.; Middelboe, M.; Dalsgaard, I.; Madsen, L.; Espejo, R. Diversity of Flavobacterium psychrophilum and the potential use of its phages for protection against bacterial cold water disease in salmonids. J. Fish Dis. 2012, 35, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.; Blackshields, G.; Brown, N.; Chenna, R.; Mcgettigan, P.; Mc William, H.; Valentin, F.; Wallace, I.; Wilm, A.; López, R.; et al. Clustal W and Clustal X version 2. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple Alignment of Conserved Genomic Sequence with Rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Maxwell, C.S. Hypothesis: A Plastically Produced Phenotype Predicts Host Specialization and Can Precede Subsequent Mutations in Bacteriophage. mBio 2018, 9. [Google Scholar] [CrossRef]
- Lorenz, N.; Reiger, M.; Toro-Nahuelpan, M.; Brachmann, A.; Poettinger, L.; Plener, L.; Lassak, J.; Jung, K. Identification and Initial Characterization of Prophages in Vibrio campbellii. PLoS ONE 2016, 11, e0156010. [Google Scholar] [CrossRef]
- Zabala, B.; García, K.; Espejo, R.T. Enhancement of UV Light Sensitivity of a Vibrio parahaemolyticus O3:K6 Pandemic Strain Due to Natural Lysogenization by a Telomeric Phage. Appl. Environ. Microbiol. 2009, 75, 1697–1702. [Google Scholar] [CrossRef] [PubMed]
- Lan, S.F.; Huang, C.H.; Chang, C.H.; Liao, W.C.; Lin, I.H.; Jian, W.N.; Wu, Y.G.; Chen, S.Y.; Wong, H.C. Characterization of a New Plasmid-Like Prophage in a Pandemic Vibrio parahaemolyticus O3:K6 Strain. Appl. Environ. Microbiol. 2009, 75, 2659–2667. [Google Scholar] [CrossRef] [PubMed]
- Faruque, S.M.; Asadulghani, M.; Rahman, M.M.; Waldor, M.K.; Sack, D.A. Sunlight-Induced Propagation of the Lysogenic Phage Encoding Cholera Toxin. Infect. Immun. 2000, 68, 4795–4801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espeland, E.M.; Huq, A.; Lipp, E.K.; Colwell, R.R. Polylysogeny and prophage induction by secondary infection in Vibrio cholerae. Environ. Microbiol. 2004, 6, 760–763. [Google Scholar] [CrossRef] [PubMed]
- Gödeke, J.; Paul, K.; Lassak, J.; Thormann, K.M. Phage-induced lysis enhances biofilm formation in Shewanella oneidensis MR-1. ISME J. 2011, 5, 613–626. [Google Scholar] [CrossRef]
- Nanda, A.M.; Heyer, A.; Kramer, C.; Grünberger, A.; Kohlheyer, D.; Frunzke, J. Analysis of SOS-induced spontaneous prophage induction in Corynebacterium glutamicum at the single-cell level. J. Bacteriol. 2014, 196, 180–188. [Google Scholar] [CrossRef]
- Christiansen, R.H.; Madsen, L.; Dalsgaard, I.; Castillo, D.; Kalatzis, P.G.; Middelboe, M. Effect of Bacteriophages on the Growth of Flavobacterium psychrophilum and Development of Phage-Resistant Strains. Microb. Ecol. 2016, 71, 845–859. [Google Scholar] [CrossRef]
- Li, X.Y.; Lachnit, T.; Fraune, S.; Bosch, T.C.G.; Traulsen, A.; Sieber, M. Temperate phages as self-replicating weapons in bacterial competition. J. R. Soc. Interface 2017, 14, 20170563. [Google Scholar] [CrossRef] [Green Version]
- Harrison, E.; Brockhurst, M.A. Ecological and Evolutionary Benefits of Temperate Phage: What Does or Doesn’t Kill You Makes You Stronger. BioEssays 2017, 39, 1700112. [Google Scholar] [CrossRef]
- Davies, E.V.; James, C.E.; Kukavica-Ibrulj, I.; Levesque, R.C.; Brockhurst, M.A.; Winstanley, C. Temperate phages enhance pathogen fitness in chronic lung infection. ISME J. 2016, 10, 2553–2555. [Google Scholar] [CrossRef]
- Burns, N.; James, C.E.; Harrison, E. Polylysogeny magnifies competitiveness of a bacterial pathogen in vivo. Evol. Appl. 2015, 8, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Castillo, D.; Espejo, R.; Middelboe, M. Genomic structure of bacteriophage 6H and its distribution as prophage in Flavobacterium psychrophilum strains. FEMS Microbiol. Lett. 2014, 351, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Castillo, D.; Rørbo, N.; Jørgensen, J.; Lange, J.; Tan, D.; Kalatzis, P.G.; Svenningsen, S.L.; Middelboe, M. Phage defense mechanisms and their genomic and phenotypic implications in the fish pathogen Vibrio anguillarum. FEMS Microbiol. Ecol. 2019, 95, 4. [Google Scholar] [CrossRef] [PubMed]
- Gentile, G.M.; Wetzel, K.S.; Dedrick, R.M.; Montgomery, M.T.; Garlena, R.A.; Jacobs-Sera, D.; Hatfull, G.F. More Evidence of Collusion: A New Prophage-Mediated Viral Defense System Encoded by Mycobacteriophage Sbash. mBio 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Wendling, C.C.; Piecyk, A.; Refardt, D.; Chibani, C.; Hertel, R.; Liesegang, H.; Bunk, B.; Overmann, J.; Roth, O. Tripartite species interaction: Eukaryotic hosts suffer more from phage susceptible than from phage resistant bacteria. BMC Evol. Biol. 2017, 17, 98. [Google Scholar] [CrossRef]
- Luria, S.E.; Human, M.L. A nonhereditary, host-induced variation of bacterial viruses. J. Bacteriol. 1952, 64, 557–569. [Google Scholar]
- Loenen, W.A.M.; Dryden, D.T.F.; Raleigh, E.A.; Wilson, G.G.; Murray, N.E. Highlights of the DNA cutter: A short history of the restriction enzymes. Nucleic Acids Res. 2013, 42, 3–19. [Google Scholar] [CrossRef]
- Ferris, M.T.; Joyce, P.; Burch, C.L. High Frequency of Mutations That Expand the Host Range of an RNA Virus. Genetics 2007, 176, 1013–1022. [Google Scholar] [CrossRef] [Green Version]
- Krüger, D.H.; Bickle, T.A. Bacteriophage survival: Multiple mechanisms for avoiding the deoxyribonucleic acid restriction enzymes of their hosts. Microbiol. Rev. 1983, 47, 345–360. [Google Scholar]
- Fortier, L.C.; Sekulovic, O. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence 2013, 4, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Kalatzis, P.G.; Carstens, A.B.; Katharios, P.; Castillo, D.; Hansen, L.H.; Middelboe, M. Complete Genome Sequence of Vibrio anguillarum Nontailed Bacteriophage NO16. Microbiol. Resour. Announc. 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, K.; Ackermann, H.W.; Halden, R.U.; Jiao, N.; Chen, F. Searching for a “hidden” prophage in a marine bacterium. Appl. Environ. Microbiol. 2010, 76, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Kalatzis, P.G.; Bastías, R.; Kokkari, C.; Katharios, P. Isolation and Characterization of Two Lytic Bacteriophages, φSt2 and φGrn1; Phage Therapy Application for Biological Control of Vibrio alginolyticus in Aquaculture Live Feeds. PLoS ONE 2016, 11, e0151101. [Google Scholar] [CrossRef] [PubMed]
- Rørbo, N.; Rønneseth, A.; Kalatzis, P.G.; Rasmussen, B.B.; Engell-Sørensen, K.; Kleppen, H.P.; Wergeland, H.I.; Gram, L.; Middelboe, M. Exploring the effect of phage therapy in preventing Vibrio anguillarum infections in cod and turbot larvae. Antibiotics 2018, 7, 42. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Moon, B.Y.; Park, J.W.; Thornton, J.A.; Park, Y.H.; Seo, K.S. Genetic engineering of a temperate phage-based delivery system for CRISPR/Cas9 antimicrobials against Staphylococcus aureus. Sci. Rep. 2017, 7, 44929. [Google Scholar] [CrossRef] [PubMed]
- Yosef, I.; Manor, M.; Kiro, R.; Qimron, U. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc. Natl. Acad. Sci. USA 2015, 112, 7267–7272. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Fouts, D.E.; DePew, J.; Stevens, R.H. Genetic modifications to temperate Enterococcus faecalis phage Ef11 that abolish the establishment of lysogeny and sensitivity to repressor, and increase host range and productivity of lytic infection. Microbiology 2013, 159, 1023–1035. [Google Scholar] [CrossRef]
| Group | Phage | Prophage Host | Induction Type | Genome Size kb (Prophage) | Proliferation Host (s) |
|---|---|---|---|---|---|
| I | Va_4299_p10 | 4299 | Mitomycin C | 9.7 | T265; Ba35 |
| II | Va_91-11-286_p16 | 90-11-286 | Mitomycin C | 41.2 | T265; Ba35 |
| III | Va_VIB93_p41 | VIB93 | Mitomycin C; spontaneous | 53.1 | T265; Ba35 |
| Va_90-11-287_p41 | 91-11-287 | Mitomycin C; spontaneous | 53.1 | T265; Ba35 | |
| Va_51-82-2_p41 | 51-82-2 | Mitomycin C; spontaneous | 53.1 | T265; Ba35 | |
| Va_87-9-116_p41 | 87-9-116 | Spontaneous | 53.1 | Ba35 | |
| Va_91-7-154_p41 | 91-7-154 | Mitomycin C; spontaneous | 53.1 | T265; Ba35 | |
| Va_601/90_p41 | 601/90 | Mitomycin C; spontaneous | 53.1 | T265; Ba35 | |
| Va_178/90_p41 | 178/90 | Mitomycin C; spontaneous | 53.1 | T265; Ba35 | |
| Va_9014/8_p41 | 9014/8 | Mitomycin C; spontaneous | 53.1 | T265; Ba35 | |
| Va_NB10_p41 | NB10 | Mitomycin C | 53.1 | T265; Ba35 | |
| IV | Va_BA35_p44 | Ba35 | Mitomycin C | 9.2 | T265; Ba35 |
| V | Va_PF430-3_p42 | PF430-3 | Mitomycin C | 52.9 | T265; Ba35 |
| VI | Va_PF7_p40 | PF7 | Mitomycin C | 19.0 | T265; Ba35 |
| VII | Va_PF7 | PF7 | Spontaneous | ND | T265 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castillo, D.; Andersen, N.; Kalatzis, P.G.; Middelboe, M. Large Phenotypic and Genetic Diversity of Prophages Induced from the Fish Pathogen Vibrio anguillarum. Viruses 2019, 11, 983. https://doi.org/10.3390/v11110983
Castillo D, Andersen N, Kalatzis PG, Middelboe M. Large Phenotypic and Genetic Diversity of Prophages Induced from the Fish Pathogen Vibrio anguillarum. Viruses. 2019; 11(11):983. https://doi.org/10.3390/v11110983
Chicago/Turabian StyleCastillo, Daniel, Nana Andersen, Panos G. Kalatzis, and Mathias Middelboe. 2019. "Large Phenotypic and Genetic Diversity of Prophages Induced from the Fish Pathogen Vibrio anguillarum" Viruses 11, no. 11: 983. https://doi.org/10.3390/v11110983
APA StyleCastillo, D., Andersen, N., Kalatzis, P. G., & Middelboe, M. (2019). Large Phenotypic and Genetic Diversity of Prophages Induced from the Fish Pathogen Vibrio anguillarum. Viruses, 11(11), 983. https://doi.org/10.3390/v11110983






